Abstract
The identification of new virus species is a key issue for the study of infectious disease but is technically very difficult. We developed a system for large-scale molecular virus screening of clinical samples based on host DNA depletion, random PCR amplification, large-scale sequencing, and bioinformatics. The technology was applied to pooled human respiratory tract samples. The first experiments detected seven human virus species without the use of any specific reagent. Among the detected viruses were one coronavirus and one parvovirus, both of which were at that time uncharacterized. The parvovirus, provisionally named human bocavirus, was in a retrospective clinical study detected in 17 additional patients and associated with lower respiratory tract infections in children. The molecular virus screening procedure provides a general culture-independent solution to the problem of detecting unknown virus species in single or pooled samples. We suggest that a systematic exploration of the viruses that infect humans, “the human virome,” can be initiated.
Keywords: bioinformatics, nucleotide sequencing, respiratory tract infection, virus
Virus infections impose an enormous disease burden on humanity, but our knowledge of the viruses that infect humans is still incomplete. Because of the elusive nature of viruses, most studies are limited to the investigation of already known viruses, whereas the discovery of an unknown virus and production of the first diagnostic reagent is very difficult and remains a rare occurrence. The majority of viruses known today were first identified by animal experiments or virus replication in tissue culture. We know that there are viruses that cannot be replicated in the laboratory, and some of these have been identified by molecular methods (1-4). No generally applicable method for the identification of such viruses has been available, and, in fact, a very large number of unidentified human viruses may exist (2). Thus, numerous acute and chronic diseases with unknown etiology may be caused by unidentified viruses, and the systematic search for unknown viruses is an urgent scientific task (2).
Lower respiratory tract infection (LRTI) is a leading cause for hospitalization of infants and young children and accounts for ≈250,000 hospitalizations a year in the United States alone (5). The most important viral agent in this group of patients is respiratory syncytial virus (RSV). Other important agents are influenza viruses, parainfluenza viruses, adenoviruses, rhinoviruses, coronaviruses, and human metapneumovirus (6-8). Human metapneumovirus was cultured and characterized in 2001, and, more recently, a renewed interest in coronaviruses after the severe acute respiratory syndrome epidemic has led to the characterization of two coronavirus species associated with LRTI (9-11). In comprehensive studies of the etiology of LRTI, no etiologic agent has been found in 12-39% of cases, which suggests that additional unknown agents may be involved in the etiology of LRTI (6-8).
The identification of viruses in the respiratory tract could have implications for diseases other than respiratory tract infections. Viruses have limited options for transmission between host organisms. Many viruses primarily associated with nonrespiratory symptoms are nevertheless transmitted through the respiratory tract and can be detected there, e.g., herpes viruses, enteroviruses, and parvovirus B19 (8, 12). The respiratory tract may therefore harbor unknown viruses associated with trivial and severe symptoms and is a good starting point for an attempt to systematically explore the human virus flora. In this paper, we present a general strategy for molecular virus screening of clinical samples and the systematic screening of a set of respiratory tract samples resulting in the discovery and characterization of a human parvovirus.
Methods
Virus Screening Library Construction. The samples included in the study were randomly selected nasopharyngeal aspirates submitted to the Karolinska University Laboratory, Stockholm, Sweden, for diagnostics of respiratory tract infections. Standard diagnostics included immunofluorescence (IF) analysis as requested by the clinician (in general for influenza and/or RSV) and virus culture. Upon arrival in the laboratory, the nasopharyngeal aspirates (aspirated material diluted in 0.9% NaCl during the sampling process) were centrifuged at 1,500 rpm for 10 min in a Sigma 204 table-top centrifuge as part of the routine diagnostics to collect cells for IF. The cell-free supernatants were collected for the present study, anonymized, and stored at -80°C until