Abstract
Myosin V is a double-headed molecular motor involved in organelle transport. Two distinctive features of this motor, processivity and the ability to take extended linear steps of ≈36 nm along the actin helical track, depend on its unusually long light chain-binding domain (LCBD). The LCBD of myosin V consists of six tandem IQ motifs, which constitute the binding sites for calmodulin (CaM) and CaM-like light chains. Here, we report the 2-Å resolution crystal structure of myosin light chain 1 (Mlc1p) bound to the IQ2–IQ3 fragment of Myo2p, a myosin V from Saccharomyces cerevisiae. This structure, combined with FRET distance measurements between probes in various CaM–IQ complexes, comparative sequence analysis, and the previously determined structures of Mlc1p-IQ2 and Mlc1p-IQ4, allowed building a model of the LCBD of myosin V. The IQs of myosin V are distributed into three pairs. There appear to be specific cooperative interactions between light chains within each IQ pair, but little or no interaction between pairs, providing flexibility at their junctions. The second and third IQ pairs each present a light chain, whether CaM or a CaM-related molecule, bound in a noncanonical extended conformation in which the N-lobe does not interact with the IQ motif. The resulting free N-lobes may engage in protein–protein interactions. The extended conformation is characteristic of the single IQ of myosin VI and is common throughout the myosin superfamily. The model points to a prominent role of the LCBD in the function, regulation, and molecular interactions of myosin V.
Keywords: calmodulin, IQ motif, x-ray crystallography, FRET
Myosin V is a molecular motor involved in a range of organelle-transporting functions, including the transport of melanosomes and synaptic vesicles in mammals and vacuoles and mRNA in yeast (1–4). Myosin V is composed of two identical heavy chains and 12 light chains. Each heavy chain consists of an N-terminal motor domain, containing the actin-binding and ATP catalytic sites, followed by the light chain-binding domain (LCBD), formed by six IQ motifs in tandem, and the tail domain, composed of regions of coiled-coil and a globular domain involved in cargo binding. The coiled-coil regions mediate the association of the heavy chains into dimers. The IQ motifs are ≈25-aa segments, centered around the consensus sequence IQxxxRGxxxR, and constitute the binding sites for the light chains, which can be either calmodulin (CaM) or CaM-related molecules (5, 6).
A number of features distinguish myosin V from other myosin families. Myosin V has a high duty cycle, defined as the property to remain attached to actin for a large fraction of the mechanochemical cycle (7–9). The high duty cycle of myosin V is explained by a slow rate of ADP release, which becomes the rate-limiting step in the ATPase cycle (7). This kinetic adaptation allows myosin V to take multiple steps without dissociating from the actin filament, that is, myosin V is a processive motor (10–12). Linked with processivity is the ability of myosin V to take large steps of ≈36 nm (10), a distance equal to the helical repeat of the actin filament. Such a step size allows myosin V to walk in a straight line on the actin filament, in a hand-over-hand fashion (13–16). These characteristics seem to adapt myosin V for its cellular function, the transport of large cargoes atop the actin filament while avoiding collisions with cellular structures (3). Yet, central to this motor's uniqueness is its unusually long LCBD (4). A number of laboratories have recently established a direct connection between the length and structural integrity of the LCBD and the step size and processivity of myosin V (4, 17–20). These studies focus on the role of the LCBD as a passive structural device whose function is to amplify small nucleotide-dependent motions originating in the motor domain, which explains why this domain is often referred to as the lever arm or neck domain. By analogy with some class II myosins, for which the lever arm plays a regulatory function, it is plausible that the LCBD of myosin V also serves a regulatory role. In agreement with this view, a number of reports uncover a regulatory effect of Ca2+ and CaM on the conformation and activity of myosin V that could involve the LCBD directly (21–25). Inhibition of the ATPase activity of myosin V after increases in Ca2+ concentration has been attributed to the dissociation of CaM from the heavy chain (26) and/or a conformational change in CaM (27).
Here, we report the results of a comprehensive study of the structure of the LCBD of Myo2p, a myosin V from Saccharomyces cerevisiae (28), including the determination of the 2.0-Å resolution crystal structure of the tandem IQ repeat IQ2–IQ3 of Myo2p complexed with two molecules of myosin light chain 1 (Mlc1p), a Myo2p-specific light chain (29), and FRET distance measurements between probes in various representative IQ–CaM complexes. The results, combined with the previously determined structures of complexes of Mlc1p with IQ2 and IQ4 (30, 31) and sequence analysis, allowed building a model of the LCBD of myosin V. The results have important implications for our understanding of the structure–function relationship of the lever arm of myosin V.
Materials and Methods
Preparation of Proteins and Peptides. Mlc1p (UniProt accession no. P53141), cloned into vector pAED4 under the control of the T7 promoter system, was expressed in Escherichia coli strain BL21 (DE3). The protein was purified by ion exchange chromatography using a Whatman DE52 column. Human CaM (UniProt accession no. P62158), cloned into vector pAED4, was expressed by using E. coli strain BL21 (DE3) and purified on a DEAE-Sephacel column (Amersham Pharmacia) followed by purification on a PhenylSepharose column (Amersham Pharmacia). Mutants of Mlc1p (Ile-64–Met) and CaM (Asp-50–Cys) were generated by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). WT Mlc1p contains a single Met residue at position 109. Mlc1p mutant Mlc1pIle64Met, containing two Met residues, was made to facilitate the determination of the structures of Mlc1p–IQ complexes by using the anomalous signal from a Se-Met-substituted protein. CaM mutant CaMAsp50Cys was designed to allow for labeling with the FRET acceptor 4-dimethylaminophenylazophenyl-4′-maleimide (DABMI) to monitor the conformation of complexes of CaM with IQ peptides.
