Abstract
Chimpanzees are the closest evolutionary cousins of humans, sharing >99% identity in most protein sequences. Plasmodium falciparum is the major worldwide cause of malaria mortality. Plasmodium reichenowi, a morphologically identical and genetically very similar parasite, infects chimpanzees but not humans. Conversely, experimental P. falciparum infection causes brief moderate parasitization and no severe infection in chimpanzees. This surprising host specificity remains unexplained. We modified and enhanced traditional methods for measuring sialic acid (Sia)-dependent recognition of glycophorins by merozoite erythrocyte-binding proteins, eliminating interference caused by endogenous Sias on transfected cells, and by using erythroleukemia cells to allow experimental manipulation of Sia content. We present evidence that these remarkable differences among such closely related host-parasite pairs is caused by species-specific erythrocyte-recognition profiles, apparently related to the human-specific loss of the common primate Sia N-glycolylneuraminic acid. The major merozoite-binding protein erythrocyte-binding antigen-175 of P. falciparum apparently evolved to take selective advantage of the excess of the Sia N-acetylneuraminic acid (the precursor of N-glycolylneuraminic acid) on human erythrocytes. The contrasting preference of P. reichenowi erythrocyte-binding antigen-175 for N-glycolylneuraminic acid is likely the ancestral condition. The surprising ability of P. falciparum to cause disease in New World Aotus monkeys (geographically isolated from P. falciparum until arrival of peoples from the Old World) can be explained by parallel evolution of a human-like Sia expression pattern in these distantly related primates. These results also have implications for the prehistory of hominids and for the genetic origins and recent emergence of P. falciparum as a major human pathogen.
Keywords: human origins, Plasmodium, sialic acids, primates, Aotus
Of the four Plasmodium species that cause malaria in humans, Plasmodium falciparum causes almost all malaria mortality (>1 million deaths per year, mostly among children and pregnant women) (1, 2). Although other species such as Plasmodium vivax can cause severe anemia and morbidity, only P. falciparum infection causes the most serious symptoms such as metabolic acidosis, respiratory distress, and cerebral malaria, which is characterized by progressing coma, convulsions, and frequently death (1, 2). P. falciparum shared a remote common ancestor with avian malarial parasites and is related only distantly to the other three human malaria species or to parasites of Old World monkeys (3-8). Its only known close relative is the morphologically indistinguishable great-ape parasite Plasmodium reichenowi (9), first described between 1917 and 1920 by Reichenow (10), in blood smears from wild chimpanzees and gorillas in Cameroon.
The morphological similarity of this ape parasite initially made it indistinguishable from P. falciparum, and it was suggested that apes might be a natural reservoir for P. falciparum. However, in vivo experiments performed at that time (and unthinkable today) showed that humans could not be infected with parasitized chimpanzee blood (11, 12). Conversely, chimpanzees were not successfully infected with blood from humans suffering from P. falciparum parasitemia (12). These results justified the original designation by a different species name, P. reichenowi. This distinction was confirmed more recently by genetic comparisons (3, 7, 13). Also, it has been observed that captive chimpanzees in Gabon did not get infected with P. falciparum in the face of a high rate of attack among their keepers and despite being exposed to the same mosquito-containing environment (14). Indeed, even after splenectomy to increase parasite survival, experimentally infected chimpanzees do not develop the overwhelming parasitemia and severe “malignant tertian malaria” that is seen in humans (9, 15).
Chimpanzees are the closest evolutionary relatives of humans, sharing a common ancestor ≈6-7 million years ago (mya) (16-19). P. falciparum and P. reichenowi are estimated to have shared a common ancestor at about the same time (3, 7) but have since clearly diverged to become host-specific. This highly species-specific behavior of closely related parasites and hosts is intriguing and begs an explanation.
Multiple proteins expressed by Plasmodium merozoites are involved in the invasion of erythrocytes. Merozoite invasion ligands fall into two broad superfamilies: the erythrocyte-binding-like (EBL, or more appropriately the Duffy-binding-like) and reticulocyte-binding-like proteins, members of which are loosely conserved across human, simian, and rodent Plasmodium species (2). Although the P. falciparum genome contains six EBL paralogs, only erythrocyte-binding antigen (EBA) 175 was originally identified on the basis of its ability to bind erythrocytes (20) rather than through homology searching. EBA-175 recognizes sialic acids (Sias) presented on clusters of O-linked glycans attached to erythrocyte surface glycophorin A (GYPA) (21), with a 30-aa stretch of the polypeptide carrying 11 O-glycans being critical for optimal binding (22, 23). The Cys-rich region II domain of EBA-175 (EBA-175-RII) is responsible for this recognition (22).
Although some P. falciparum strains can invade erythrocytes by using sialidase (neuraminidase)-resistant pathways (24), invasion under such conditions is almost always less efficient than into untreated erythrocytes, suggesting that such “alternative” pathways play accessory rather than primary roles in the invasion process. Furthermore, although no member of the P. falciparum EBL gene family has been shown thus far to be essential for invasion in vitro, all EBLs tested to date can be knocked out in some strains, and their relative importance in vivo has not been determined. Regardless, it seems that P. falciparum (Pf)EBA-175 plays an important role in invasion, even in strains that are able to use alternate pathways. When EBA-175 is knocked out in the W2-mef strain, alternative pathways are only used for invasion after the knockout, indicating that EBA-175 is the dominant invasion ligand before the knockout (25). Indeed, even in the 3D7 strain that naturally uses alternative pathways for invasion, knocking out EBA-175 affected the profile of erythrocyte receptors used for invasion, suggesting that EBA-175 still routinely played a role in erythrocyte invasion in this strain before the knockout despite alternative pathways being available (25). In contrast, knockout of other P. falciparum EBL family members, EBA-140/BAEBL (which binds Sias on glycophorin C/D) or EBA-181/JESEBL, did not cause major changes in the patterns of receptor usage after the knockout (26, 27). Thus, even in strains in which it is not essential, EBA-175 is still likely playing a role in invasion, but the same cannot be said for EBA-140/BAEBL or EBA-181/JSEBL.
