Abstract
Yersinia pestis is a facultative intracellular bacterial pathogen that can replicate in macrophages. Little is known about the mechanism by which Y. pestis replicates in macrophages, and macrophage defense mechanisms important for limiting intracellular survival of Y. pestis have not been characterized. In this work, we investigated the ability of Y. pestis to replicate in primary murine macrophages that were activated with IFN-γ. Y. pestis was able to replicate in macrophages that were activated with IFN-γ after infection (postactivated). A region of chromosomal DNA known as the pigmentation (pgm) locus was required for replication in postactivated macrophages, and this replication was associated with reduced nitric oxide (NO) levels but not with reduced inducible NO synthase (iNOS) expression. Y. pestis Δpgm replicated in iNOS-/- macrophages that were postactivated with IFN-γ, suggesting that killing of Δpgm Y. pestis is NO-dependent. A specific genetic locus within pgm, which shares similarity to a pathogenicity island in Salmonella, was shown to be required for replication of Y. pestis and restriction of NO levels in postactivated macrophages. These data demonstrate that intracellular Y. pestis can evade killing by macrophages that are exposed to IFN-γ and identify a potential virulence gene encoded in the pgm locus that is required for this activity.
Keywords: intracellular replication, nitric oxide
The genus Yersinia includes three pathogens of humans; Yersinia pestis; and two enteropathogens, Yersinia pseudotuberculosis and Yersinia enterocolitica. Y. pseudotuberculosis and Y. enterocolitica cause a broad range of gastrointestinal diseases, including enteritis and mesenteric lymphadenitis, and more rarely cause septicemia. Y. pestis is the causative agent of plague, a fatal disease that has decimated millions of people in three major pandemics. Although the number of confirmed plague cases that occur worldwide has stabilized at ≈1,800 in recent years, outbreaks still occur in some countries, mostly in Africa, Asia, and South America (1).
Y. pestis primarily infects wild rodents, and accidental transmission to humans occurs primarily via the bite of an infected flea. The bacteria multiply at the site of the infection and spread to the lymph nodes, which swell and give rise to characteristic buboes. The infection spreads to the bloodstream, causing septicemia and growth in internal organs. Occasionally, the lungs become involved, leading to secondary pneumonic plague; this severe form of the disease can be transmitted from human to human via respiratory droplets (1).
To disseminate within the host, Y. pestis produces multiple virulence factors, several of which are encoded on plasmids. The ≈70-kb virulence plasmid (pCD1) that is common to all three Yersinia pathogens (referred to as pYV in the enteric Yersinia) and the plasmids pPCP1 (≈9.5 kb) and pMT1 (≈101 kb) that are specific to Y. pestis. These plasmids play important roles for tissue invasion (pPCP1) and for survival of plague in the flea vector (pMT1) (2-5). Factors encoded on pCD1 play a role in the modulation of the host immune response, including down-regulation of proinflammatory cytokine production (6-8).
Another important aspect of the Y. pestis pathogenesis is its ability to acquire iron from mammalian iron-binding proteins. The yersiniabactin (Ybt) siderophore-dependent iron transport system plays a critical role at an early stage of infection (9-12). The Ybt system is encoded on the high pathogenicity island (HPI) that is common to the highly pathogenic species, Y. pestis, Y. pseudotuberculosis serotypes O1 and O3, and Y. enterocolitica biotype 1B (13, 14). In Y. pestis, the HPI (≈34 kb) is part of a 102-kb chromosomal region known as the pigmentation (pgm) locus. The pgm locus is comprised of the HPI and a second ≈68-kb region called the pigmentation segment (see Fig. 5A). The pigmentation segment encodes the hemin storage genes (hmsHFRS) that are required for temperature-dependent hemin and Congo red binding (15, 16) and enable plague transmission by blocking the proventriculus of the flea via formation of a biofilm (17-21). The pgm locus undergoes spontaneous deletion at high frequency, and Δpgm and Ybt- mutants of Y. pestis lacking specific Ybt biosynthetic or transport functions are completely avirulent from peripheral infection routes (10-12, 22). Whether other genes within the pigmentation segment also contribute to the pathogenesis of plague has not been addressed.
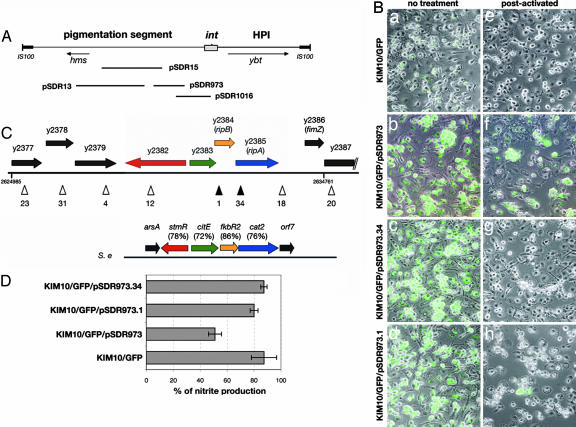
Fig. 5.