Peptides corresponding to Myo2p (UniProt accession no. P19524) IQ1 (amino acids 783–805), IQ2 (amino acids 806–830), IQ3 (amino acids 831–853), IQ4 (amino acids 854–878), IQ5 (amino acids 879–901), IQ6 (amino acids 902–926), and IQ2–IQ3 (amino acids 806–853) were synthesized on an Applied Biosystems 431 peptide synthesizer and then purified by HPLC. In addition, peptides corresponding to IQ1, IQ3, IQ4, and IQ6, containing an extra Cys residue at the N terminus (referred to as Cys-IQ peptides) were synthesized to allow for labeling with the fluorescent probe N-iodoacetyl-N′-(5-sulfo-1-naphthyl)ethylenediamine (1,5-IAEDANS) (see below). To prevent double labeling, the Cys residue at position 835 of IQ3 was substituted with Ser in the Cys-IQ3 peptide. In this way, all of the Cys-IQ peptides were labeled at structurally equivalent positions, i.e., nine amino acids N-terminal to the conserved Ile at position 1 of the canonical IQ motif.
Crystallization, Data Collection, and Structure Determination. The complex of Mlc1p with IQ2–IQ3 of Myo2p (referred to as Mlc1p–IQ2,3) was crystallized, and its structure was determined from the anomalous dispersion signal of a seleno-methionine derivative as described (31). Refinement was carried out with the program cns (32) (Table 1).
Table 1. Refinement statistics.
| Space group | P21212 |
| Unit cell parameters | |
| a, b, c, Å | 80.02, 64.24, 72.79 |
| α, β, γ, ° | 90.0, 90.0, 90.0 |
| Resolution range, Å | 2.00-41.27 (2.00-2.13) |
| Completeness, % | 99.3 (97.1) |
| Rfactor, % | 20.8 (27.9) |
| Free Rfactor, % | 25.7 (34.3) |
Values in parentheses correspond to last resolution shell. Rfactor = Σ|Fo — Fc|/Σ|Fc|, where Fo and Fc are observed and calculated structure factors. Free Rfactor — Rfactor is calculated for a subset of reflections (5%) omitted during the refinement.
Labeling of IQ Peptides and CaM. The Cys-IQ peptides (Cys-IQ1, Cys-IQ3, Cys-IQ4, and Cys-IQ6) were reduced with 20 mM DTT for ≈30 min. DTT was then removed on a Sephadex G-15 column (Amersham Pharmacia), using 20 mM Hepes, pH 7.5 and 100 mM NaCl as running buffer. The concentration of free SH groups was determined by the 5,5′-dithiobis(2-nitrobenzoic acid) method (33), followed by labeling overnight (in the dark, at 4°C) with 20:1 molar excess 1,5-IAEDANS (Molecular Probes). Labeled peptides were isolated by HPLC, using a C18 reverse-phase column (Waters), and their concentration was determined with fluorescamine (34). The purity of the Cys-IQ-labeled peptides was verified by MS. Labeling of CaMAsp50Cys with the nonfluorescent FRET acceptor DABMI was carried out in a similar way. The concentration of labeled CaMAsp50Cys was determined by Micro BCA assay (Pierce).
Fluorescence Measurements. All fluorescence measurements were carried out at 20°C in a solution containing 100 mM NaCl, 20 mM Hepes (pH 7.5), and 0.5 mM EGTA. The steady-state fluorescence emission spectra (λem = 400–600 nm) were recorded on a Cary Eclipse fluorometer (Varian) with excitation at λex = 337 nm. Fluorescence lifetime measurements were carried out on a TimeMaster T-4 stroboscope lifetime fluorometer from Photon Technology International, Lawrenceville, NJ. The scans were collected within a 75-ns window, with 0.5 ns per channel at λem = 500 nm. The concentration of the 1,5-IAEDANS-labeled IQ peptides was 2 μM and that of WT CaM or DABMI-labeled CaMAsp50Cys (when present) was 4 μM. The data were fitted with one, two, and three exponentials with felix32 software (Photon Technology International). The distance R between donor and acceptor was calculated by using the equation: R = R0 (τda/(τd – τda))1/6, where τda is the fluorescence lifetime for a donor–acceptor pair and τd is the lifetime for the donor alone (1,5-IAEDANS-labeled IQ peptide complexed with unlabeled WT CaM). The critical transfer distance R0 was calculated for each peptide from the relation R0 = R0′ (τd/τd′)1/6, using R0′ = 39.9 Å and τd′ = 13.5 ns as reference values for the pair 1,5-IAEDANS–DABMI (35). Implicit in this calculations is the use of κ2 = 2/3, corresponding to a random orientation of the probes.