The extent to which EBA-175 independent pathways are used in wild isolates is not yet clear. However, the only study of multiple P. falciparum isolates from Africa (where humans and chimpanzees coexist) suggested that EBA-175 is the primary invasion ligand in the majority of isolates (28). EBA-175-RII is also the only EBL domain that has been shown to be under significant positive selection pressure (29, 30). Furthermore, the polypeptide sequence of its primary target molecule (the Sia-capped glycoprotein GYPA) also shows evidence of rapid evolution (30) and extensive polymorphisms in humans (31-34). Overall, the weight of population and in vitro data suggests that EBA-175 plays a central, if not dominant, role in erythrocyte invasion in the majority of P. falciparum strains. Thus, we chose to focus on the binding specificity of PfEBA-175 and P. reichenowi (Pr)EBA-175 as a potential explanation for the striking host specificities of P. falciparum and P. reichenowi by using the corresponding Pf/PrEBA-140 molecules as controls. A major biochemical change in Sia biology occurred in the human lineage after divergence from our common ancestor with chimpanzees (35, 36). The most common mammalian Sias are N-glycolylneuraminic acid (Neu5Gc) and N-acetylneuraminic acid (Neu5Ac), Neu5Ac being the metabolic precursor of Neu5Gc. Humans cannot produce Neu5Gc from Neu5Ac because of a mutation in the CMAH gene (35). Thus, although human erythrocytes show no overall loss of Sias, they express no Neu5Gc and a great excess of the precursor, Neu5Ac (37). In contrast, chimpanzee erythrocytes carry a mixture of both Sias, with Neu5Gc being dominant (37).
Homologs of the EBLs have been identified in P. reichenowi (38, 39), but nothing is known about their Sia dependence for erythrocyte binding. We hypothesized that the binding preference of PfEBA-175 and PrEBA-175 for Neu5Ac or Neu5Gc is a primary factor responsible for species-specific binding to human and chimpanzee glycophorins and, given the central role of EBA-175 in invasion, may explain the marked host specificity of each parasite.
Methods
Expression Constructs of RII from PfEBA-175, PrEBA-175, PfEBA-140, and EBA-140. RII of PfEBA-140 from clone Dd2/Nmwasakindgift from L. Miller (National Institutes of Health, Bethesda) and was cloned into the T8 vector, which adds a signal for a glycosylphosphatidylinositol anchor, resulting in attachment to the surface of COS cells (40). RIIs from PfEBA-175, PrEBA-175, and PrEBA-140 were cloned and transferred into the NotI and EcoRI sites in the T8 vector. The PfEBA-175-RII region from the Camp strain was originally cloned in the pRE4 vector (22) and was also kindly provided by L. Miller. Both PrEBA-175 and PrEBA-140 were obtained from genomic DNA (39) and originally cloned in pCRScript (Promega) by using the SrfI site. All of the sequences were prepared for insertion into the T8 vector by using PCR primers generating the same restriction sites: PfEBA-175, forward (5′-ATTGCGGCCGCGGAAGAAATACTTCATCTAATAA) and reverse (5′-AGCCGCGAATTCCGAAGTTTGTTCATTATTTCTTATTATAG); PrEBA-175, forward (5′-ATTTGCGGCCGTGGAAGAGTTATGGAACTC) and reverse (5′-CGCTGAATTCGCTTCTGTTTTGAATCACGCCAG); and PrEBA-140, forward (5′-ATTATGCGGCCGCAATAAAAAGAAAATCAATAAAATG) and reverse (5′-GCCGGCTTAAAGATAAAATTATAGA-ACACATAGGCAAC). PCR amplifications were performed by using the high-fidelity enzyme AccuPrime Pfx (Invitrogen) and following manufacturer recommendations [briefly, a hot start at 94°C for 2 min, 30 cycles at 94°C for 15 s, 55°C for 30 s, and 68°C for 2 min]. pcDNA3.1 (Invitrogen) was used as negative control for sham transfection.
Cell Culture and Transfection. COS cells (American Type Culture Collection) were cultured in DMEM with 10% FCS (Invitrogen) at 37°C in 5% CO2. For erythrocyte-binding assays, cells were seeded 24 h before transfection onto a 12-well plate to obtain a 90% confluence. K562 cells (American Type Culture Collection) were incubated in Iscove's modified Dulbecco's medium (Invitrogen) containing 10% heat-inactivated human AB-type serum (Pel-Freez Biologicals). For K562 cell-binding assays, the same number of COS cells per well was seeded on two-well chamber slides (Nunc). Transfection was performed by using 4 μl per well of Lipofectamine 2000 (Invitrogen) and 0.8 μg of plasmid DNA diluted in Optimem (Invitrogen). Five hours after transfection, 20% FCS containing DMEM was added to each well for a final concentration of 10% FCS. COS-cell transfection rates were ≈70-75% in control experiments. All experiments were also controlled by parallel “sham” (vector-alone) transfections.
Anticoagulated Blood Samples. Human blood was obtained from healthy volunteers at the University of California at San Diego with institutional review board approval. Chimpanzee blood was kindly provided by the Yerkes National Primate Research Center (Atlanta). Owl monkey (Aotus nancymai) blood was obtained from Vanderbilt University (Nashville, TN), courtesy of Jon Kaas. Primate blood samples were shipped overnight on ice, and human samples were collected and stored overnight on ice to match the shipped samples. After centrifugation at 500 × g, erythrocytes were washed four times with PBS and maintained at 4°C in Elsevier's solution.
Erythrocyte-Binding Assay. Binding assays were performed 2 days after transfection of COS cells. Thirty microliters of compacted erythrocytes were resuspended in 470 μl of 0.05 M Hepes/140 mM NaCl (pH 6.9) and incubated with or without 20 milliunits of Arthrobacter ureafaciens sialidase (EY Laboratories) for 1 h at 37°C. COS7 cells were washed with 0.05 M Hepes/140 mM NaCl (pH 6.9) containing 1 mM CaCl2 and 1 mM MgCl2 and incubated for 1 h at 37°C with or without 5 milliunits per well of A. ureafaciens sialidase.