analyzed. Samples were included regardless of the results obtained in subsequent diagnostic IF and virus culture assays. One hundred microliters per sample was saved separately for confirmatory analysis and sequencing after the screening. The rest of the cell-free supernatants (100-1,000 μl per sample) were pooled in a single tube. Pool 1 was first ultracentrifuged at 41,000 rpm in an sw41 rotor (Beckman) for 90 min, after which the resulting pellet was dissolved in 200 μl of 2% human blood donor serum in molecular-biology-grade water, and filtered through a 0.22-μm spin filter (Ultrafree-MC, Millipore) at 3,000 rpm in an Eppendorf 5415C microcentrifuge. Pool 2 was divided into two aliquots that were first filtered through a 0.22-μm or 0.45-μm disk filter (Millex GV/HV, Millipore) respectively, and then ultracentrifuged at 41,000 rpm in an sw41 rotor (Beckman) for 90 min. The resulting pellet was dissolved in 200 μl of 2% filtered human blood donor serum in molecular-biology-grade water. After this stage, both pools were handled identically. DNase I (100 units, Stratagene) was added, and the samples were incubated for 2 h at 37°C (4). An additional 200 μl of molecular-biology-grade water was added, and each sample was divided into two aliquots. DNA was extracted from one aliquot with the QIAamp Blood Mini Kit (Qiagen, Hilden, Germany) and eluted in 50 μl of the supplied elution buffer. RNA was extracted from the other aliquot with TRIzol LS reagent (Invitrogen). The final RNA pellet was dissolved in 10 μl of RNase-free water with 1 mM DTT and 4 units of recombinant RNase inhibitor (Promega). Extracted DNA and RNA were amplified separately by “random PCR” (4, 13). Extracted DNA (20 μl) was mixed with 2.5 μl of 10× Ecopol buffer (100 mM Tris·HCl, pH 7.5/50 mM MgCl2/75 mM DTT) (New England Biolabs), a 1-μl solution containing each dNTP at 10 mM, and 2 μl of primer FR26RV-N (GCCGGAGCTCTGCAGATATCNNNNNN) at 10 μM. The reaction was incubated at 94°C for 2 min and on ice for 2 min, after which 2.5 units (0.5 μl) of 3′-5′ exo- Klenow DNA polymerase (New England Biolabs) was added, and the reaction was incubated at 37°C for 1 h. This denaturation-annealing-elongation cycle was repeated once and then followed by an enzyme inactivation step at 75°C for 10 min. The extracted RNA was handled similarly. All 10 μl of RNA was mixed with 2 μl of primer FR26RV-N at 10 μM, incubated at 65°C for 5 min, and chilled on ice. A reaction mix of 7.7 μl containing 4 μl of 5× First-Strand buffer (250 mM Tris·HCl, pH 8.3/375 mM KCl/15 mM MgCl2) (Invitrogen), 2 μl of 100 mM DTT, a 1-μl solution containing each dNTP at 10 mM, 8 units (0.2 μl) of recombinant RNase inhibitor (Promega), and 100 units (0.5 μl) of SuperScript II reverse transcriptase (Invitrogen) was added. The reaction was incubated at 25°C for 10 min and 42°C for 50 min. After a denaturation step at 94°C for 3 min and chilling on ice, 2.5 units (0.5 μl) of 3′-5′ exo- Klenow DNA polymerase (New England Biolabs) was added, and the reaction was incubated at 37°C for 1 h, followed by an enzyme inactivation step at 75°C for 10 min. Of each reaction mix, 5 μl was used as a template in a subsequent PCR. The 50-μl reaction mix consisted of 1× GeneAmp PCR buffer II (100 mM Tris·HCl, pH 8.3/500 mM KCl) (Applied Biosystems), 2.5 mM MgCl2, each dNTP at 0.2 mM, 40 pmol of the primer FR20RV (GCCGGAGCTCTGCAGATATC), and 2.5 units of AmpliTaq Gold DNA polymerase (Applied Biosystems). After 10 min at 94°C, 40 cycles of amplification (94°C for 1 min, 65°C for 1 min, and 72°C for 2 min) were performed.
The amplification products were purified by using a QIAquick PCR Purification Kit (Qiagen), and digested with EcoRV to remove the amplification primers. Products were then separated on an agarose gel and fragments between ≈600 and 1,500 bp in length were excised and extracted by QIAquick Gel Extraction Kit (Qiagen). Five microliters of the eluted, purified PCR product was ligated to the vector pCR-Blunt and introduced into chemically competent E. coli TOP-10 according to the manufacturer's instructions (Zero Blunt PCR Cloning Kit, Invitrogen). Bacteria were plated on Luria-Bertani agar plates containing 50 μg/ml kanamycin.
Sequencing. Sequencing templates were produced directly from colonies in a 96-well format by rolling circle amplification using Templiphi (Amersham Biosciences). Sequencing was performed with DYEnamic ET Dye Terminator Cycle Sequencing reagents (Amersham Biosciences) and Megabace 1000 sequencers in one-eighth reactions. Otherwise, reaction conditions were according to the manufacturer's instructions. The reads were base-called with phred and assembled by using phrap.