Results and Discussion
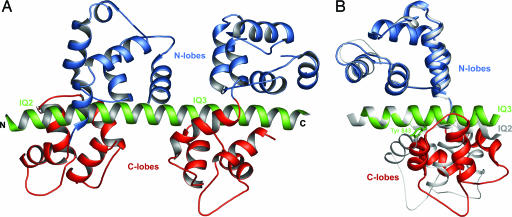
The Structure of Mlc1p–IQ2,3 and the Relationship Between IQ Sequence and Light Chain Conformation. The structure of the complex of Mlc1p with IQ2–IQ3 of Myo2p (Mlc1p–IQ2,3) reveals two molecules of Mlc1p, one per IQ motif, bound in the canonical compact conformation (Fig. 1A). Like other members of the CaM superfamily, Mlc1p presents a dumbbell-shaped structure with two homologous domains, the N- and C-terminal lobes, connected by a flexible linker. Each lobe is formed by two EF-hand-like motifs, each consisting of a pair of α-helices linked by a loop. In contrast to CaM, the EF-hands of Mlc1p do not bind Ca2+, because of substitutions of some of the amino acids involved in the coordination of Ca2+ in CaM. The compact conformation characteristic of the two Mlc1p molecules in this complex has been observed in a number of light chain–IQ structures (30, 36–39). The N-lobe in these structures is fully closed (hydrophobic core nonexposed) and interacts with the C-terminal half of the IQ motif (GxxxR), mainly through electrostatic contacts. The C-lobe assumes a semiopen conformation, with its hydrophobic core only partially exposed (39), and interacts with the N-terminal half of the IQ motif (IQxxxR), primarily through hydrophobic contacts.
Fig. 1.
Structure of Mlc1p–IQ2,3. (A) Ribbon diagram representation of the structure (N-lobes, blue; C-lobes, red; heavy chain, green). (B) Superimposition of the Mlc1p–IQ2 (gray) and Mlc1p–IQ3 (colored as in A) portions of the structure. The side chain of Tyr-843, which forces the opening of the C-lobe in Mlc1p–IQ3, is shown.
Although the complexes are generally similar, important differences exist between the two compact light chain–IQ complexes that form Mlc1p–IQ2,3. Because the light chain is the same in both complexes, sequence variations within the IQ motifs are most likely responsible for these differences. Indeed, other factors, such as crystal contacts and light chain–light chain interactions across neighboring IQs, can be ruled out because independently determined structures of Mlc1p with IQ2 (30) and IQ3 (this 3.0-Å resolution structure is not described here) are very similar to the corresponding portions of ternary complex Mlc1p–IQ2,3 (Cα rms deviations of 0.89 and 0.91 Å, respectively).
A better understanding of the relationship between IQ sequence and light chain conformation can be obtained from a superimposition of the Mlc1p–IQ2 and Mlc1p–IQ3 halves of the structure. Superimposing the IQ portions of these complexes positions the N-lobes on top of each other, but the C-lobes diverge significantly (Fig. 1B). A similar observation can be made of other light chain–IQ complexes (30, 36–39); i.e., a superposition of the IQ portions of these structures leads to a good superimposition of the N-lobes, whereas the C-lobes diverge broadly (data not shown). It appears, therefore, that the conformational differences observed among compact structures of light chain–IQ complexes are caused mostly by variations within the N-terminal IQxxxR portion of the motif, which interacts with the C-lobe (Fig. 2).
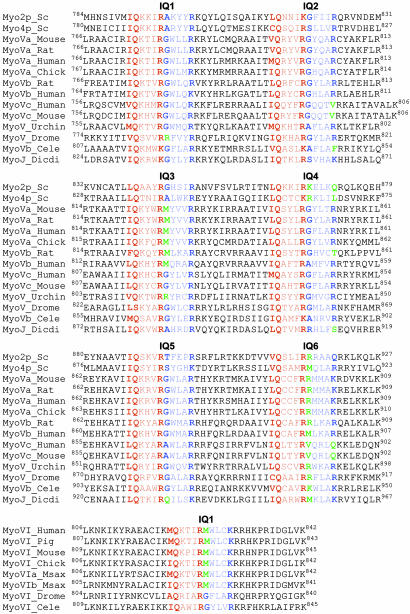
Fig. 2.
Alignment of the LCBD domains of class V and class VI myosins. The IQ motif is based on the canonical sequence IQxxxRGxxxR. The N-terminal portion of the IQ motif (IQxxxR), which binds the C-lobe, is colored red. The C-terminal part (GxxxR), which binds the N-lobe, is colored blue. UniProt accession numbers are: Myo2p_Sc, P19524; Myo4p_Sc, P32492; MyoVa_Mouse, Q99104; MyoVa_Rat, Q9QYF3; MyoVa_Human, Q9Y4I1; MyoVa_Chick, Q02440; MyoVb_Rat, P70569; MyoVb_Human, Q9ULV0; MyoVc_Human, Q9NQX4; MyoVc_Mouse, Q8BWY8; MyoV_Urchin, Q9NBH3; MyoV_Drome, O97417; MyoVb_Cele, Q17383; MyoJ_Dicdi, P54697; MyoVI_Human, Q9UM54; MyoVI_Pig, Q29122; MyoVI_Mouse, Q64331; MyoVI_Chick, Q9I8D1; MyoVIa_Msax, Q9PWF6; MyoVIb_Msax, Q9PWF5; MyoVI_Drome, Q01989l; and MyoVI_Cele, Q9TZI9.
The C-lobe covers approximately three helical turns of the IQ motif, with the side chains at positions 1 and 5 of the IQ motif being always directed toward the hydrophobic core of the C-lobe. These positions are typically occupied by small, branched aliphatic amino acids (Ile, Leu, and Val). However, it is not uncommon to find bulkier aromatic side chains such as Phe at position 1 and Phe, Tyr, and Trp at position 5 (Fig. 2). The presence of these amino acids is typically associated with additional opening or rearrangement of the C-lobe, as exemplified by IQ3 of Myo2p, which presents a Tyr residue at position 5. Because of its polarity, the hydroxyl group of Tyr-843 of IQ3 cannot be accommodated within the hydrophobic core of the C-lobe, which opens slightly so that the side chain of Tyr-843 can be directed toward the solvent (Fig. 1B). Similarly, the presence of a Phe residue at position 1 of the first IQ of smooth muscle (37) and scallop myosin II (39) leads to local rearrangement of the C-lobes of the bound essential light chains.