For binding assays, erythrocytes were washed twice in PBS and resuspended in RAS buffer (DMEM containing 0.25% BSA) up to ≈0.25% hematocrit. COS7 cells were also washed, and 500 μl of RAS buffer were added per well. Five-hundred microliters of the erythrocyte suspension were added to each well and allowed to interact with COS cells for 30 min at 37°C. Nonbound erythrocytes were removed by three PBS washes, and bound erythrocytes were examined immediately under an inverted microscope (Olympus CK2, Olympus, Melville, NY). Photos from 35-50 fields per well were taken by using a digital camera (Sony DKC-5000, Sony, Tokyo). The total number of erythrocytes per field was counted in 15 of these images, randomly selected, and the low background seen with sham transfection was subtracted.
K562 Cell Culture and Neu5Gc Loading. The human erythroleukemia-like cell line K562 (American Type Culture Collection) was maintained in Iscove's modified DMEM (Invitrogen) containing 10% heat-inactivated AB human serum (Pel-Freez Biologicals) to ensure that no Neu5Gc was present (41). Rabbit anti-human GYPA and anti-glycophorin C (Santa Cruz Biotechnology) were used to check glycophorin expression by flow cytometry. Some cells were incubated with 3 mM free Neu5Gc for 24 h to cause high expression of Neu5Gc on the cell surface (41). Other loading experiments were performed by incubation for 3 days in 100 μM of the per-O-acetylated precursor N-glycolylmannosamine (41, 42), resulting in a lower expression of Neu5Gc on the cell surface.
Binding Assay with Fluorescent K562 Cells. K562 cells (5 × 105) were desialylated following the same procedure as for erythrocytes. After three washings with PBS, they were resuspended into 1 ml of PBS and loaded with the vital fluorescent dye SNARF1-AM (Molecular Probes/Invitrogen) by adding 1 μl of a 1.5 mM stock solution in DMSO and incubating at 37°C for 30 min in the dark. After washing, cells were resuspended into RAS buffer, and 500 μl were added to transfected and desialylated COS7 cells. After 30 min, wells were washed three times with PBS, and the bound K562 cells were fixed with 1% paraformaldehyde in PBS for 10 min. The chamber walls were removed, and slides were mounted by using mounting medium for examination under a fluorescence microscope (Zeiss HBO 100). The low background seen with sham-transfected COS cells was subtracted.
Determination of Neu5Gc Content. Sias were released by mild acid, derivatized with 1,2-diamino-4,5-methylene dioxybenzene (Sigma), and analyzed by HPLC to determine the percentage of Neu5Gc in total Sias (41).
Statistical Analysis. Erythrocyte- or K562 cell-binding assay data from at least two different experiments were analyzed by using the T test in Microsoft excel. Data are expressed as mean ± SE.
Results
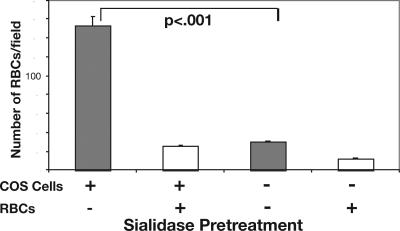
Cell-surface Sias of COS cells can interfere with the Sia-dependent binding of transfected P. falciparum and P. reichenowi EBAs to erythrocytes. Binding of erythrocytes to EBA-transfected COS cells is commonly used to test EBA-RII domain interactions with their natural receptors. Sia-binding EBA-RII domains from P. falciparum (PfEBA-175-RII) or P. reichenowi (PrEBA-175-RII) were transfected into COS cells as glycosylphosphatidylinositol-anchored proteins and assayed for interactions with human and chimpanzee erythrocytes. Because Plasmodium do not express or synthesize Sias (43, 44), EBAs naturally expressed on the parasite surface would not be blocked by any plasmodial ligand. However, as with any other mammalian cell, the surface of COS cells is rich in Sias. Traditional binding assays were therefore modified by introducing a sialidase pretreatment of the transfected COS cells. This is an approach that we have found to be important in similar studies of unrelated Sia-binding proteins to reduce competition by endogenous Sias on the COS cell surface (45). As predicted, sialidase pretreatment of COS cells expressing PfEBA-175-RII gave a marked increase in erythrocyte binding (see Fig. 1). Thus although traditional erythrocyte-binding assays (22) remain valid, the current approach maximizes interactions by using sialidase pretreatment of COS cells. The same principle was used to optimize a new assay involving recognition of glycophorins on human erythroleukemia (K562) cells (see below).
Fig. 1.
Sialidase pretreatment of transfected COS cells improves erythrocyte binding. COS7 cells were transfected with PfEBA-175-RII 2 days before the assay. Erythrocytes (RBCs), COS cells, or both were treated with sialidase or sham incubated in the same buffer without enzyme. Erythrocytes and COS cells then were allowed to interact for 30 min at 37°C, and total bound erythrocytes in 10 microscope fields were counted. Data are mean ± SEM.
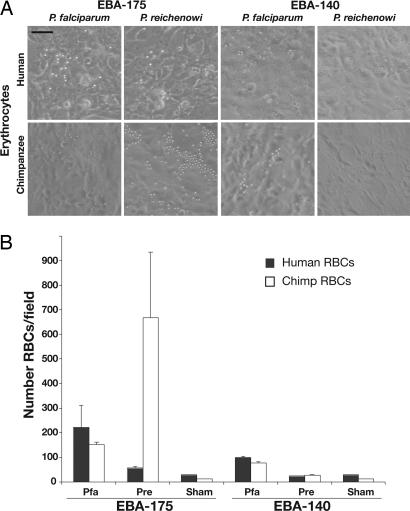
Recombinant PfEBA-175 and PrEBA-175 RIIs Show Sia-Dependent Species-Specific Preferential Binding of Erythrocytes. Binding assays using human and chimpanzee erythrocytes were performed as described above to test the specificity of the Sia-binding RII domains from both P. falciparum and P. reichenowi. As shown in Fig. 2, PfEBA-175-RII preferentially bound human erythrocytes, with a significant difference (P < 0.005 to P < 0.0005) found in comparing four independent pairs of human and chimpanzee erythrocytes. Conversely, PrEBA-175-RII bound chimpanzee erythrocytes much more efficiently than human ones (Fig. 2). Sialidase treatment of erythrocytes eliminated all binding (data not shown), establishing that Sias are critical to the interactions of both EBA-175s with both types of erythrocytes. In keeping with its presumed minor role in invasion, EBA-140 binding was much poorer. Although PfEBA-140-RII bound a few human and chimpanzee erythrocytes, even fewer were observed with transfected PrEBA-140-RII. Neither of the EBA-140s showed statistically significant species specificity in their weak binding.