Automated Sequence Editing and Database Searches. A set of C++ and Perl programs were written to automate the process of quality trimming, clustering, GenBank/SwissProt searches, sorting and formatting of the sequence reads. The output was a sorted list of the best database hits for nucleotide and translated sequences. A MisEd module was used for quality trimming (14), phrap for clustering. GenBank and SwissProt databases were downloaded and searched locally with blast (15).
Phylogenetic Analysis. All sequences were downloaded from GenBank and SwissProt with a BioPerl script. Accession numbers are available upon request. Multiple alignments and bootstrapped (1,000 replicates) neighbor-joining trees were generated by clustalx (1.83) and njplot (16, 17).
Diagnostic PCR for Human Bocavirus (HBoV). The experiments were performed in a diagnostic laboratory setting, ensuring that necessary precautions to avoid contamination were taken. Samples were screened in pools of 10, and, for PCR-positive pools, samples were extracted and amplified individually. Positive and negative controls were included in each experiment. DNA was extracted by QIAamp DNA Blood Mini Kit (Qiagen). Extracted DNA (5 μl) was used as template for the PCR. The 50-μl reaction mix consisted of 1× GeneAmp PCR buffer II (100 mM Tris·HCl, pH 8.3/500 mM KCl) (Applied Biosystems), 2.5 mM MgCl2, each dNTP at 0.2 mM, 20 pmol each of the primers 188F(GACCTCTGTAAGTACTATTAC) and 542R (CTCTGTGTTGACTGAATACAG), and 2.5 units of AmpliTaq Gold DNA polymerase (Applied Biosystems). After 10 min at 94°C, 35 cycles of amplification (94°C for 1 min, 54°C for 1 min, and 72°C for 2 min) were performed. Products were visualized on an agarose gel. The expected product size was 354 bp. All PCR products were sequenced to confirm that they were specific for HBoV.
Results
A Procedure for Molecular Virus Screening of Clinical Samples. A pipeline for sequence-based detection of known and unknown viruses in clinical samples was set up as described in detail under Methods. The main components were systematic collection and pooling of excess material from clinical samples, virus concentration by ultracentrifugation, depletion of contaminating nucleic acids by filtration and DNase treatment (4), amplification by random PCR (a procedure using a generic primer sequence with a random 3′ end) (13), cloning of the PCR products, large-scale sequencing of the clones, and automated editing and database searches of the sequencing results. To obtain a virus detection sensitivity estimate for the procedure, we analyzed dilutions of serum samples with known titers of hepatitis B and C viruses (a DNA virus and an RNA virus; virus titers were determined by commercial diagnostic assays) (4). The ultracentrifugation virus concentration step was not included in that analysis. For both viruses, 105 virus copies were detected in 50 μl of serum by analysis of 96 clones. Thus, minimum reproducible detection level was estimated to ≈106 virions per ml in serum or 105 virions in total.
Molecular Virus Screening of Respiratory Tract Samples. Two pools of centrifuged, cell-free supernatants of nasopharyngeal aspirates were analyzed initially. Both pools were made from randomly selected diagnostic samples so that samples positive for known viruses would serve as positive controls for the screening procedure. However, exact matching with diagnostic results of individual samples was not possible because the samples were anonymized. The first pool was of 28 supernatants collected in November and December 2003, and the second pool was of 20 supernatants collected in March 2004. Of the 48 samples, 38 were from pediatric patients. Each pool was treated for enrichment of viruses and removal of nonviral nucleic acids by ultracentrifugation, microfiltration, and DNase treatment. The remaining DNA and RNA was extracted from separate aliquots, amplified, cloned in a PCR-Blunt vector, and sequenced. A total of 480 clones (192 clones derived from RNA and 288 clones derived from DNA) from the first library and 384 clones (192 each from RNA and DNA) from the second library were sequenced bidirectionally. A linked set of computer programs was used for automated quality trimming, clustering, database searches, sorting, and formatting of the sequence reads. The output was a sorted list of the best database hits for nucleotide and translated sequences. After the vector sequence and low-quality sequence were automatically discarded, 343 (71%) and 306 (80%) clones remained from the respective library. By using automated nucleotide and translated blast searches (15), the clones were categorized into human, bacterial, phage, virus, and unknown sequences (Table 1). In total, 20% of clones analyzed showed significant (E < 10-5) similarity to viral sequences. The sequences matched seven different virus species, of which four were RNA viruses and three were DNA viruses. The sequences matching influenza A virus, RSV, metapneumovirus, and adenovirus, were due to high sequence similarity considered to represent previously known virus species. The TT-virus-like sequences were not analyzed further, despite moderate homology to known sequence, because this group of viruses is known to be ubiquitous and highly heterogeneous (3). Coronavirus-like sequences were found in both libraries and were all similar to group 2 coronaviruses, in particular to murine hepatitis virus. A virus genome highly similar to our sequences was recently identified in Hong Kong and published as a new species named coronavirus HKU1 (10). Parvovirus-like sequences were found in both libraries. These sequences showed no significant similarity to database sequences at the nucleotide level in the blast search. The deduced amino acid sequence significantly matched bovine parvovirus (BPV) and canine minute virus (MVC; also known as minute virus of canines), two related members of the Parvoviridae family, subfamily Parvovirinae, genus Bocavirus.