Within the IQxxxR portion of the IQ motif, the strongest constraint is toward the conservation of the Gln residue at position 2. In complexes with light chains this amino acid is buried, forming three strong hydrogen-bonding contacts with main-chain atoms of the C-lobe. The only other amino acid known to substitute for Gln at position 2 is Ser (30). Most likely, the hydroxyl group of Ser coordinates a water molecule that accounts for some of the hydrogen-bonding contacts of Gln at position 2. Position 6 of the IQ motif is also highly conserved, occupied by Arg, and sometimes Lys. The residue at position 7 is also typically involved in hydrogen-bonding contacts with main-chain atoms of the C-lobe. The remaining amino acids of the N-terminal part of the IQ motif, positions 3 and 4, are partially solvent-exposed and vary more widely, with limited effect on the conformation of the light chain.
Compact and Extended Conformation of CaM–IQ Complexes. Although the compact structures of light chain–IQ complexes reveal the importance of the N-terminal half of the IQ motif in determining the conformation of the light chain, departure from the canonical sequence within the C-terminal GxxxR half of the motif can have an even greater effect. Thus, substitutions of the Gly residue at position 7 of the IQ motif by bulky side chains such as Arg, Lys, and Met, and/or the loss of the Arg at position 11 result in a noncanonical, extended conformation of the bound light chain. The extended conformation was originally observed in the structure of the complex of Mlc1p with IQ4 of Myo2p, which presents a Lys at position 7 (30). Sedimentation velocity analysis further demonstrated that the compact complex of Mlc1p–IQ2 becomes extended when the Gly at position 7 is replaced by Lys. Conversely, the extended complex of Mlc1p–IQ4 was converted into a compact complex by a double mutation, which introduced a Gly at position 7 and an Arg at position 11. Note that in this case introducing a Gly at position 7 alone was insufficient to produce a compact conformation. Based on these results it was concluded that the presence of a bulky side chain at position 7 sterically hinders the interaction of the N-lobe of the light chain with the IQ motif, precluding a compact conformation (30). Moreover, a compact conformation also requires the presence of an Arg, or possibly a Lys, at position 11, as only these two amino acids can form the hydrogen-bonding interactions with the N-lobe that characterize this conformation. Based on these results and sequence analysis of the myosin superfamily (Fig. 2), it can be predicted that the extended conformation is widespread. For instance, the last IQ motif of myosin V and the single IQ motif of myosin VI typically present amino acids at positions 7 and 11 that are consistent with the extended light chain conformation. However, the most abundant light chain in myosin V is CaM (1, 21). Will the complexes of CaM with IQ motifs assume similar compact and extended conformations as those observed in Mlc1p? To answer this question, a series of FRET distance measurements between probes attached to CaM and a representative group of IQ motifs were carried out.
First, two models, compact and extended, of CaM–IQ complexes were built by using the structures of Mlc1p–IQ2 and Mlc1p–IQ4 as reference. The models were used to determine the best location for the fluorescence donor and acceptor that, although sensitive to the conformation of CaM, would not interfere with the structure of the complexes. Based on this information, Asp-50 in the N-lobe of CaM was mutated to Cys (CaMAsp50Cys) and labeled with the acceptor probe DABMI. A group of four representative IQs of Myo2p were synthesized with addition of an extra Cys at their N termini: Cys-IQ1, Cys-IQ3, Cys-IQ4, and Cys-IQ6. The position of the extra Cys is identical in all four IQ peptides, nine amino acids N-terminal to the first canonical amino acid of the motif. In this way, the IQ peptides were labeled at structurally equivalent positions with the fluorescence donor 1,5-IAEDANS. Within this group of IQ motifs, IQ3 is canonical and should form a compact complex, IQ4 and IQ6 present noncanonical substitutions at positions 7 and 11 and are predicted to form extended complexes. IQ1 represents a group of IQ motifs that present the conserved Arg at position 11, but have undergone substitutions at position 7 that incorporate small amino acids such as Ala, Ser, and Thr (Fig. 2). Using the compact and extended models of CaM–IQ complexes, the distances between probes attached to these IQs and CaM were predicted to be ≈22 and ≈35 Å for the compact and extended conformations, respectively.
The results of the lifetime distance measurements are in good agreement with the distances predicted from the models (Table 2) and indicate that the complexes of CaM–IQ1 and CaM–IQ3 are compact, whereas those of CaM–IQ4 and CaM–IQ6 are extended. This finding is evidence that Ca2+-free CaM adopts two different conformations when bound to different kind of IQ motifs, compact and extended, which are structurally similar to those in the crystal structures of Mlc1p–IQ2,3 and Mlc1p–IQ4, respectively. This result is not surprising because despite the evolutionary distance that separates CaM and the myosin light chains, most of the structural elements involved in the binding of IQ motifs are conserved (30). More surprising, however, is the finding that the complex of CaM–IQ1 is also compact, and possibly even more so than the complex of CaM–IQ3, despite the fact that the canonical Gly at position 7 has been replaced by Ala in IQ1. This result allows refining our understanding of the sequence determinants of the IQ motif associated with the compact and extended conformations. Thus, a compact conformation requires the presence of a positively charged amino acid (Arg or Lys) at position 11 and Gly or a small amino acid (Ala, Ser, Thr, Val) at position 7 of the IQ motif. On the other hand, the presence of a bulky side chain at position 7 (Lys, Arg, Met) and/or the absence of a positively charged amino acid at position 11 causes the conformation of the bound light chain, whether CaM or a specific myosin light chain, to be extended.