Identification of rip genes in the pigmentation segment. (A) Map of the pgm locus. The pgm locus contains the pigmentation segment that encodes the hemin storage locus (hms) and the HPI that encodes the yersiniabactin locus (ybt) and carries a P4-like integrase gene (int). Positions of inserts in recombinant plasmids are indicated below the map. (B) BMM were infected with the indicated strains, treated or not with IFN-γ, and processed for microscopy, as in the legend to Fig. 1. (C) ORF map of the insert in pSDR973 and comparison with SPI6. The insert in pSDR973 corresponds to nucleotides 2624985-2634761 of the KIM chromosome (48). ORFs shown as arrows are named according to the nomenclature of Deng et al. (48) or according to our nomenclature for ripA and ripB. In the right end of the insert, black parallel lines indicate a truncation of Y2387 in the insert. Positions and identification numbers of transposon insertions in the insert are indicated as arrowheads below the ORF map. Filled arrowheads represent insertions that prevented Y. pestis replication in postactivated macrophages. The SPI6 (S.e.) is compared with the rip region (percent identities in parentheses). (D) The concentration of nitrite in the supernatant of postactivated macrophages infected with the indicated strains was assessed 25 h after infection as described in the legend to Fig. 3A. Each bar represents the average of values obtained in single infections from three independent experiments. Error bars show the standard deviations.
The majority of the virulence factors identified to date in Y. pestis are critical for an extracellular phase of Y. pestis growth during infection. However, Y. pestis is considered a facultative intracellular pathogen, because it grows in naïve macrophages and nonphagocytic cells in vitro (4, 23-25), and an intracellular phase of growth may play a role in the infection process. Macrophages are professional phagocytes involved at all stages of the immune response. Several pathogenic bacteria (e.g., Mycobacteria, Legionella, and Salmonella) have developed successful strategies to counteract the macrophage defense mechanisms (26, 27).
Macrophages can be activated to express increased bactericidal activity by exposure to cytokines such as IFN-γ. IFN-γ is secreted by T lymphocytes and natural killer (NK) cells and functions to up-regulate the production of reactive nitrogen intermediates by macrophages (28). Nitric oxide (NO) is produced by the inducible NO synthase (iNOS), which catalyzes the oxidation of l-arginine into NO and l-citrulline. NO has been shown to be important for killing or controlling the proliferation of many intracellular pathogens, such as Leishmania major (29), Toxoplasma gondii (30), and Mycobacterium tuberculosis (31).
The ability of Y. pestis to survive and replicate in naïve macrophages in vitro has been well documented (4, 23-25). The plasmids pCD1 and pPCP1 are dispensable for in vitro replication in naïve macrophages (25, 32). The response regulator PhoP is required for survival and replication in naïve macrophages, and a PhoP- mutant shows a modest reduction in virulence (33, 34). Beyond this, little is known about the genetic basis for intracellular growth of Y. pestis or its importance for the pathogenesis of bubonic and pneumonic plague. In addition, it is not known whether Y. pestis can survive in activated macrophages. Here, we investigated the effect of IFN-γ treatment on the ability of Y. pestis to replicate in macrophages. We found that Y. pestis is able to replicate in macrophages that are exposed to IFN-γ after infection, and that a gene encoded within the pgm locus is required for this activity. These findings have important implications for understanding both the protective host immune response to plague and the molecular basis for Y. pestis pathogenesis.
Methods
Bacterial Strains and Growth Conditions. The Y. pestis strains used in this study are listed in Table 1, which is published as supporting information on the PNAS web site. KIM10+ and KIM10 were obtained from Kathleen McDonough (Wadsworth Center, New York State Department of Health, Albany, NY). Y. pestis strains were cultivated at 26°C on heart infusion (HI) agar plates for 50 h or in HI broth with aeration for 20 h. Escherichia coli strains DH5α (35), NovaBlue (Novagen), and S17-1 lysogenized with λpir (36) were grown as described (32). The Congo red (CR)-binding or Hms+ phenotype of Y. pestis strains was assessed on CR agar plates (37) incubated at 26°C. When needed, the bacterial growth medium was supplemented with ampicillin at 100 μg/ml, chloramphenicol at 10 μg/ml, kanamycin at 25 μg/ml, or tetracycline at 10 μg/ml.
DNA Amplification by PCR and Sequencing. The oligonucleotides used are listed in Table 2, which is published as supporting information on the PNAS web site, and were purchased from Invitrogen. Amplification of Y. pestis DNA was performed as described (32) with an annealing temperature determined by using MacVector (Accelrys, San Diego). Plasmid DNA and PCR products were sequenced by using an Applied Biosystems PRISM BigDye Terminator cycle sequencing kit (Applied Biosystems) and an Applied Biosystems 3100 automatic genetic analyzer.
Plasmid and Strain Constructions. The plasmids listed in Table 1 were introduced into Y. pestis by electroporation (38) or conjugation (39). Random transposition mutagenesis of pSDR973 was performed by using the EZ::TN<TET-1> Insertion kit, following a procedure supplied by the manufacturer (Epicentre, Beaverton, OR). The product of the reaction was electroporated into DH5α. Tetracycline-resistant (TcR) and kanamycin-resistant (KmR) transformants were isolated, and the locations of transposon insertions in the insert region of pSDR973 were mapped by using PCR and appropriate pairs of primers listed in Table 2 (see Fig. 5C). The resulting plasmids were introduced into KIM10/GFP by electroporation. The exact positions of the transposon insertions in ORFs Y2384 and Y2385 (subsequently named RipB and RipA, respectively) were determined by sequence analysis of PCR products by using TET-1 FP-1 and TET-1 RP-1 primers. For complementation analysis, the ripA ORF was amplified by PCR by using the oligonucleotides ripA-EcoRI-F2 and ripA-XbaI-R2 and whole KIM10+ extract as DNA template. Purified PCR product was introduced into pMMB67EH between the EcoRI and XbaI sites. The resulting plasmid pripA+ was then introduced by electroporation into S17λpir. A fosmid library harboring DNA from KIM10+ strain was constructed by using a fosmid library production kit (Epicentre) following the manufacturer's guidelines.