Table 1. Categorization by blast search of the sequenced clones derived from two pools of respiratory tract samples.
| Category | Library 1 (%) | Library 2 (%) |
|---|---|---|
| Human | 84 (24) | 110 (36) |
| Bacterial | 202 (59) | 65 (21) |
| Phage | 6 (2) | 2 (1) |
| Unknown | 22 (6) | 33 (11) |
| Virus | 29 (8) | 99 (32) |
| Influenza A virus | 18 | 0 |
| Adenovirus | 6 | 0 |
| Respiratory syncytial virus | 0 | 10 |
| Metapneumovirus | 0 | 1 |
| TT virus | 2 | 0 |
| Coronavirus | 1 | 26 |
| Parvovirus | 2 | 62 |
| Total | 343 | 309 |
Genome Analysis of HBoV. The individual source samples in the respective screening pool were identified by specific PCR targeting of the sequence of the first detected clones. By using these samples as templates, we determined the complete coding consensus sequence of both index isolates: Stockholm 1 (ST1; 5,217 nt; accession no. DQ000495) and Stockholm 2 (ST2; 5,299 nt; accession no. DQ000496). The complete coding sequences were determined by assembly of the clones derived from the screening procedure, combined with PCR amplification and sequencing of the connecting regions and regions covered by less than three clones. Finally, the terminal sequences were amplified by a modified protocol for RACE. Despite the RACE experiments, the expected terminal hairpin sequences were most likely not completely determined for either isolate.
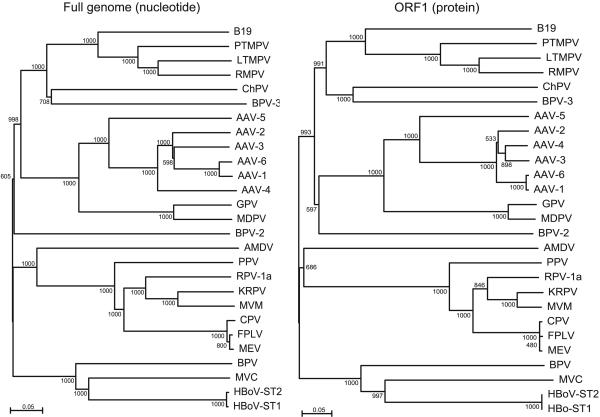
Phylogenetic trees were constructed based on alignments of the isolates ST1 and ST2 and the viruses in the Parvovirinae subfamily. Results from full-length nucleotide sequences as well as nucleotide and deduced amino acid sequences of the two major ORFs were consistent and confirmed that the isolates ST1 and ST2 group with MVC and BPV, as expected from the blast results (Fig. 1 and data not shown). It has previously been recognized that MVC and BPV form a separate clade within the Parvovirinae (18, 19), and the International Committee on Taxonomy of Viruses has recently assigned a separate genus with the name Bocavirus to BPV and MVC (International Committee on Taxonomy of Viruses Virus Index Database, www.danforth-center.org/iltab/ictvnet). The new virus is clearly separate from BPV and MVC, having only 43% amino acid identity to the nearest neighbor MVC in both major ORFs. The distance to BPV is remarkably similar: 42% amino acid identity in both major ORFs. We therefore conclude that the isolates ST1 and ST2 represent a previously uncharacterized species of the genus Bocavirus, and we propose the provisional name “human bocavirus” for the new virus.