Fig. 2.
EBA-175-RII from Plasmodium species preferentially bind to their natural host erythrocytes. Binding assays (as in Fig. 1) were performed by using human and chimpanzee erythrocytes (RBCs) on COS cells transfected with EBA-175 or EBA-140 RIIs from P. falciparum and P. reichenowi.(A) Representative photos from 50 microscope fields are shown. (Scale bar, 1 μm.) (B) Mean erythrocyte numbers in 15 microscope fields, selected randomly. Sham, sham-transfected cells; Pfa, P. falciparum homolog; Pre, P. reichenowi homolog. Data are mean ± SE.
Quantitative Binding Using Fluorescent K562 Cells. The distinctive species preferences of PfEBA-175-RII and PrEBA-175-RII for erythrocytes could be explained by the presence or absence of Neu5Gc and/or by sequence differences in the underlying GYPA backbone that presents the sialic-bearing O-glycans. To determine the relative importance of Sia-specificity, we experimentally changed the Sia profile while keeping the glycophorin protein backbone constant. Human cells growing in human serum express only Neu5Ac but can metabolically incorporate free Neu5Gc from the medium and eventually express it on their membrane glycoconjugates (41, 46, 47). The human erythroleukemia cell line K562 is known to express GYPA (48), and we found that it also expresses glycophorin C [both were detected by specific antibody staining and flow cytometry (data not shown)]. Thus, these cells express natural glycophorin ligand carriers for both PfEBA-175 and PfEBA-140 while permitting experimental manipulation of their Neu5Ac versus Neu5Gc content. This approach allowed the development of a novel assay for Sia- and glycophorin-dependent EBA binding.
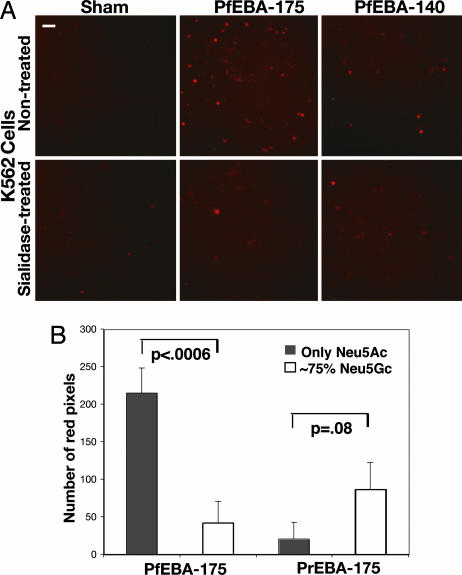
Preloading K562 cells with the fluorescent vital dye SNARF1 allowed an accurate quantitation of binding. As with erythrocyte binding, sialidase pretreatment of transfected COS cells was used to eliminate interference with optimal binding. Fluorescent K562 cells bound to sialidase-pretreated COS cells transfected with PfEBA-175-RII or PfEBA-140-RII (Fig. 3A). Very few or no cells bound to sham-transfected COS cells. Treatment of K562 cells with sialidase significantly reduced interactions (P < 0.006), indicating that binding is mediated by Sias on the surface of the K562 cells, presumably carried by the glycophorins. Although sialidase treatment of the K562 cells did not abrogate binding completely, quantitation showed that treatment was incomplete, a finding typical for treatment of nucleated cells with this enzyme. Thus, residual Sia can explain the binding of a few sialidase-treated K562 cells to the PfEBAs.
Fig. 3.
Neu5Gc expression on K562 cells reduces binding to PfEBA-175. (A) K562 cells were loaded with SNARF1-AM after treatment with or without sialidase as described for Fig. 1 and then incubated with desialylated COS cells expressing RII of PfEBA-175 or PfEBA-140 for 30 min at 37°C. After washing and fixation, bound K562 cells were analyzed by fluorescence microscopy. (Scale bar, 1 μm.) (B) Binding of desialylated COS cells expressed PfEBA-175 or PrEBA-175 to K562 cells expressing only cell-surface Neu5Ac (gray bars) or 75% Neu5Gc (open bars). Bound cells were quantitated by using Adobe photoshop to measure the number of red pixels from 15 microscope fields. (Scale bar, 1 μm.) Data are expressed as mean ± SE.
Neu5Gc Expression on Human GYPA Markedly Reduces Binding of PfEBA-175. Analysis by 1,2-diamino-4,5-methylene dioxybenzene HPLC confirmed that Neu5Ac was the only Sia present when K562 cells were grown in human serum [i.e., they are identical in Sia composition to human erythrocytes (data not shown)]. To “chimpanize” the K562 cells, we added Neu5Gc to the medium, allowing for its metabolic incorporation into surface glycoconjugates (41). After 24 h in 3 mM Neu5Gc, the cells were similar to chimpanzee erythrocytes with regard to Sia content [≈75% of total Sias in the form of Neu5Gc (data not shown)]. A separate incubation with a metabolic precursor of Neu5Gc (N-glycolylmannosamine, per-O-acetylated form) gave a lower expression of Neu5Gc (only ≈10%) (42). The pathways by which such metabolic incorporation occurs are known (41, 42). With both approaches, the resulting cell-surface Neu5Gc appears exactly as if it had been originally synthesized by the human cell.