Construction of a Chromosomal Mutation and Complemented Strains in Y. pestis. The ripA gene was inactivated in KIM6+ using the pKOBEG-sacB system (40). For this purpose, the pKOBEG-sacB plasmid was first introduced into KIM6+ by electroporation. The resulting strain (KIM6+/pKOBEG-sacB) was grown and treated to prepare electrocompetent cells (40). Linear DNA was prepared by PCR by using pSDR973.34 as a template and the primer pair ripA-F1 and ripA-R1. Approximately 1 μg of purified PCR product was used to electroporate KIM10+/pKOBEG-sacB or KIM6+/pKOBEG-sacB cells. Chloramphenicol-resistant (CmR) and tetracycline-resistant (TcR) transformants harboring the mutated allele were screened by PCR, and selected clones were plated onto LB plates containing 10% sucrose. CmS TcR transformants were isolated and conjugated with S17λpir containing p67GFP3.1 or pripA+ (see above and Table 1). Transconjugants were selected on Yersinia selective agar plates (Oxoid, Hampshire, U.K.) containing ampicillin.
Macrophage Infections, Colony-Forming Units (cfu) Assay, and Microscopy. All tissue culture reagents were purchased from Life Technologies, Grand Island, NY, unless specified. Bone marrow macrophages (BMM) were isolated from female C57BL/6 WT or congenic iNOS-/- female mice (The Jackson Laboratory) and infected as described (32, 41), with the exception that when needed, LPS (Sigma, E. coli 026:B6; 100 ng/ml final concentration) and IFN-γ (100 units/ml final concentration; Roche Molecular Biochemicals, Indianapolis) were added to BMM 20 min after infection. cfu assay and detection of intracellular bacteria by microscopy were carried out as described (32).
Measurement of NO Levels. NO levels in IFN-γ-activated macrophages were determined by measuring the accumulation of nitrite (NO-2), a stable metabolite of the reaction of NO with oxygen, using the Griess reaction assay (42) as follows. Conditioned medium was collected 25 h after infection and centrifuged (13,000 × g) for 10 min at room temperature. An aliquot of the supernatant was mixed with an equal volume of Griess reagent [0.5% sulfanilamide; 0.05% N-(1-naphthyl)ethylenediamide in 2.5% acetic acid] and incubated 10 min at room temperature. The samples were measured spectrophotometrically at 550 nm, and the concentration of NO-2 was calculated by using a standard curve prepared with sodium nitrite.
Western Blot Analysis. BMM were cultured in 24-well plates and infected as described above. Twenty-five hours after infection, the cells were washed with PBS, lysed in Laemmli buffer 1× supplemented with a mixture of protease inhibitors (Roche), and the lysates were analyzed by immunoblotting following standard procedures (43). The anti-iNOS antibody and the mouse secondary antibody conjugated to horseradish peroxidase were, respectively, from Transduction Laboratories, Lexington, KY, and Jackson ImmunoResearch, and were used as recommended by the suppliers. The anti-src kinase-associated protein homologue (SKAP-Hom) antibody was used as described (44).
Results
Survival and Replication of Y. pestis in IFN-γ-Activated Macrophages. We recently confirmed that Y. pestis KIM10+, which lacks pCD1 and pPCP1, replicates in naïve murine macrophages (32). However, whether Y. pestis can survive in activated macrophages is still unknown. Macrophages become activated after bacterial infection in vivo when cytokine-activated T lymphocytes or natural killer (NK) cells secrete IFN-γ. Although pCD1-encoded factors act to reduce IFN-γ production during infection (6), recent experiments suggest that infection of mice with pCD1+ Y. pestis does stimulate expression of IFN-γ mRNA at low levels in NK cells (7). Thus, it is likely that pCD1+ Y. pestis encounters activated macrophages during infection. To mimic this in vivo response, unactivated BMM were infected with KIM10+/GFP and then activated with IFN-γ (postactivated) and bacterial survival and growth assessed via GFP expression (see Methods). Interestingly, similar numbers of GFP-positive bacteria were detected after 25 h of infection in unactivated and postactivated BMM (Fig. 1 a, b, e, and f; see also Fig. 6 c-f, which is published as supporting information on the PNAS web site). To determine whether macrophages were activated under these conditions, expression of the iNOS, a marker for macrophage activation, was examined by immunoblotting. The iNOS was expressed under these conditions (see Fig. 3B, lane 4), showing that Y. pestis can survive and replicate in postactivated macrophages.