Fig. 1.
Phylogenetic analysis: bootstrapped neighbor-joining tree based on full-length nucleotide sequences (Left) and ORF1 amino acid sequences (Right)of HBoV and the Parvovirinae. Bootstrap values are indicated at each branching point. Analysis of capsid gene nucleotide and amino acid sequences yielded highly similar results (data not shown). B19, erythrovirus B19; PTMPV, pig-tailed macaque parvovirus; LTMPV, long-tailed macaque parvovirus; RMPV, rhesus macaque parvovirus; ChPV, chipmunk parvovirus; AAV, adeno-associated virus; GPV, goose parvovirus; MDPV, Muscovy duck parvovirus; AMDV, Aleutian mink disease virus; PPV, porcine parvovirus; RPV-1a, rat parvovirus-1a; KRPV, Kilham rat parvovirus; MVM, minute virus of mice; CPV, canine parvovirus; FPLV, feline panleukopenia virus; MEV, mink enteritis virus.
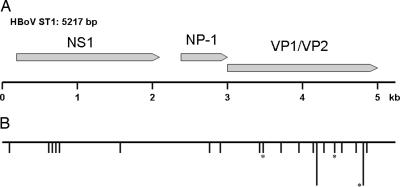
The genomic organization of HBoV closely resembles that of the other known bocaviruses BPV and MVC (Fig. 2). Like in all members of the Parvovirinae subfamily, there are two major ORFs encoding a nonstructural protein (NS1) and at least 2 capsid proteins (VP1 and VP2), respectively. However, like MVC and BPV, HBoV also has a third, middle ORF. In MVC and BPV, this ORF encodes a nonstructural protein with unknown function, named NP-1 (19, 20). The mid-ORF product of HBoV is homologous to NP-1, having 47% amino acid identity to NP-1 of MVC and BPV. This fact further supports the classification of HBoV as a Bocavirus. The two isolates ST1 and ST2 were closely related, differing at only 26 nucleotide positions. Eighteen of these differences, including the only three nonsynonymous substitutions, occur in the capsid gene (Fig. 2).
Fig. 2.
Map of the HBoV genome. (A) Schematic map of isolate ST1 of HBoV showing the three ORFs as arrows: NS1, 1,920 nt (nucleotides 183-2102), 639 aa; NP-1, 660 nt (nucleotides 2340-2999), 219 aa; and VP1/VP2, 2,016 nt (nucleotides 2986-5001), 671 aa. (B) A map showing the location of the 26 nucleotide differences that were detected between two isolates of HBoV. The horizontal line represents the sequence of ST1, and each vertical line represents a nucleotide difference to ST2. In two cases where several differences were located close together, a longer vertical line representing four differences was used. The asterisks mark the three differences that resulted in a predicted amino acid change.
Incidence and Symptoms of HBoV Infection. To estimate the prevalence of HBoV in respiratory tract samples and the clinical picture associated with HBoV infection, a series of PCR screening experiments was performed. As a first overview, 378 culture-negative nasopharyngeal aspirate samples drawn from November 2003 through September 2004 were screened for HBoV by a PCR assay targeting 354 base pairs in the NP-1 gene. These samples came from various clinics served by the Karolinska University Laboratory (266 samples from pediatric patients and 112 samples from adult patients). Seven samples were positive for HBoV DNA, and all seven came from infants and children. Therefore, a more detailed retrospective study was performed in the pediatric infectious diseases ward at the Karolinska University Hospital. All 540 available nasopharyngeal aspirates drawn in the ward (hospitalized patients only) from November 2003 through October 2004 were investigated, including some of the samples included in the first screening. Samples from 17 different patients (3.1%) were positive. The HBoV specificity of the PCR products was confirmed by sequencing. Fourteen HBoV-positive samples were negative for other viruses investigated (by IF and virus culture), whereas HBoV was detected along with another virus in three cases (two RSV and one adenovirus). Morbidity from LRTI is highest in the winter season, which was reflected by sampling frequency and by findings of HBoV (Table 2).