The presence of ≈75% Neu5Gc markedly reduced binding of K562 cells by PfEBA-175 (P < 0.0006; see Fig. 3B). Indeed, only ≈10% Neu5Gc content was enough to reduce the binding by 40% relative to control (data not shown; P < 0.02). Because the GYPA peptide backbone remains unchanged, these findings indicate a strong preference of PfEBA-175 for Neu5Ac over Neu5Gc. Conversely, the presence of ≈75% Neu5Gc enhanced binding by PrEBA-175 (see Fig. 3B; because of scatter in values, the difference only approached significance at P = 0.08). The much greater specificity of PrEBA-175 for chimpanzee erythrocytes (Fig. 2) must therefore be caused by additional factors such as differences in presentation of Sias by the chimpanzee GYPA peptide backbone. EBA-140-RIIs from both species were also tested with this approach. As with erythrocytes, PrEBA-140 showed very little binding to K562 cells. PfEBA-140-RII binding was also weak, although increased Neu5Gc did reduce the weak binding (at ≈75% Neu5Gc but not at ≈10%). Taken together, these data indicate that the preferences of PfEBA-175s and PrEBA-175s for human and chimpanzee erythrocytes can be explained by corresponding preferences for Neu5Ac and Neu5Gc, with possible additional contributions by differences in the glycophorin backbone.
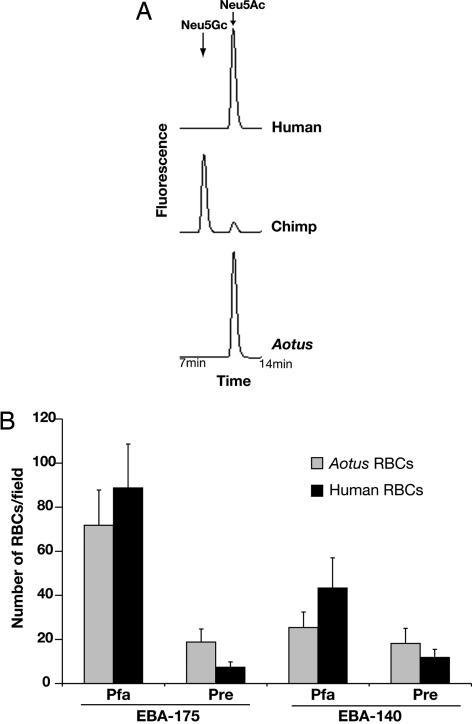
Independent Corroboration of the P. falciparum Preference for Neu5Ac: The Aotus Monkey Case. Unlike the chimpanzee, the Aotus (owl) monkey (a much more distantly related primate) is a susceptible model for experimental P. falciparum malaria (49-51) despite the fact that Aotus monkeys are thought to have been isolated from P. falciparum infection for >30 mya until Europeans or Africans brought this malarial species to the New World in the 16th century. We hypothesized that erythrocytes from Aotus might, like humans, preferentially express Neu5Ac. Indeed, erythrocyte Sias from A. nancymai (one of the species used for experimental P. falciparum malaria) expressed primarily Neu5Ac and no detectable Neu5Gc [Fig. 4A; similar results are seen with other Aotus species (data not shown)]. Thus, although these New World monkeys are evolutionarily very distant from humans (sharing a most recent common ancestor ≈30-50 mya), parallel evolution has led to terminal Sias on Aotus erythrocytes more similar to those of humans than of chimpanzees. We previously suggested that rapid evolution of glycans across taxa could result in exactly such instances of “glycan homoplasy” (52).
Fig. 4.
Aotus erythrocytes are more similar to human erythrocytes than to those of chimpanzees. (A) Sias released from human, chimpanzee, or Aotus erythrocytes (RBCs) were derivatized with 1,2-diamino-4,5-methylene dioxybenzene and profiled by HPLC. The elution position of Neu5Ac and Neu5Gc standards are indicated. (B) COS7 cells transfected with PfEBA-175, PrEBA-175, PfEBA-140, or PrEBA-140 were desialylated and used to test binding of Aotus or human erythrocytes as described for Fig. 2. Data are mean ± SE. Pfa, P. falciparum homolog; Pre, P. reichenowi homolog.
In keeping with the utility of Aotus as a disease model, erythrocytes from this primate bound to COS7 cells expressing either PfEBA-175-RII or PfEBA-140-RII (Fig. 4B). This occurred even though the Aotus glycophorin peptide sequences are likely to be rather different, indicating again that it is the terminal Sias on the erythrocyte surface that are the main factor in determining EBA binding. Although binding to human erythrocytes seemed better than to Aotus erythrocytes, this was not statistically different (P = 0.3). Although PrEBA-175-RII bound better to Aotus erythrocytes than to the human ones (P < 0.002), the number of bound Aotus erythrocytes was still much lower than that seen with chimpanzee erythrocytes (see Fig. 2), suggesting that, just as with human erythrocytes, the lack of Neu5Gc on Aotus erythrocytes precludes a strong interaction with PrEBA-175.
Discussion
P. falciparum and P. reichenowi are two morphologically indistinguishable Plasmodium species that nevertheless exhibit strong host preferences for humans and chimpanzees, respectively (11, 12, 15). Recent phylogenetic sequence studies have confirmed the genetic distinctness of these species. However, no comparative functional studies had been done until now. From the present experiments, we conclude that the strong host species preferences of P. falciparum and P. reichenowi could be explained on the basis of differences in host erythrocyte Sias. Our data show not only that PfEBA-175 prefers Neu5Ac but also that Neu5Gc interferes with this binding, likely explaining why P. falciparum is unable to successfully infect healthy chimpanzees. Conversely, PrEBA-175 strongly prefers Neu5Gc, perhaps explaining why P. reichenowi failed to infect human subjects in old studies (11, 12). The impact of merozoite invasion ligands on host species specificity has been confirmed recently in the simian malaria, Plasmodium knowlesi, in which knockout of P. knowlesi DBP-α, an invasion ligand related to EBA-175, renders that parasite unable to invade human erythrocytes (53).