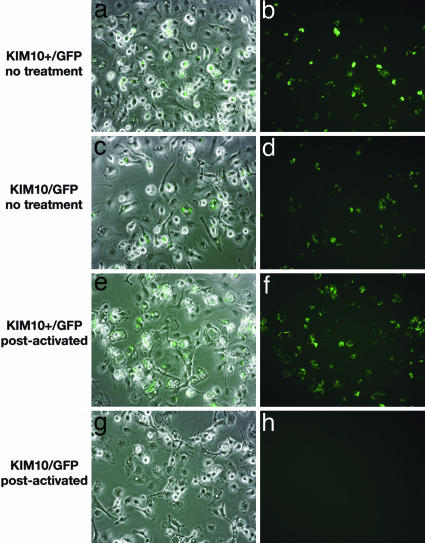
Fig. 1.
Δpgm Y. pestis does not survive in postactivated macrophages. BMM were infected with KIM10+/GFP (pgm+)(a, b, e, and f) or KIM10/GFP (Δpgm) (c, d, g, and h) for 20 min. Gentamicin was then added to prevent survival of extracellular bacteria. The BMM were then left untreated (a- d) or exposed to IFN-γ (e- h; postactivated). After 24 h of infection, isopropyl β-d-thiogalactoside was added to the infected cells to induce GFP expression. One hour later, the samples were fixed and examined by fluorescence or phase microscopy. Representative images were captured by digital photography. Overlays of GFP expression and phase-contrast images are shown (a, c, e, and g).
Fig. 3.
Reduction of NO levels in postactivated macrophages infected with pgm+ Y. pestis is not controlled at the level of iNOS expression. (A) BMM infected with KIM10+/GFP or KIM10/GFP were exposed to IFN-γ. Uninfected BMM were exposed to LPS and IFN-γ. Twenty-five hours after infection or LPS/IFN-γ exposure, the nitrite concentration in the supernatant of the macrophages was measured. Nitrite levels in infected cells were normalized relative to the nitrite levels in uninfected macrophages exposed to LPS/IFN-γ. Each bar represents the average of five infections from two independent experiments. (B) Uninfected BMM exposed to LPS, or BMM infected with KIM10+/GFP or KIM10/GFP, were incubated for 25 h in the presence or absence of IFN-γ. Lysates of the infected macrophages were analyzed by immunoblotting with an anti-iNOS antibody or with an anti-SKAP-HOM antiserum to control for loading.
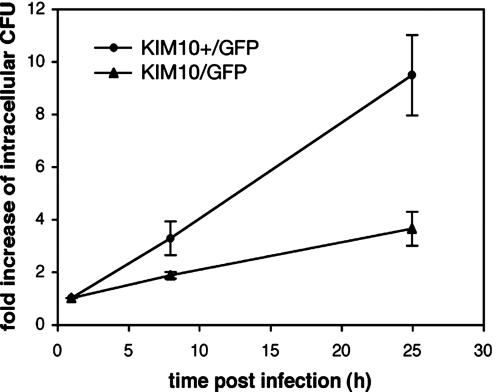
The pgm Locus Is Required for Replication of Y. pestis in Postactivated Macrophages. To determine whether the pgm locus played a role in the survival of Y. pestis in postactivated BMM, we compared two strains of Y. pestis, KIM10+/GFP (pgm+) and KIM10/GFP (Δpgm), for their ability to replicate in unactivated or postactivated macrophages. Both KIM10+/GFP and KIM10/GFP were able to replicate in unactivated macrophages (Fig. 1 b and d), as expected from earlier work (4, 23-25). However, a cfu assay revealed that KIM10/GFP replicated with slower kinetics than KIM10+/GFP (Fig. 2). The number of KIM10+/GFP cfu increased on average by 3.3-fold 8 h after infection and 9.5-fold after 25 h (Fig. 2). For KIM10/GFP, the number of cfu increased only ≈3.6-fold after 25 h (Fig. 2). Furthermore, when KIM10+/GFP and KIM10/GFP were compared for replication in postactivated BMM, a striking difference was observed: KIM10/GFP was completely defective for intracellular replication (Fig. 1 g and h). Similar numbers of KIM10+/GFP and KIM10/GFP were internalized by macrophages (data not shown), suggesting that the pgm locus is important for optimal replication of Y. pestis in naïve macrophages and is required for the survival of Y. pestis in postactivated macrophages.
Fig. 2.
Role of the pgm locus in the replication of Y. pestis in naïve macrophages. Unactivated BMM were infected with KIM10+/GFP or KIM10/GFP for 20 min before gentamicin addition to the medium (0 h postinfection). At the indicated times postinfection, intracellular bacteria were recovered and enumerated by cfu assay. Fold increase values are normalized relative to the 1-h value and represent averages of triplicate samples from one representative experiment. Error bars show the standard deviations.
Replication of pgm+Y. pestis in Postactivated Macrophages Correlates with Decreased NO Levels. To determine whether replication of pgm+ Y. pestis in macrophages is associated with reduced levels of NO, BMM were infected with KIM10+/GFP or KIM10/GFP in the presence or absence of IFN-γ as described (see Methods). Twenty-five hours after infection, the supernatants of macrophage cultures were collected. The concentration of nitrite, an end product of the oxidation of NO, was measured in the supernatants by using the Griess reaction assay (Methods). Uninfected BMM were in parallel incubated with a mixture of IFN-γ and LPS to determine the level of NO produced by activated BMM. Postactivated macrophages infected with KIM10/GFP had a similar amount of NO as uninfected activated macrophages (Fig. 3A). In contrast, the level of NO in postactivated macrophages infected with KIM10+/GFP was on average 40% lower compared with the uninfected control (Fig. 3A).