Table 2. Fluorescence lifetimes of CaM–IQ complexes.
| Experiment | τ1, ns | α1 | τ2, ns | α2 | τ3, ns | α3 | χ2 | R0, Å | R, Å |
|---|---|---|---|---|---|---|---|---|---|
| IQ1—CaMwt | 13.4 | 1.00 | — | — | — | — | 0.71 | 39.9 | 21.3 ± 1.7 |
| IQ1—CaMDABMI | 11.9 | 0.30 | 2.29 | 0.23 | 0.20 | 0.47 | 0.96 | ||
| IQ3—CaMwt | 12.4 | 1.00 | — | — | — | — | 0.77 | 39.5 | 23.4 ± 1.3 |
| IQ3—CaMDABMI | 10.5 | 0.40 | 0.50 | 0.60 | — | — | 1.04 | ||
| IQ4—CaMwt | 12.1 | 1.00 | — | — | — | — | 0.93 | 39.2 | 30.5 ± 0.8 |
| IQ4—CaMDABMI | 11.4 | 0.71 | 2.85 | 0.29 | — | — | 0.88 | ||
| IQ6—CaMwt | 12.4 | 1.00 | — | — | — | — | 0.84 | 39.3 | 33.8 ± 1.0 |
| IQ6—CaMDAMBI | 12.6 | 0.67 | 3.84 | 0.33 | — | — | 0.91 |
τ1-3 and α1-3 are, respectively, lifetimes and fractional contributions to fluorescence decay. Two- or three-exponential fits were carried out to minimize χ2. Lifetimes in bold were used to calculate the distances.
Light Chain–Light Chain Interactions as a Function of the Spacing Between IQ Motifs. The light chains in the structure of Mlc1p–IQ2,3 do not interact with each other. This lack of interaction results from the 14-aa spacing between the last canonical residue of IQ2 (Arg-824) and the first canonical residue of IQ3 (Leu-839), and from the straight conformation of the 13-turn α-helix encompassing IQ2–IQ3 (Fig. 1A). In contrast, in myosin II where the spacing between IQs is 15 aa, the N-lobe of the essential light chain bound to IQ1 interacts with the C-lobe of the regulatory light chain bound to IQ2 (36, 39). This interaction results from a ≈30° bend in the heavy chain segment that separates the two IQs. Such a bend appears to be a consequence of the noncanonical nature of IQ2 and the constrained structure of the regulatory light chain that binds to it. Indeed, IQ2 of myosin II lacks the C-terminal GxxxR half, which is replaced by a second 90° bend in the heavy chain, followed by a hydrophobic sequence that binds the N-lobe of the regulatory light chain in a fully open conformation. The direct interaction between light chains in myosin II has been linked to the Ca2+-mediated regulation of scallop myosin (39) and could play a similar role in the phosphorylation-mediated regulation of smooth muscle myosin. In myosin V, a spacing of 14 aa between IQ2 and IQ3, and between IQ4 and IQ5, leads to the lack of stabilizing interactions between some of the contiguous light chains. Under load the LCBD of myosin V may bend at the junctions between these IQs. Interestingly, bending of the LCBD of the leading head of myosin V has been observed by electron microscopy (13) and has been suggested to function as a symmetry braking mechanism facilitating the communication between heads for processive movement (4).
The 14-aa spacing between IQ2 and IQ3, and between IQ4 and IQ5, effectively separates the LCBD of myosin V into three semiindependent pairs of IQ motifs: IQ1–IQ2, IQ3–IQ4, and IQ5–IQ6 (Fig. 2). The spacing between IQs within each pair is only 12 aa. Despite multiple attempts, a structure of a 12-aa-spaced pair could not be obtained, yet models of the pairs were relatively simple to build by using the structures now available. First, a model was built for the IQ3–IQ4 pair, based on the structures of Mlc1p–IQ2,3 and Mlc1p–IQ4 (30). A heavy chain fragment corresponding to IQ3–IQ4 was built by using IQ2–IQ3 as a reference, by turning the portion of the heavy chain corresponding to IQ4 two amino acids backward along the α-helical path. The position of IQ4 thus obtained was then used to superimpose the structure of Mlc1p–IQ4. Another model was built for IQ1–IQ2, based on the structures of Mlc1p–IQ2,3 and chicken myosin Va motor–essential light chain complex (38), which includes one light chain bound to IQ1. Despite the fact that the models of IQ1–IQ2 and IQ3–Q4 were built independently, they superimpose well, except for the N-lobe of the light chain bound to IQ4, which is in the extended conformation.
Although there is no crystal structure of any part of IQ5–IQ6, the FRET distance measurements performed here indicate that the light chain bound to IQ6 is in the extended conformation, and because the spacing between IQ5 and IQ6 is also 12 aa, the structure of this pair is likely similar to that of the IQ3–IQ4 pair. Yet, IQ5 of Myo2p is the only IQ motif among all myosins of class V that presents a Pro residue (Pro-896) within its canonical core (Fig. 2). IQ5 of Myo2p is therefore unique, and unlikely to be fully functional (see below), which may be connected with the fact that Myo2p is one of two myosins of class V known to be nonprocessive (40) (the other being Myo4p). Because our goal is to build a general model of the LCBD of myosin V, Pro-896 of Myo2p was replaced by Ala, which is the most common amino acid at this position. A model of IQ5–IQ6 was then obtained by duplication of the model of IQ3–IQ4 described above.