At first glance, it may seem surprising that only 10% Neu5Gc can have such a large negative impact on the interaction between PfEBA-175-RII and GYPA. A possible explanation may come from our prior suggestion of “clustered saccharide patches” as novel epitopes for protein-glycan interactions (54). The GYPA domain carrying the cluster of sialylated O-glycans (23) shows selective evidence of rapid evolution, including many human polymorphisms. If EBA-175 recognizes a complex clustered patch of ≈22 Sias displayed on 11 glycans, then the inclusion of even a few N-glycolyl groups in place of N-acetyl groups could alter some critical aspect of the epitope patch, profoundly affecting the binding. A crystal structure of the binding domain in complex with its natural ligand is needed to explore this hypothesis further. It is notable that the sialyltransferases that add Sias to GYPA are not easily subject to selection, because they are expressed in many other cell types, modify many other glycoproteins, and have other distinct functions. Moreover, it is the combination of Sias and their specific clustering on GYPA that generates optimal binding by EBA-175. Thus, changing the scaffold polypeptide of GYPA is the simplest way for the host to evade the parasite.
Although PfEBA-175 clearly binds preferentially to Neu5Ac over Neu5Gc, infection of splenectomized chimpanzees by P. falciparum can be readily explained, because chimpanzee erythrocytes do carry both types of Sias. P. falciparum-infected erythrocytes are rapidly cleared and destroyed by the spleen. In its absence, intraerythrocytic parasites are allowed to develop into merozoites, which then invade new erythrocytes. However, the resulting parasitemia still evolves more slowly than expected, presumably the result of impaired binding of PfEBA-175 to the Neu5Gc-rich chimpanzee GYPA. The fact that P. falciparum malaria can cause severe disease in some species of the distantly related New World Aotus monkey can also be explained by our findings. We show here that, like humans, Aotus monkeys only have Neu5Ac on their erythrocytes. Thus, even if the GYPA sequence differs from that of humans, the overall aspect of the O-glycans and their terminal Neu5Ac Sias decorating the protein must be similar enough in both species to allow the infection.
A study comparing 280 genes among Old World primates showed that the GYPA gene exhibited the strongest evidence for rapid evolution (30), likely reflecting strong malarial parasite-mediated selection pressure. PfEBA-175-RII also seems to be evolving rapidly (30). It is reasonable to link both phenomena, because the interaction between these two proteins seems to be a key step affecting the efficiency of malaria propagation. A scenario wherein PfEBA-175 is rapidly evolving to track changes in GYPA, which, in turn, is rapidly evolving to escape from the former, represents a classical example for the “red-queen effect” operating on protein-glycan recognition (52).
A brief comparison with other human malarial parasites is in order. P. vivax does have a merozoite-stage homolog of EBA-175, the Duffy-binding protein, PvDBP. However, it is does not recognize Sias, binding instead to a sulfated tyrosine in the context of a peptide backbone of the extracellular domain of the Duffy antigen receptor for chemokines (55). No EBA-175-like sequences have been identified in the other two hominid malaria species, Plasmodium ovale and Plasmodium malariae, but only a handful of genes have been sequenced from these species.
P. falciparum species history seems to be a succession of demographic bottlenecks, apparently related to our own hominid prehistory and also to changes in climate and the emergence of new Anopheles mosquito hosts. Severe bottlenecks followed by population-growth events dated 50,000 years ago and 5,000-10,000 years ago have been associated with the expansion of modern Homo sapiens and the rise of Neolithic agriculture, respectively (7, 56-58). It is tempting to now propose an earlier bottleneck for the P. falciparum ancestor caused by the loss of Neu5Gc in the human lineage. Given the widespread occurrence of Neu5Gc in apes and evidence for occurrence of P. reichenowi in gorillas, it is reasonable to suggest that the falciparum/reichenowi common ancestor was more like P. reichenowi, preferring Neu5Gc over Neu5Ac for binding. Thus, the loss of Neu5Gc in the human lineage sometime after the initial mutation ≈3 mya (36) might have provided our emerging Homo ancestors with temporary relief from this form of malaria. However, the loss of Neu5Gc expression also led to a marked increase in its precursor, Neu5Ac (35). Thus, we suggest that P. falciparum emerged later, through selective evolution of its EBA-175, toward preferentially recognizing the Neu5Ac-rich erythrocytes of humans (i.e., P. falciparum evolved from a strain of ancestral P. reichenowi that adapted to this radical change in the human “sialome”).
This hypothesis may seem to be at odds with current estimates of the divergence of P. falciparum and P. reichenowi of ≥6-7 mya (3, 13). However, this estimate is based on only one P. reichenowi isolate, likely from a Central or East African chimpanzee subspecies (Pan troglodytes troglodytes or Pan troglodytes schweinfurthii) (9). Meanwhile, there is substantial evidence that multiple subspecies of chimpanzees may have been separated >1 mya (16, 17). It is interesting to note that there is other evidence of distinct pathogen regimes for each of these subspecies, including hepatitis B (59) and HIV (60, 61).
We suggest that sequencing of P. reichenowi isolates from other regions in which chimpanzees have been present for a long time (Western, Central, and East Africa) may reveal unexpected divergence, including discovery of a strain more closely related to P. falciparum, potentially a more recent ancestor of P. falciparum. Such studies eventually may allow a better reconstruction of the history of this major human pathogen.
Note Added in Proof. While this paper was under review, a 3D structure of PfEBA-175 in complex with 3′-sialyllactose was published (62). Notably, amino acids that contact glycans in two of the binding sites are divergent in PrEBA-175.
Acknowledgments
We thank Louis Miller (National Institutes of Health) and Jon Kaas (Vanderbilt University) for providing valuable materials and Tasha Altheide for helpful discussions. This work was supported by National Institutes of Health Grant GM32373 (to A.V.) and the G. Harold and Leila Y. Mathers Charitable Foundation.