Because these results suggested that pgm+ Y. pestis might be interfering with the production of NO in postactivated macrophages, we investigated whether there was a similar effect on iNOS protein production. As shown in Fig. 3B, Western blot analysis using total cell extracts of BMM and an anti-iNOS antibody revealed slightly higher levels of iNOS in postactivated macrophages infected with KIM10+/GFP (lane 4) than in postactivated macrophages infected with KIM10/GFP (lane 6) or in uninfected BMM stimulated with LPS/IFN-γ (lane 2). Therefore, the reduced level of NO by postactivated macrophages infected with KIM10+/GFP could not be attributed to a reduced expression of the iNOS protein.
To assess whether killing of Δpgm Y. pestis in postactivated macrophages is NO-dependent, replication of KIM10+/GFP and KIM10/GFP was compared in postactivated macrophages obtained from iNOS-/- mice (Methods). Unlike iNOS+/+ macrophages, iNOS-/- macrophages were not able to prevent replication of Δpgm Y. pestis (Fig. 4). These data indicate that NO in postactivated macrophages is bactericidal for Δpgm Y. pestis, and that pgm+ Y. pestis can replicate in postactivated macrophages by reducing the level of NO.
Fig. 4.
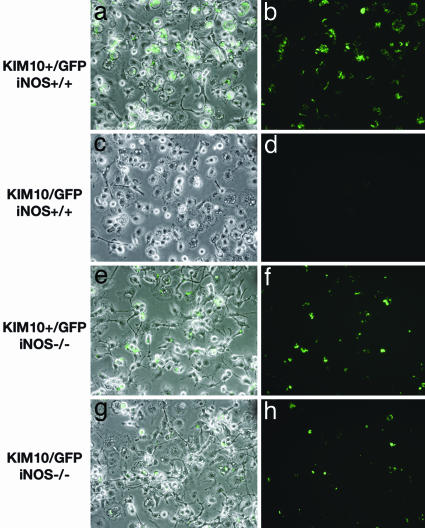
Killing of Δpgm Y. pestis in postactivated macrophages depends on iNOS expression. BMM from iNOS+/+ (a- d) or iNOS-/- (e- h) mice were infected with KIM10+/GFP (a, b, e, and f) or KIM10/GFP (c, d, g, and h), exposed to IFN-γ, and processed for phase and fluorescence microscopy as described in the legend of Fig. 1.
Identification of Genes in the pgm Locus That Are Required for Replication of Y. pestis in Postactivated Macrophages. To define which genes within the pgm locus are required for Y. pestis replication in postactivated macrophages, we took advantage of Yersinia strains with varying pgm locus genotypes (45). A serogroup O2 Y. pseudotuberculosis strain, which carries the pigmentation segment but lacks the HPI, replicated in postactivated macrophages (data not shown). In contrast, a biotype 1B Y. enterocolitica strain, carrying the HPI but lacking the pigmentation segment, does not replicate in postactivated macrophages (46). These results suggest that the determinants required for replication in postactivated macrophages are encoded within the pigmentation segment (see Fig. 5A). To further define the region involved in this phenotype, we introduced a fosmid library of Y. pestis DNA into KIM10/GFP and screened for Congo red-binding (Hms+) transformants (Methods). None of the Hms+ isolates carrying fosmids with ≈40-kb insertions were able to replicate in postactivated macrophages (data not shown). Thus, the critical genes are not located in the left half of the pigmentation segment (see Fig. 5A). Next, recombinant plasmids containing inserts that encompass the right half of the pigmentation segment (47) (Fig. 5A) were introduced into KIM10/GFP or KIM10 (see Table 1), and the resulting strains were then tested by microscopy and cfu assays for their ability to replicate in postactivated macrophages. One plasmid, pSDR973, was able to complement KIM10/GFP for growth in postactivated macrophages (Fig. 5B and data not shown).
Sequence analysis of pSDR973 revealed nine putative ORFs (Y2377-Y2387) encoded within the insert (Fig. 5C) (48). To identify ORFs in this insert that are required for KIM10/GFP/pSDR973 to replicate in postactivated macrophages, random mutagenesis by in vitro transposition was performed (Methods). Two derivatives of pSDR973, which contained single tetR transposon insertions in either Y2384 (KIM10/GFP/pSDR973.1) or Y2385 (KIM10/GFP/pSDR973.34), displayed a clear defect in survival after 25 h of infection in postactivated macrophages (Fig. 5 B and C). Strains with insertions in other ORFs or in intragenic regions had a phenotype similar to KIM10/GFP/pSDR973 (data not shown). GFP-positive bacteria were occasionally found in postactivated macrophages infected with KIM10/GFP/pSDR973.1 (Fig. 5B), suggesting that the y2384::tetR insertion mutant was not completely defective. The greatly decreased ability of the mutants with insertions in Y2384 and Y2385 to survive in postactivated macrophages correlated with an inability to decrease NO levels (Fig. 5D). ORFs Y2384 and Y2385 are hereafter referred to as ripB and ripA (required for intracellular proliferation), respectively.