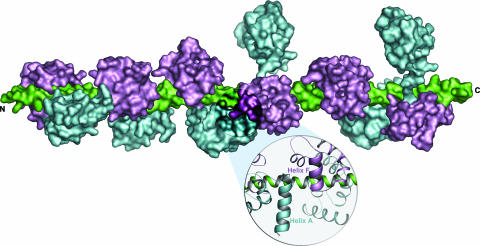
The models of the three IQ pairs of myosin V reveal a specific interaction between the light chains that form each pair (Fig. 3). This interaction involves the first α-helix of the N-lobe (equivalent to CaM helix A) of the light chain bound to the first IQ and the second α-helix of the C-lobe (CaM helix F) of the light chain bound to the second IQ. The interacting α-helices run antiparallel for ≈1.5 helical turns. The amino acids involved in this interaction (Glu-15 to Asp-25, Glu-105 to Lys-116 in CaM; and Asp-9 to Asp-19, Asp-105 to Lys-116 in Mlc1p) are among the most highly conserved regions between Mlc1p and CaM. Ca2+ binding to the EF-hands of CaM will most likely affect this interaction. Moreover, because of this interaction, the binding of light chains to 12-aa-spaced pairs is most likely cooperative.
Fig. 3.
Model of the LCBD of myosin V. The light chains, which can be either CaM or CaM-related molecules such as Mlc1p are colored cyan (N-lobes) and magenta (C-lobes), and the six-IQ fragment of the heavy chain is colored green. The LCBD of myosin V can be conceptually subdivided into three semiindependent pairs of IQ motifs, with little or no interactions between pairs. The linkers between neighboring IQ pairs are 14 aa long, whereas the linkers between IQs in a pair are 12 aa long. An enlargement illustrates the interaction between light chains in a pair.
A Model of the LCBD of Myosin V. The models of the IQ1–IQ2 and IQ3–IQ4 pairs of Myo2p are naturally connected by the structure of Mlc1p–IQ2,3. Because the spacing between IQ4 and IQ5 is also 14 aa, the structure of Mlc1p–IQ2,3 can also be used to connect the IQ5–IQ6 pair to the first two IQ pairs, yielding a complete model of the LCBD of myosin V (Fig. 3). However, this model does not take into account the distribution of CaM and light chains among the IQ motifs of myosin V. It is generally believed that the LCBD of myosin V binds between four and five CaMs and one or two specific light chains (1, 21, 28, 29). Because IQ1 is thought to bind a light chain (1, 41), CaM is most likely bound to at least four of the remaining IQs. To check this possibility, the affinities of the six IQs of Myo2p for yeast CaM and Mlc1p were measured by using the Trp fluorescence of mutants CaMPhe90Trp and Mlc1pPhe90Trp (data not shown). Although the measurements indicate a preference of Mlc1p for IQ2 and CaM for IQ6, the remaining IQs bind CaM and Mlc1p with similar affinities. IQ5, which is unique in that it presents a Pro residue within its core region, binds both CaM and Mlc1p with very low affinities and is unlikely to be fully functional. The affinity measurements for isolated IQs cannot account for potential cooperativity in the binding of light chains to neighboring IQs. Considering, in addition, that the distribution of CaM and light chains may be subject to regulation and could vary from myosin to myosin, this study leaves the question of CaM vs. light chain distribution unresolved.
The model proposed in Fig. 3 should be regarded as a general model of the LCBD of myosin V and not a specific model of the LCBD of Myo2p. In addition to the general notion that the LCBD of myosin V is divided into three semiindependent IQ pairs, the model reflects the fact that the light chain bound to IQ6 is always extended, whereas a second extended light chain is associated with either IQ3 or IQ4 (Fig. 2). In three of the myosins (MyoVc_Human, MyoVc_Mouse, and MyoVb_Cele) the second extended light chain occurs in IQ2, whereas MyoV_Drome is the only myosin of class V where the second extended light chain occurs in IQ1. MyoVc_Human and MyoVc_Mouse are unique in that they present three amino acid inserts in the linker region that separates the first and second IQ pairs. At least two myosins, MyoVb_Rat and MyoJ_Dicdi, present three light chains in the extended conformation and combine two extended light chains in a single IQ pair. As illustrated by the model (Fig. 3) and the alignment of the sequences (Fig. 2), the extended light chain conformation is present in ≈35% of the light chains of myosin V. Interestingly, myosin VI presents a single IQ motif, which according to its sequence (Fig. 2), is predicted to bind a light chain in the extended conformation in most members of this family. Myosin VI is the only myosin known to move backward, toward the minus end of the actin filament (42), and despite a relatively short lever arm, generates large steps and appears to be processive like myosin V (43). This observation raises the question as to whether the extended light chain conformation plays a role in determining processivity and step size. The crystal structure of myosin VI, including two CaM molecules, a Ca2+-free CaM bound to the IQ motif and a Ca2+-loaded CaM bound to a unique reverse gear insert present in the converter domain of this myosin, has just been reported (44). However, the CaM molecule bound to the IQ motif is mostly disordered and its conformation could not be unambiguously determined.
In summary, the LCBD of myosin V can be conceptually subdivided into three pairs of IQ motifs. There are extensive interactions between light chains within pairs, but little or no interaction exist between light chains of neighboring pairs, allowing for the LCBD to flex under load. Flexing of the LCBD has been observed by electron microscopy (13) and may play a role in the communication between heads during processive movement (4).