Author contributions: M.J.M., J.C.R., and A.V. designed research; M.J.M. and P.G. performed research; M.J.M., J.C.R., and J.W.B. contributed new reagents/analytic tools; M.J.M., J.C.R., P.G., J.W.B., and A.V. analyzed data; and M.J.M., J.C.R., P.G., J.W.B., and A.V. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: mya, million years ago; EBL, erythrocyte-binding-like; EBA, erythrocyte-binding antigen; Sia, sialic acid; GYPA, glycophorin A; RII, region II; Pf, P. falciparum; Pr, P. reichenowi; Neu5Gc, N-glycolylneuraminic acid; Neu5Ac, N-acetylneuraminic acid.
References
- 1.Rasti, N., Wahlgren, M. & Chen, Q. (2004) FEMS Immunol. Med. Microbiol. 41, 9-26. [DOI] [PubMed] [Google Scholar]
- 2.Miller, L. H., Baruch, D. I., Marsh, K. & Doumbo, O. K. (2002) Nature 415, 673-679. [DOI] [PubMed] [Google Scholar]
- 3.Escalante, A. A. & Ayala, F. J. (1994) Proc. Natl. Acad. Sci. USA 91, 11373-11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Escalante, A. A., Barrio, E. & Ayala, F. J. (1995) Mol. Biol. Evol. 12, 616-626. [DOI] [PubMed] [Google Scholar]
- 5.McCutchan, T. F., Kissinger, J. C., Touray, M. G., Rogers, M. J., Li, J., Sullivan, M., Braga, E. M., Krettli, A. U. & Miller, L. H. (1996) Proc. Natl. Acad. Sci. USA 93, 11889-11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kedzierski, L., Escalante, A. A., Isea, R., Black, C. G., Barnwell, J. W. & Coppel, R. L. (2002) Infect. Genet. Evol. 1, 297-301. [DOI] [PubMed] [Google Scholar]
- 7.Rich, S. M. & Ayala, F. J. (2003) Adv. Parasitol. 54, 255-280. [DOI] [PubMed] [Google Scholar]
- 8.Qari, S. H., Shi, Y. P., Pieniazek, N. J., Collins, W. E. & Lal, A. A. (1996) Mol. Phylogenet. Evol. 6, 157-165. [DOI] [PubMed] [Google Scholar]
- 9.Coatney, G. R., Collins, W. E., Warren, M. & Contacos, P. G. (1971) The Primate Malarias (US Government Printing Office, Washington, DC).
- 10.Reichenow, E. (1920) Zentralbl. Bakteriol. 85, 207-216. [Google Scholar]
- 11.Blacklock, B. & Adler, S. (1922) Ann. Trop. Med. Parasitol. 160, 99-106. [Google Scholar]
- 12.Rodhain, J. (1939) Ann. Soc. Belg. Med. Trop. 19, 563-572. [Google Scholar]
- 13.Tanabe, K., Sakihama, N., Hattori, T., Ranford-Cartwright, L., Goldman, I., Escalante, A. A. & Lal, A. A. (2004) J. Mol. Evol. 59, 687-694. [DOI] [PubMed] [Google Scholar]
- 14.Ollomo, B., Karch, S., Bureau, P., Elissa, N., Georges, A. J. & Millet, P. (1997) Am. J. Trop. Med. Hyg. 56, 440-445. [DOI] [PubMed] [Google Scholar]
- 15.Bray, R. S. (1958) Am. J. Trop. Med. Hyg. 7, 20-24. [DOI] [PubMed] [Google Scholar]
- 16.Morin, P. A., Moore, J. J., Chakraborty, R., Jin, L., Goodall, J. & Woodruff, D. S. (1994) Science 265, 1193-1201. [DOI] [PubMed] [Google Scholar]
- 17.Gagneux, P., Wills, C., Gerloff, U., Tautz, D., Morin, P. A., Boesch, C., Fruth, B., Hohmann, G., Ryder, O. A. & Woodruff, D. S. (1999) Proc. Natl. Acad. Sci. USA 96, 5077-5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodman, M. (1999) Am. J. Hum. Genet. 64, 31-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olson, M. V. & Varki, A. (2003) Nat. Rev. Genet. 4, 20-28. [DOI] [PubMed] [Google Scholar]
- 20.Camus, D. & Hadley, T. J. (1985) Science 230, 553-556. [DOI] [PubMed] [Google Scholar]
- 21.Tomita, M., Furthmayr, H. & Marchesi, V. T. (1978) Biochemistry 17, 4756-4770. [DOI] [PubMed] [Google Scholar]
- 22.Sim, B. K., Chitnis, C. E., Wasniowska, K., Hadley, T. J. & Miller, L. H. (1994) Science 264, 1941-1944. [DOI] [PubMed] [Google Scholar]
- 23.Orlandi, P. A., Klotz, F. W. & Haynes, J. D. (1992) J. Cell Biol. 116, 901-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitchell, G. H., Hadley, T. J., McGinniss, M. H., Klotz, F. W. & Miller, L. H. (1986) Blood 67, 1519-1521. [PubMed] [Google Scholar]
- 25.Duraisingh, M. T., Maier, A. G., Triglia, T. & Cowman, A. F. (2003) Proc. Natl. Acad. Sci. USA 100, 4796-4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maier, A. G., Duraisingh, M. T., Reeder, J. C., Patel, S. S., Kazura, J. W., Zimmerman, P. A. & Cowman, A. F. (2003) Nat. Med. 9, 87-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilberger, T. W., Thompson, J. K., Triglia, T., Good, R. T., Duraisingh, M. T. & Cowman, A. F. (2003) J. Biol. Chem. 278, 14480-14486. [DOI] [PubMed] [Google Scholar]
- 28.Baum, J., Pinder, M. & Conway, D. J. (2003) Infect. Immun. 71, 1856-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baum, J., Thomas, A. W. & Conway, D. J. (2003) Genetics 163, 1327-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, H. Y., Tang, H., Shen, C. K. & Wu, C. I. (2003) Mol. Biol. Evol. 20, 1795-1804. [DOI] [PubMed] [Google Scholar]