To further demonstrate that ripA is required for replication of Y. pestis in postactivated macrophages, we introduced the mutated allele ripA::tetR into the chromosome of a pgm+ strain (KIM6+) by allelic exchange (Methods). A plasmid expressing ripA under the control of an isopropyl β-d-thiogalactoside inducible promoter (pripA+) was constructed and introduced into the ripA::tetR mutant (KIM6ripA) for complementation (Methods). KIM6+, KIM6ripA, and KIM6ripA/pripA+ were assayed for replication in postactivated macrophages, using immunofluorescence microscopy and anti-Yersinia antiserum. As shown in Fig. 7 A and B, which is published as supporting information on the PNAS web site, KIM6ripA replicated in unactivated but not in postactivated macrophages. Complemented strain KIM6ripA/pripA+ regained the ability to replicate in postactivated macrophages (Fig. 7 A and B).
Analysis of the Region Encoding for ripA and ripB. A blastp search (49) of the GenBank protein database performed using the gene products of ripB and ripA revealed that ripB is predicted to encode a protein with similarity to a monoamine oxidase and ripA to a CoA transferase. Interestingly, two proteins similar to RipB and RipA (FkbR2 and Cat2, respectively; Fig. 5C) were recently shown to be encoded in the Salmonella enterica serovar enteritidis pathogenicity island 6 (SPI6) that is required for bacterial infection of chicken macrophages [GenBank accession no. AF376036 (50)]. SPI6 is also conserved in S. enterica serovar typhimurium (GenBank accession no. NC003197). Moreover, the region encompassing ripB and ripA in Y. pestis is similar to the SPI6 in gene organization and sequence (Fig. 5C). Upstream of ripB are genes encoding a putative citrate lyase β subunit (Y2383 for Y. pestis and citE for Salmonella enteritidis) and a putative transcriptional regulator (Y2382 for Y. pestis and stmR for S. enteritidis) (Fig. 5C). In contrast, the ORFs upstream of Y2382/stmR and downstream of ripA/cat2 are divergent (Fig. 5C). Y2383-ripBA appears to be organized as an operon, which we refer to as the rip operon. Because the phenotype of the ripB mutant could be due to a polar effect on expression of ripA, we can only conclude that ripA is required for replication in activated macrophages at this time. Together, these data show that Y. pestis harbors a putative operon similar to an operon in the SPI6 of S. enteritidis and S. typhimurium, and that at least one gene within this operon (ripA) is essential for Y. pestis to survive in activated macrophages.
Discussion
The ability of Y. pestis to survive and/or replicate inside macrophages is likely to be important for pathogenesis in the human or animal host. Y. pestis survives and replicates in macrophages in vitro (4, 23-25), but the defense mechanisms of macrophages that function to counteract intracellular Y. pestis infection have not been studied in detail. We show that Y. pestis is able to resist the enhanced killing activity of macrophages exposed to IFN-γ, and that a putative operon that is similar to SPI6 of S. typhimurium and S. enteritidis and located in the pgm segment is required for this process.
The pigmentation segment harbors a large number of genes (16, 51) with unknown functions. The hmsHFRS locus, which is required for biofilm formation and transmission from fleas to mammals (17, 21), is not required for Y. pestis replication in postactivated macrophages, and we have identified a previously uncharacterized gene, ripA, that is required for this process. Analysis of the region encompassing ripA and ripB revealed that, together with Y2383, ripA and ripB constitute a putative operon similar to an operon in the SPI6. SPI6 is required for S. enteritidis infection of chicken macrophages (50). Interestingly, an RNA microarray analysis performed on S. typhimurium while growing inside murine macrophages showed that the genes corresponding to ripA, ripB, and Y2383 are highly up-regulated from 4 to 12 h postinfection, with a maximum level at 12 h (52). These findings are consistent with the idea that the rip genes would be expressed by Y. pestis inside macrophages.
Replication in postactivated macrophages by pgm+ bacteria correlated with a reduced accumulation of NO and the ability of Δpgm bacteria to replicate in iNOS-/- macrophages demonstrated that the NO in activated macrophages was bactericidal to Δpgm Y. pestis. Interestingly, the inability of ripA and ripB mutants to replicate in postactivated macrophages also correlated with an inability to reduce NO levels. Because reduced NO levels did not result from decreased iNOS expression, pgm+ Y. pestis may detoxify NO or counteract the reaction by reducing or eliminating the iNOS substrate. For example, Helicobacter pylori inhibits NO production via the expression of an endogenous l-arginase (RocF), which competes with iNOS for the common substrate l-arginine (53). However, the products of the rip operon are not predicted to function as l-arginases, and no clear l-arginase homologue is encoded in the KIM genome, so the mechanism of NO reduction by Y. pestis may be mechanistically different from competition for iNOS substrate.