Table 2. Findings of HBoV in nasopharyngeal aspirate samples drawn in the pediatric infectious diseases unit November 2003 to October 2004, distributed per month.
| Month | Tested | Positive |
|---|---|---|
| November | 28 | 0 |
| December | 125 | 4 |
| January | 100 | 5 |
| February | 110 | 4 |
| March | 85 | 1 |
| April | 43 | 2 |
| May | 12 | 0 |
| June | 4 | 1 |
| July | 11 | 0 |
| August | 3 | 0 |
| September | 12 | 0 |
| October | 7 | 0 |
| Total | 540 | 17 |
The medical records of the 14 patients infected with HBoV only were reviewed (Table 3). All 14 children were admitted from home with respiratory distress of 1- to 4-day duration. Seven children had a history of wheezing bronchitis/asthma and were under daily treatment with inhaled beta-2-stimulans and steroids. Four of them had previously been hospitalized for wheezing bronchitis. Two children had chronic lung disease that originated in the neonatal period, and five patients had no history of previous respiratory tract problems. All patients had variable degree of respiratory distress, and fever was prevalent. Chest x-ray demonstrated interstitial bilateral infiltrates in six of seven cases. Gastrointestinal symptoms, conjunctivitis, or rash was not recorded in any case.
Table 3. Clinical characteristics of 14 patients hospitalized for LRTI and positive for HBoV DNA by PCR.
| Age | Sex | Comorbidity | Hypoxia | Tachypnea | Fever >38.5°C | Pathological chest x-ray | Days hospitalized |
|---|---|---|---|---|---|---|---|
| 8 mo | M | Asthma | NA | + | + | ND | 1 |
| 11 mo | M | Asthma | − | + | + | + | 3 |
| 17 mo | F | No | + | + | − | + | 3 |
| 4 yr | M | No | + | NA | + | + | 2 |
| 12 mo | M | No | − | − | + | ND | 2 |
| 15 mo | M | Asthma | − | + | + | ND | 1 |
| 2 yr | M | Asthma | + | + | + | ND | 2 |
| 14 mo | F | No | − | + | − | + | 3 |
| 12 mo | M | CLD | − | + | + | ND | 4 |
| 5 mo | M | No | + | + | + | − | 4 |
| 2 yr | F | Asthma | + | + | + | ND | 2 |
| 3 yr | M | Asthma | + | + | + | + | 3 |
| 13 mo | F | Asthma | − | − | − | ND | 2 |
| 6 mo | M | CLD | + | + | − | + | 8 |
Hypoxia was defined as oxygen saturation by pulse oxymetry consistently <90% without oxygen treatment. Tachypnea was defined as respiratory rate >60 per min for children 0-12 months (mo) old and >50 per min for children >12 months old. Pathological chest x-ray findings were bilateral interstitial infiltrates in all cases. CLD, cronic lung disease; NA, data not available; ND, not done; M, male; F, female.
To establish that HBoV was the likely etiologic agent of the observed symptoms and not just a coincidental finding, we investigated how findings of HBoV correlated to findings of other likely etiologic agents. In the 540 samples analyzed, a known viral pathogen (mainly influenza A virus or RSV) was identified by standard diagnostics (IF and virus culture) in 258 of the 540 patients (48%), and no virus was found by standard diagnostics in 282 patients (52%). Of the 17 HBoV findings, 14 were in the latter group. Thus, HBoV was primarily found in samples negative for other viruses (P < 0.01, Fisher's exact test), suggesting that it is a likely etiologic agent of LRTI in our patients.
Discussion
The Parvoviridae family (“parvoviruses”) is divided into two subfamilies, Densovirinae, which infects arthropods, and Parvovirinae, which infects birds and mammals. There is no sequence homology between the Densovirinae and Parvovirinae. Accordingly, our phylogenetic analysis included only the Parvovirinae. The viruses in this subfamily have recently been reclassified into five genera by the International Committee on Taxonomy of Viruses: Parvovirus, Erythrovirus, Dependovirus, Amdovirus, and Bocavirus. The previously known human parvoviruses are the well known pathogen parvovirus B19 (Erythrovirus) and the presumably apathogenic adeno-associated viruses (Dependovirus). The present study reports a human virus of the genus Bocavirus, and the HBoV is most likely the second known parvovirus species pathogenic to humans.