- 31.Rearden, A., Phan, H., Kudo, S. & Fukuda, M. (1990) Biochem. Genet. 28, 209-222. [DOI] [PubMed] [Google Scholar]
- 32.Storry, J. R., Coghlan, G., Poole, J., Figueroa, D. & Reid, M. E. (2000) Vox Sang. 78, 52-56. [DOI] [PubMed] [Google Scholar]
- 33.Huang, C. H., Xie, S. S., Socha, W. & Blumenfeld, O. O. (1995) J. Mol. Evol. 41, 478-486. [DOI] [PubMed] [Google Scholar]
- 34.Ugorski, M., Blackall, D. P., Påhlsson, P., Shakin-Eshleman, S. H., Moore, J. & Spitalnik, S. L. (1993) Blood 82, 1913-1920. [PubMed] [Google Scholar]
- 35.Varki, A. (2002) Yearb. Phys. Anthropol. 44, 54-69. [Google Scholar]
- 36.Chou, H. H., Hayakawa, T., Diaz, S., Krings, M., Indriati, E., Leakey, M., Paabo, S., Satta, Y., Takahata, N. & Varki, A. (2002) Proc. Natl. Acad. Sci. USA 99, 11736-11741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muchmore, E. A., Diaz, S. & Varki, A. (1998) Am. J. Phys. Anthropol. 107, 187-198. [DOI] [PubMed] [Google Scholar]
- 38.Ozwara, H., Kocken, C. H., Conway, D. J., Mwenda, J. M. & Thomas, A. W. (2001) Mol. Biochem. Parasitol. 116, 81-84. [DOI] [PubMed] [Google Scholar]
- 39.Rayner, J. C., Huber, C. S. & Barnwell, J. W. (2004) Mol. Biochem. Parasitol. 138, 243-247. [DOI] [PubMed] [Google Scholar]
- 40.Mayer, D. C., Mu, J. B., Feng, X., Su, X. Z. & Miller, L. H. (2002) J. Exp. Med. 196, 1523-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bardor, M., Nguyen, D. H., Diaz, S. & Varki, A. (2005) J. Biol. Chem. 280, 4228-4237. [DOI] [PubMed] [Google Scholar]
- 42.Collins, B. E., Fralich, T. J., Itonori, S., Ichikawa, Y. & Schnaar, R. L. (2000) Glycobiology 10, 11-20. [DOI] [PubMed] [Google Scholar]
- 43.Schauer, R., Wember, M. & Tschesche, H. (1984) Hoppe Seylers Z. Physiol. Chem. 365, 419-426. [DOI] [PubMed] [Google Scholar]
- 44.Howard, R. F., Varki, A. & Reese, R. T. (1984) Prog. Clin. Biol. Res. 155, 45-61. [PubMed] [Google Scholar]
- 45.Angata, T. & Varki, A. (2000) J. Biol. Chem. 275, 22127-22135. [DOI] [PubMed] [Google Scholar]
- 46.Tangvoranuntakul, P., Gagneux, P., Diaz, S., Bardor, M., Varki, N., Varki, A. & Muchmore, E. (2003) Proc. Natl. Acad. Sci. USA 100, 12045-12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin, M. J., Muotri, A., Gage, F. & Varki, A. (2005) Nat. Med. 11, 228-232. [DOI] [PubMed] [Google Scholar]
- 48.Gahmberg, C. G., Jokinen, M. & Andersson, L. C. (1979) J. Biol. Chem. 254, 7442-7448. [PubMed] [Google Scholar]
- 49.Miller, L. H. (1969) Am. J. Trop. Med. Hyg. 18, 860-865. [DOI] [PubMed] [Google Scholar]
- 50.Schmidt, R. E. (1978) J. Med. Primatol. 7, 274-318. [DOI] [PubMed] [Google Scholar]
- 51.Baruch, D. I., Gamain, B., Barnwell, J. W., Sullivan, J. S., Stowers, A., Galland, G. G., Miller, L. H. & Collins, W. E. (2002) Proc. Natl. Acad. Sci. USA 99, 3860-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gagneux, P. & Varki, A. (1999) Glycobiology 9, 747-755. [DOI] [PubMed] [Google Scholar]
- 53.Singh, A. P., Ozwara, H., Kocken, C. H., Puri, S. K., Thomas, A. W. & Chitnis, C. E. (2005) Mol. Microbiol. 55, 1925-1934. [DOI] [PubMed] [Google Scholar]
- 54.Varki, A. (1994) Proc. Natl. Acad. Sci. USA 91, 7390-7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choe, H., Moore, M. J., Owens, C. M., Wright, P. L., Vasilieva, N., Li, W., Singh, A. P., Shakri, R., Chitnis, C. E. & Farzan, M. (2005) Mol. Microbiol. 55, 1413-1422. [DOI] [PubMed] [Google Scholar]
- 56.Volkman, S. K., Barry, A. E., Lyons, E. J., Nielsen, K. M., Thomas, S. M., Choi, M., Thakore, S. S., Day, K. P., Wirth, D. F. & Hartl, D. L. (2001) Science 293, 482-484. [DOI] [PubMed] [Google Scholar]
- 57.Conway, D. J. (2003) Trends Genet. 19, 671-674. [DOI] [PubMed] [Google Scholar]
- 58.Hartl, D. L. (2004) Nat. Rev. Microbiol. 2, 15-22. [DOI] [PubMed] [Google Scholar]
- 59.Hu, X., Javadian, A., Gagneux, P. & Robertson, B. H. (2001) Virus Res. 79, 103-108. [DOI] [PubMed] [Google Scholar]
- 60.Prince, A. M., Brotman, B., Lee, D. H., Andrus, L., Valinsky, J. & Marx, P. (2002) AIDS Res. Hum. Retroviruses 18, 657-660. [DOI] [PubMed] [Google Scholar]
- 61.Sharp, P. M., Shaw, G. M. & Hahn, B. H. (2005) J. Virol. 79, 3891-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tolia, N. H., Enemark, E. J., Sim, B. K. & Joshua-Tor, L. (2005) Cell 122, 183-193. [DOI] [PubMed] [Google Scholar]