About 35% of the light chains of myosin V, whether CaM or CaM-like molecules, are bound in a noncanonical extended conformation in which the N-lobes do not interact with the IQ motifs. What is the cellular function of the extended light chain conformation? The extended light chain conformation could play a role in myosin localization and/or binding to targets and effectors. Chicken brain MyoVa, for example, coimmunoprecipitates with CaM-dependent protein kinase II (CaMKII), is a substrate of CaMKII, and activates both the autophosphorylation and MyoVa phosphorylation activities of CaMKII, possibly by delivering CaM molecules directly to CaMKII (45).
In class V myosins, the extended light chains are found mainly within the second and third pairs of IQ motifs, one in each pair. More notably, the last light chain of the LCBD of all myosins V and the single light chain of most myosins VI, are predicted to be extended. Because of the unique characteristics of these two myosin classes, including processivity and their unusually long step size, it is important to understand the cellular functions of the extended light chain conformation. The structural determinants of light chain conformation revealed by this study provide a framework for the design of in vivo mutants to address this question.
Acknowledgments
This work was supported by National Institutes of Health Grants AR46524 and AR41637. Use of the Advanced Photon Source (Argonne, IL) was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract W-31-109-Eng-38. Use of the BioCARS facilities at the Argonne National Laboratory (Argonne, IL) was supported by National Institutes of Health Grant RR07707. Use of the Cornell High Energy Synchrotron Source (Ithaca, NY) and the Macromolecular Crystallography Facility at the Cornell High Energy Synchrotron Source was supported by National Science Foundation Award DMR 97-13424 and National Institutes of Health Award RR-01646, respectively.
Author contributions: Z.G. and R.D. designed research; M.T., G.R., and R.D. performed research; R.C.L. contributed new reagents/analytic tools; M.T., G.R., Z.G., and R.D. analyzed data; and R.D. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CaM, calmodulin; LCBD, light chain-binding domain; Mlc1p, myosin light chain 1; 1,5-IAEDANS, N-iodoacetyl-N′-(5-sulfo-1-naphthyl)ethylenediamine; DABMI, 4-dimethylaminophenylazophenyl-4′-maleimide.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1N2D).
References
- 1.Reck-Peterson, S. L., Provance, D. W., Jr., Mooseker, M. S. & Mercer, J. A. (2000) Biochim. Biophys. Acta 1496, 36–51. [DOI] [PubMed] [Google Scholar]
- 2.Mehta, A. (2001) J. Cell Sci. 114, 1981–1998. [DOI] [PubMed] [Google Scholar]
- 3.Vale, R. D. (2003) J. Cell Biol. 163, 445–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tyska, M. J. & Mooseker, M. S. (2003) Trends Cell Biol. 13, 447–451. [DOI] [PubMed] [Google Scholar]
- 5.Espindola, F. S., Suter, D. M., Partata, L. B., Cao, T., Wolenski, J. S., Cheney, R. E., King, S. M. & Mooseker, M. S. (2000) Cell Motil. Cytoskeleton 47, 269–281. [DOI] [PubMed] [Google Scholar]
- 6.Bahler, M. & Rhoads, A. (2002) FEBS Lett. 513, 107–113. [DOI] [PubMed] [Google Scholar]
- 7.De La Cruz, E. M., Wells, A. L., Rosenfeld, S. S., Ostap, E. M. & Sweeney, H. L. (1999) Proc. Natl. Acad. Sci. USA 96, 13726–13731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De La Cruz, E. M., Sweeney, H. L. & Ostap, E. M. (2000) Biophys. J. 79, 1524–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore, J. R., Krementsova, E. B., Trybus, K. M. & Warshaw, D. M. (2001) J. Cell Biol. 155, 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mehta, A. D., Rock, R. S., Rief, M., Spudich, J. A., Mooseker, M. S. & Cheney, R. E. (1999) Nature 400, 590–593. [DOI] [PubMed] [Google Scholar]
- 11.Veigel, C., Wang, F., Bartoo, M. L., Sellers, J. R. & Molloy, J. E. (2002) Nat. Cell Biol. 4, 59–65. [DOI] [PubMed] [Google Scholar]
- 12.Bahloul, A., Chevreux, G., Wells, A. L., Martin, D., Nolt, J., Yang, Z., Chen, L. Q., Potier, N., Van Dorsselaer, A., Rosenfeld, S., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 4787–4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walker, M. L., Burgess, S. A., Sellers, J. R., Wang, F., Hammer, J. A., 3rd, Trinick, J. & Knight, P. J. (2000) Nature 405, 804–807. [DOI] [PubMed] [Google Scholar]
- 14.Forkey, J. N., Quinlan, M. E., Shaw, M. A., Corrie, J. E. & Goldman, Y. E. (2003) Nature 422, 399–404. [DOI] [PubMed] [Google Scholar]