The genes ripA and ripB encode for a putative acetyl CoA transferase and a putative monoamine oxidase (or dehydrogenase) (MaoC), respectively. MaoC catalyzes the oxidation of a monoamine into an aldehyde, plus ammonium and H2O2. Acetyl CoA transferase converts acetyl-CoA to acetate. The first ORF in the rip operon (Y2383) encodes a protein with similarity to the citrate lyase β subunit. Y2383 shows 33% identity (96 of 286 amino acids) to the citrate lyase β subunit of Klebsiella pneumoniae (GenBank accession no. S60775). Citrate lyase (composed of α, β, and γ subunits) is a fermentative enzyme that cleaves citrate into oxaloacetate and acetate. Additional studies will be required to determine how the rip gene products, which most likely function as metabolic enzymes, promote replication of Y. pestis in activated macrophages.
The Ybt iron transport system, encoded by the HPI, is essential for virulence from peripheral routes of infection, as shown by inactivation of specific ybt genes (12, 22). However, the previously undescribed rip locus may also contribute to the virulence of pgm+ Y. pestis, and studies to answer this question are underway.
Supplementary Material
Acknowledgments
We thank K. McDonough (Wadsworth Center, New York State Department of Health, Albany) for sending the Y. pestis strains and G. Viboud and other members of the J.B.B. laboratory for discussions and careful reading of the manuscript. We are grateful to A. Derbise (Pasteur Institute, Paris) for providing the pKOBEG plasmid and for helpful technical suggestions. This work was supported by Grants P01 AI55621 and R01 AI48507 (to J.B.B.) from the National Institutes of Health.
Author contributions: C.P. and J.B.B. designed research; C.P. performed research; J.P.G. and R.D.P. contributed new reagents/analytic tools; C.P. and J.B.B. analyzed data; and C.P., R.D.P., and J.B.B. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Ybt, yersiniabactin; HPI, high pathogenicity island; pgm locus, pigmentation locus; iNOS, inducible NO synthase; cfu, colony-forming unit(s); BMM, bone marrow macrophage; SPI6, Salmonella enterica serovar enteritidis pathogenicity island 6.
References
- 1.Inglesby, T. V., Dennis, D. T., Henderson, D. A., Bartlett, J. G., Ascher, M. S., Eitzen, E., Fine, A. D., Friedlander, A. M., Hauer, J., Koerner, J. F., et al. (2000) J. Am. Med. Assoc. 283, 2281-2290. [DOI] [PubMed] [Google Scholar]
- 2.Brubaker, R. R., Beesley, E. D. & Surgalla, M. J. (1965) Science 149, 422-424. [DOI] [PubMed] [Google Scholar]
- 3.Sodeinde, O. A., Subrahmanyam, Y. V., Stark, K., Quan, T., Bao, Y. & Goguen, J. D. (1992) Science 258, 1004-1007. [DOI] [PubMed] [Google Scholar]
- 4.Cavanaugh, D. C. & Randall, R. (1959) J. Immunol. 85, 348-363. [PubMed] [Google Scholar]
- 5.Hinnebusch, B. J., Rudolph, A. E., Cherepanov, P., Dixon, J. E., Schwan, T. G. & Forsberg, A. (2002) Science 296, 733-735. [DOI] [PubMed] [Google Scholar]
- 6.Nakajima, R. & Brubaker, R. R. (1993) Infect. Immun. 61, 23-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kerschen, E. J., Cohen, D. A., Kaplan, A. M. & Straley, S. C. (2004) Infect. Immun. 72, 4589-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornelis, G. R. (2002) Nat. Rev. Mol. Cell. Biol. 3, 742-752. [DOI] [PubMed] [Google Scholar]
- 9.Burrows, T. W. & Jackson, S. (1956) Br. J. Exp. Pathol. 37, 577-583. [PMC free article] [PubMed] [Google Scholar]
- 10.Une, T. & Brubaker, R. R. (1984) Infect. Immun. 43, 895-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bearden, S. W., Fetherston, J. D. & Perry, R. D. (1997) Infect. Immun. 65, 1659-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bearden, S. W. & Perry, R. D. (1999) Mol. Microbiol. 32, 403-414. [DOI] [PubMed] [Google Scholar]
- 13.Carniel, E., Mercereau-Puijalon, O. & Bonnefoy, S. (1989) Infect. Immun. 57, 1211-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Almeida, A. M., Guiyoule, A., Guilvout, I., Iteman, I., Baranton, G. & Carniel, E. (1993) Microb. Pathog. 14, 9-21. [DOI] [PubMed] [Google Scholar]
- 15.Burrows, T. W. & Jackson, S. (1956) Br. J. Exp. Pathol. 37, 570-576. [PMC free article] [PubMed] [Google Scholar]
- 16.Perry, R. D., Pendrak, M. L. & Schuetze, P. (1990) J. Bacteriol. 172, 5929-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hinnebusch, B. J., Perry, R. D. & Schwan, T. G. (1996) Science 273, 367-370. [DOI] [PubMed] [Google Scholar]
- 18.Darby, C., Hsu, J. W., Ghori, N. & Falkow, S. (2002) Nature 417, 243-244. [DOI] [PubMed] [Google Scholar]