Confirming a causal relationship between a virus and the observed symptoms is an important but difficult issue that normally requires multiple separate studies to be ultimately resolved (21). The classical Koch postulates cannot be applied on most modern molecular discoveries, because fulfilling Koch's postulates requires in vitro culture of the agent and access to a suitable experimental animal model (21). The study of respiratory tract disease is particularly difficult, because similar symptoms can be produced by a wide range of agents, and respiratory tract samples from healthy subjects or patients without respiratory symptoms are rarely available. We addressed this issue by comparing 258 LRTI cases with known etiology to 282 cases with unresolved etiology. HBoV was primarily present in the subset of LRTI patients with unknown etiology, and this nonrandom distribution suggests that HBoV is a likely causative agent of LRTI. The three cases of double infection by HBoV and another virus do not contradict this conclusion. Double infections are frequently found in the study of LRTI (7, 8) and likely reflect the high prevalence of viral infections in infants and young children.
The prevalence estimation is highly dependent on a sensitive PCR assay capable of detecting multiple isolates. We selected the N terminus of the NP-1 gene as target sequence for the diagnostic PCR assay. This region was identical in the two isolates ST1 and ST2, and the NP-1 gene had a higher overall similarity with other Bocavirus species than had the NS-1 or VP genes. Thus, available data suggests that it is the most conserved region of the virus. However, sufficient sequence data to support this assumption are not available, and there is a risk that the true prevalence of the virus has been slightly underestimated because of a suboptimal diagnostic PCR.
The present study included only patients treated for respiratory tract symptoms. Parvoviruses are in general capable of systemic infection. Because of their need for proliferating host cells for their replication, infection of respiratory and gut epithelium, hematopoietic cells, and transplacental infection of fetuses are frequent characteristics of parvoviruses. For the same reason, different symptoms may be observed in young individuals versus adult individuals (22). The related animal bocaviruses BPV and MVC have not been extensively studied but are associated with respiratory symptoms and enteritis of young animals. Systemic infection by BPV and MVC appears likely, and there are indications that fetal infection leading to fetal death may occur (23-26). Whether HBoV was present in blood or feces of our patients could not be addressed, because neither blood nor fecal samples were available. Additional studies will be required to investigate the full pathogenic potential of HBoV.
Our results indicate that the combination of host DNA depletion, optimized nucleic acid amplification, large-scale sequencing, and bioinformatics is an efficient procedure for the identification of unknown viruses. A key issue for its general use is the virus detection sensitivity. A gross sensitivity estimation suggested that reproducible detection of 105 virus copies was possible by sequencing 96 clones. However, there is no absolute minimum detection threshold, but the probability of detection is a function of virus titer and number of clones analyzed. Sensitivity is also likely dependent on the properties of each included clinical sample as well as physical and genetic properties of the virus in question. When tested on clinical respiratory tract samples, our method identified a range of known and unknown pathogens. Apart from HBoV, the screening also identified one at that time undescribed coronavirus species, and notably, our study focused only on virus-like sequences and paid no attention to completely unknown sequences. The technology is general, unselective, and rapid. Unlike other virus discovery methods, there is no requirement for virus replication in cell culture, design of group-specific primers, or selected pairs of preinfection and postinfection samples (1, 2, 9). Therefore, it is well suited for rapid identification of an unknown or unexpected virus involved in a disease outbreak. The procedure also allows large-scale screening with a minimal hands-on effort. It can easily be scaled up only by increasing sequencing capacity. We therefore suggest that a systematic exploration of the viruses that infect humans (the human virome) can be initiated.
Acknowledgments
We thank Dr. Mats A. A. Persson for valuable support, Dr. Anders Widell for critical reading of the manuscript, Mr. Pardha S. Velandandi for assistance with phylogenetic analysis, and Mr. Hamid Darban and Mr. Daryoush Rahmani for technical assistance. The study was supported by the Swedish Research Council and Nanna Svartz' Fund. T.A. was a Clinical Fellow of the Söderberg Foundation at the Center for Molecular Medicine.
Author contributions: T.A. designed research; T.A., M.E., A.B., and A.T.-L. performed research; M.T.T. and B.A. contributed new reagents/analytic tools; T.A., M.T.T., M.E., A.B., A.T.-L., and B.A. analyzed data; and T.A., M.T.T., M.E., and B.A. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: LRTI, lower respiratory tract infection; RSV, respiratory syncytial virus; IF, immunofluorescence; HBoV, human bocavirus; MVC, minute virus of canines; BPV, bovine parvovirus, ST1, Stockholm 1; ST2, Stockholm 2.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. DQ000495 (HBoV isolate ST1) and DQ000496 (HBoV isolate ST2)].