- 15.Yildiz, A., Forkey, J. N., McKinney, S. A., Ha, T., Goldman, Y. E. & Selvin, P. R. (2003) Science 300, 2061–2065. [DOI] [PubMed] [Google Scholar]
- 16.Snyder, G. E., Sakamoto, T., Hammer, J. A., 3rd, Sellers, J. R. & Selvin, P. R. (2004) Biophys. J. 87, 1776–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell, T. J., Morris, C., Spudich, J. A. & Sweeney, H. L. (2002) Proc. Natl. Acad. Sci. USA 99, 14159–14164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schott, D. H., Collins, R. N. & Bretscher, A. (2002) J. Cell Biol. 156, 35–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakamoto, T., Wang, F., Schmitz, S., Xu, Y., Xu, Q., Molloy, J. E., Veigel, C. & Sellers, J. R. (2003) J. Biol. Chem. 278, 29201–29207. [DOI] [PubMed] [Google Scholar]
- 20.Moore, J. R., Krementsova, E. B., Trybus, K. M. & Warshaw, D. M. (2004) J. Muscle Res. Cell Motil. 25, 29–35. [DOI] [PubMed] [Google Scholar]
- 21.Cheney, R. E., O'Shea, M. K., Heuser, J. E., Coelho, M. V., Wolenski, J. S., Espreafico, E. M., Forscher, P., Larson, R. E. & Mooseker, M. S. (1993) Cell 75, 13–23. [DOI] [PubMed] [Google Scholar]
- 22.Nascimento, A. A., Cheney, R. E., Tauhata, S. B., Larson, R. E. & Mooseker, M. S. (1996) J. Biol. Chem. 271, 17561–17569. [DOI] [PubMed] [Google Scholar]
- 23.Li, X. D., Mabuchi, K., Ikebe, R. & Ikebe, M. (2004) Biochem. Biophys. Res. Commun. 315, 538–545. [DOI] [PubMed] [Google Scholar]
- 24.Krementsov, D. N., Krementsova, E. B. & Trybus, K. M. (2004) J. Cell Biol. 164, 877–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, F., Thirumurugan, K., Stafford, W. F., Hammer, J. A., 3rd, Knight, P. J. & Sellers, J. R. (2004) J. Biol. Chem. 279, 2333–2336. [DOI] [PubMed] [Google Scholar]
- 26.Cameron, L. C., Carvalho, R. N., Araujo, J. R., Santos, A. C., Tauhata, S. B., Larson, R. E. & Sorenson, M. M. (1998) Arch. Biochem. Biophys. 355, 35–42. [DOI] [PubMed] [Google Scholar]
- 27.Homma, K., Saito, J., Ikebe, R. & Ikebe, M. (2000) J. Biol. Chem. 275, 34766–34771. [DOI] [PubMed] [Google Scholar]
- 28.Brockerhoff, S. E., Stevens, R. C. & Davis, T. N. (1994) J. Cell Biol. 124, 315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stevens, R. C. & Davis, T. N. (1998) J. Cell Biol. 142, 711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terrak, M., Wu, G., Stafford, W. F., Lu, R. C. & Dominguez, R. (2003) EMBO J. 22, 362–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terrak, M., Otterbein, L. R., Wu, G., Palecanda, L. A., Lu, R. C. & Dominguez, R. (2002) Acta Crystallogr. D 58, 1882–1885. [DOI] [PubMed] [Google Scholar]
- 32.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 33.Ellman, G. L. (1959) Arch. Biochem. Biophys. 82, 70–77. [DOI] [PubMed] [Google Scholar]
- 34.Udenfriend, S., Stein, S., Bohlen, P., Dairman, W., Leimgruber, W. & Weigele, M. (1972) Science 178, 871–872. [DOI] [PubMed] [Google Scholar]
- 35.Tao, T., Lamkin, M. & Lehrer, S. S. (1983) Biochemistry 22, 3059–3066. [DOI] [PubMed] [Google Scholar]
- 36.Rayment, I., Rypniewski, W. R., Schmidt-Base, K., Smith, R., Tomchick, D. R., Benning, M. M., Winkelmann, D. A., Wesenberg, G. & Holden, H. M. (1993) Science 261, 50–58. [DOI] [PubMed] [Google Scholar]
- 37.Dominguez, R., Freyzon, Y., Trybus, K. M. & Cohen, C. (1998) Cell 94, 559–571. [DOI] [PubMed] [Google Scholar]
- 38.Coureux, P. D., Wells, A. L., Menetrey, J., Yengo, C. M., Morris, C. A., Sweeney, H. L. & Houdusse, A. (2003) Nature 425, 419–423. [DOI] [PubMed] [Google Scholar]
- 39.Houdusse, A. & Cohen, C. (1996) Structure (London) 4, 21–32. [DOI] [PubMed] [Google Scholar]
- 40.Reck-Peterson, S. L., Tyska, M. J., Novick, P. J. & Mooseker, M. S. (2001) J. Cell Biol. 153, 1121–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De La Cruz, E. M., Wells, A. L., Sweeney, H. L. & Ostap, E. M. (2000) Biochemistry 39, 14196–14202. [DOI] [PubMed] [Google Scholar]
- 42.Wells, A. L., Lin, A. W., Chen, L. Q., Safer, D., Cain, S. M., Hasson, T., Carragher, B. O., Milligan, R. A. & Sweeney, H. L. (1999) Nature 401, 505–508. [DOI] [PubMed] [Google Scholar]
- 43.Rock, R. S., Rice, S. E., Wells, A. L., Purcell, T. J., Spudich, J. A. & Sweeney, H. L. (2001) Proc. Natl. Acad. Sci. USA 98, 13655–13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Menetrey, J., Bahloul, A., Wells, A. L., Yengo, C. M., Morris, C. A., Sweeney, H. L. & Houdusse, A. (2005) Nature 435, 779–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Costa, M. C., Mani, F., Santoro, W., Jr., Espreafico, E. M. & Larson, R. E. (1999) J. Biol. Chem. 274, 15811–15819. [DOI] [PubMed] [Google Scholar]