- 19.Buchrieser, C., Prentice, M. & Carniel, E. (1998) J. Bacteriol. 180, 2321-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarrett, C. O., Deak, E., Isherwood, K. E., Oyston, P. C., Fischer, E. R., Whitney, A. R., Kobayashi, S. D., DeLeo, F. R. & Hinnebusch, B. J. (2004) J. Infect. Dis. 190, 783-792. [DOI] [PubMed] [Google Scholar]
- 21.Kirillina, O., Fetherston, J. D., Bobrov, A. G., Abney, J. & Perry, R. D. (2004) Mol. Microbiol. 54, 75-88. [DOI] [PubMed] [Google Scholar]
- 22.Fetherston, J. D., Bertolino, V. J. & Perry, R. D. (1999) Mol. Microbiol. 32, 289-299. [DOI] [PubMed] [Google Scholar]
- 23.Janssen, W. A. & Surgalla, M. J. (1969) Science 163, 950-952. [DOI] [PubMed] [Google Scholar]
- 24.Charnetzky, W. T. & Shuford, W. W. (1985) Infect. Immun. 47, 234-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Straley, S. C. & Harmon, P. A. (1984) Infect. Immun. 45, 649-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duclos, S. & Desjardins, M. (2000) Cell Microbiol. 2, 365-377. [DOI] [PubMed] [Google Scholar]
- 27.Vieira, O. V., Botelho, R. J. & Grinstein, S. (2002) Biochem. J. 366, 689-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacMicking, J., Xie, Q. W. & Nathan, C. (1997) Annu. Rev. Immunol. 15, 323-350. [DOI] [PubMed] [Google Scholar]
- 29.Liew, F. Y., Millott, S., Parkinson, C., Palmer, R. M. & Moncada, S. (1990) J. Immunol. 144, 4794-4797. [PubMed] [Google Scholar]
- 30.Scharton-Kersten, T. M., Yap, G., Magram, J. & Sher, A. (1997) J. Exp. Med. 185, 1261-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacMicking, J. D., North, R. J., LaCourse, R., Mudgett, J. S., Shah, S. K. & Nathan, C. F. (1997) Proc. Natl. Acad. Sci. USA 94, 5243-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pujol, C. & Bliska, J. B. (2003) Infect. Immun. 71, 5892-5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oyston, P. C. F., Dorrell, N., Williams, K., Li, S.-R., Green, M., Titball, R. W. & Wren, B. (2000) Infect. Immun. 68, 3419-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grabenstein, J. P., Marceau, M., Pujol, C., Simonet, M. & Bliska, J. B. (2004) Infect. Immun. 72, 4973-4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed..
- 36.Simon, R., Priefer, U. & Puhler, A. (1983) Biotechnology 1, 784-791. [Google Scholar]
- 37.Surgalla, M. J. & Beesley, E. D. (1969) Appl. Microbiol. 18, 834-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conchas, R. F. & Carniel, E. (1990) Gene 87, 133-137. [DOI] [PubMed] [Google Scholar]
- 39.Bliska, J. B. & Black, D. S. (1995) Infect. Immun. 63, 681-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Derbise, A., Lesic, B., Dacheux, D., Ghigo, J. M. & Carniel, E. (2003) FEMS Immunol. Med. Microbiol. 38, 113-116. [DOI] [PubMed] [Google Scholar]
- 41.Celada, A., Gray, P. W., Rinderknecht, E. & Schreiber, R. D. (1984) J. Exp. Med. 160, 55-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Green, L. C., Wagner, D. A., Glogowski, J., Skipper, P. L., Wishnok, J. S. & Tannenbaum, S. R. (1982) Anal. Biochem. 126, 131-138. [DOI] [PubMed] [Google Scholar]
- 43.Harlow, E. & Lane, D. (1988) Antibodies: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 44.Black, D. S., Marie-Cardine, A., Schraven, B. & Bliska, J. B. (2000) Cell. Microbiol. 2, 401-414. [DOI] [PubMed] [Google Scholar]
- 45.Carniel, E. (1999) Int. Microbiol. 2, 161-167. [PubMed] [Google Scholar]
- 46.Pujol, C. & Bliska, J. B. (2005) Clin. Immunol. 114, 216-226. [DOI] [PubMed] [Google Scholar]
- 47.Fetherston, J. D., Schuetze, P. & Perry, R. D. (1992) Mol. Microbiol. 6, 2693-2704. [DOI] [PubMed] [Google Scholar]
- 48.Deng, W., Burland, V., Plunkett, G., 3rd, Boutin, A., Mayhew, G. F., Liss, P., Perna, N. T., Rose, D. J., Mau, B., Zhou, S., et al. (2002) J. Bacteriol. 184, 4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao, Y., Jansen, R., Gaastra, W., Arkesteijn, G., van der Zeijst, B. A. & van Putten, J. P. (2002) Infect. Immun. 70, 5319-5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buchrieser, C., Rusniok, C., Frangeul, L., Couve, E., Billault, A., Kunst, F., Carniel, E. & Glaser, P. (1999) Infect. Immun. 67, 4851-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eriksson, S., Lucchini, S., Thompson, A., Rhen, M. & Hinton, J. C. D. (2003) Mol. Microbiol. 47, 103-118. [DOI] [PubMed] [Google Scholar]
- 53.Gobert, A. P., McGee, D. J., Akhtar, M., Mendz, G. L., Newton, J. C., Cheng, Y., Mobley, H. L. & Wilson, K. T. (2001) Proc. Natl. Acad. Sci. USA 98, 13844-13849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.