References
- 1.Kellam, P. (1998) Trends Microbiol. 6, 160-165. [DOI] [PubMed] [Google Scholar]
- 2.Relman, D. A. (1999) Science 284, 1308-1310. [DOI] [PubMed] [Google Scholar]
- 3.Simmonds, P. (2002) J. Med. Microbiol. 51, 455-458. [DOI] [PubMed] [Google Scholar]
- 4.Allander, T., Emerson, S. U., Engle, R. E., Purcell, R. H. & Bukh, J. (2001) Proc. Natl. Acad. Sci. USA 98, 11609-11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shay, D. K., Holman, R. C., Newman, R. D., Liu, L. L., Stout, J. W. & Anderson, L. J. (1999) J. Am. Med. Assoc. 282, 1440-1446. [DOI] [PubMed] [Google Scholar]
- 6.Iwane, M. K., Edwards, K. M., Szilagyi, P. G., Walker, F. J., Griffin, M. R., Weinberg, G. A., Coulen, C., Poehling, K. A., Shone, L. P., Balter, S., et al. (2004) Pediatrics 113, 1758-1764. [DOI] [PubMed] [Google Scholar]
- 7.Juven, T., Mertsola, J., Waris, M., Leinonen, M., Meurman, O., Roivainen, M., Eskola, J., Saikku, P. & Ruuskanen, O. (2000) Pediatr. Infect. Dis. J. 19, 293-298. [DOI] [PubMed] [Google Scholar]
- 8.Jartti, T., Lehtinen, P., Vuorinen, T., Osterback, R., van den Hoogen, B., Osterhaus, A. D. & Ruuskanen, O. (2004) Emerg. Infect. Dis. 10, 1095-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Hoek, L., Pyrc, K., Jebbink, M. F., Vermeulen-Oost, W., Berkhout, R. J., Wolthers, K. C., Wertheim-van Dillen, P. M., Kaandorp, J., Spaargaren, J. & Berkhout, B. (2004) Nat. Med. 10, 368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woo, P. C., Lau, S. K., Chu, C. M., Chan, K. H., Tsoi, H. W., Huang, Y., Wong, B. H., Poon, R. W., Cai, J. J., Luk, W. K., et al. (2005) J. Virol. 79, 884-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fouchier, R. A., Hartwig, N. G., Bestebroer, T. M., Niemeyer, B., de Jong, J. C., Simon, J. H. & Osterhaus, A. D. (2004) Proc. Natl. Acad. Sci. USA 101, 6212-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Young, N. S. & Brown, K. E. (2004) N. Engl. J. Med. 350, 586-597. [DOI] [PubMed] [Google Scholar]
- 13.Froussard, P. (1992) Nucleic Acids Res. 20, 2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tammi, M. T., Arner, E., Kindlund, E. & Andersson, B. (2003) Nucleic Acids Res. 31, 4663-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perriere, G. & Gouy, M. (1996) Biochimie 78, 364-369. [DOI] [PubMed] [Google Scholar]
- 17.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25, 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohshima, T., Kishi, M. & Mochizuki, M. (2004) Virus Genes 29, 291-296. [DOI] [PubMed] [Google Scholar]
- 19.Schwartz, D., Green, B., Carmichael, L. E. & Parrish, C. R. (2002) Virology 302, 219-223. [DOI] [PubMed] [Google Scholar]
- 20.Chen, K. C., Shull, B. C., Moses, E. A., Lederman, M., Stout, E. R. & Bates, R. C. (1986) J. Virol. 60, 1085-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fredricks, D. N. & Relman, D. A. (1996) Clin. Microbiol. Rev. 9, 18-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bloom, M. E. & Young, N. S. (2001) in Fields' Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott, Philadelphia), pp. 2361-79.
- 23.Durham, P. J., Lax, A. & Johnson, R. H. (1985) Res. Vet. Sci. 38, 209-219. [PubMed] [Google Scholar]
- 24.Carmichael, L. E., Schlafer, D. H. & Hashimoto, A. (1991) Cornell Vet. 81, 151-171. [PubMed] [Google Scholar]
- 25.Carmichael, L. E., Schlafer, D. H. & Hashimoto, A. (1994) J. Vet. Diagn. Invest. 6, 165-174. [DOI] [PubMed] [Google Scholar]
- 26.Kirkbride, C. A. (1992) J. Vet. Diagn. Invest. 4, 374-379. [DOI] [PubMed] [Google Scholar]