Abstract
Hutchinson-Gilford progeria syndrome (HGPS) is a rare genetic disorder that is characterized by dramatic premature aging and accelerated cardiovascular disease. HGPS is almost always caused by a de novo point mutation in the lamin A gene (LMNA) that activates a cryptic splice donor site, producing a truncated mutant protein termed “progerin.” WT prelamin A is anchored to the nuclear envelope by a farnesyl isoprenoid lipid. Cleavage of the terminal 15 aa and the farnesyl group releases mature lamin A from this tether. In contrast, this cleavage site is deleted in progerin. We hypothesized that retention of the farnesyl group causes progerin to become permanently anchored in the nuclear membrane, disrupting proper nuclear scaffolding and causing the characteristic nuclear blebbing seen in HGPS cells. Also, we hypothesized that blocking farnesylation would decrease progerin toxicity. To test this hypothesis, the terminal CSIM sequence in progerin was mutated to SSIM, a sequence that cannot be farnesylated. SSIM progerin relocalized from the nuclear periphery into nucleoplasmic aggregates and produced no nuclear blebbing. Also, blocking farnesylation of authentic progerin in transiently transfected HeLa, HEK 293, and NIH 3T3 cells with farnesyltransferase inhibitors (FTIs) restored normal nuclear architecture. Last, treatment of both early- and late-passage human HGPS fibroblasts with FTIs resulted in significant reductions in nuclear blebbing. Our results suggest that treatment with FTIs represents a potential therapy for patients with HGPS.
Keywords: aging, lamin, laminopathy
Hutchinson-Gilford progeria syndrome (HGPS) is an extremely rare and uniformly fatal “premature aging” disease in which all children die as a consequence of myocardial infarction or cerebrovascular accident at an average age of 12 years (range, 8-21 years). The earliest manifestations of the disease are seen at ≈12-14 months of age and include alopecia and growth retardation (Progeria Research Foundation's medical and research database). In addition to progressive atherosclerosis, HGPS is characterized by bone deformations, including craniofacial disproportion, mandibular and clavicular hypoplasia, and osteoporosis, as well as by a loss of s.c. fat, delayed dentition, sclerodermatous skin, joint stiffness, and hip dislocations (1-3).
HGPS is a sporadic autosomal dominant disease caused in nearly all cases by a de novo single-base substitution in codon 608 of exon 11 of the LMNA gene on chromosome 1 (4, 5). The LMNA gene encodes three proteins, lamin A (LA), LC, and LAΔ10, all of which are components of the nuclear lamina, a dynamic molecular interface located inside the inner nuclear membrane (6). Initially believed to be an inert scaffolding network, the lamina has now been shown to have significant roles in DNA replication, transcription, chromatin organization, nuclear shape, and cell division (7). In the LMNA gene, >180 mutations have been reported, and currently, there are eight diseases in addition to HGPS (referred to as the “laminopathies”) that are associated with various mutations in this gene (7). These diseases include disorders such as Emery-Dreifuss muscular dystrophy, mandibuloacral dysplasia, atypical Werner's syndrome, dilated cardiomyopathy type 1A, restrictive dermopathy, and Dunnigan-type familial partial lipodystrophy (8, 9).
The typical LMNA mutation in HGPS is a C-to-T nucleotide substitution at position 1824 causing no change in the encoded amino acid (G608G) but creating a cryptic splice donor site. Activation of this site results in an mRNA lacking 150 nucleotides. In turn, this mRNA is translated into a mutant protein, termed “progerin” (4), with a 50-aa internal deletion near the C terminus. LA is normally expressed by most differentiated cells, where it integrally affects both nuclear membrane structure and function (10). Progerin apparently acts in a dominant-negative manner on the nuclear function of cell types that express LA (11, 12). In addition to the potential mechanical fragility that is created by disrupting the nuclear lamina, this mutation also may affect other vital cellular processes such as gene transcription, DNA replication, and cell division.
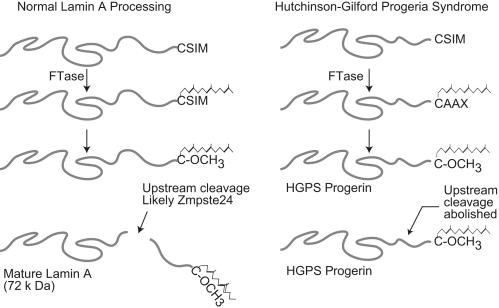
In normal cells, the prelamin A protein contains a CAAX tetrapeptide motif at the C terminus. This tetrapeptide signals the addition of a 15-carbon farnesyl isoprenoid lipid group to the cysteine by the enzyme farnesyltransferase (FTase) (13). The CAAX motif is a cysteine followed by two aliphatic amino acids and a terminal “X” residue. This final amino acid defines the specificity for the addition of an isoprenyl group with methionine, serine, glutamine, or alanine signaling modification by FTase and with leucine signaling the addition of a 20-carbon geranylgeranyl isoprenoid group catalyzed by the structurally related enzyme geranylgeranyltransferase (GGTase) I (14). For LA, the CAAX motif is CSIM. Farnesylation, together with subsequent CAAX-signaled modifications, promote prelamin A association with the nuclear membrane (15). After farnesylation, the terminal three AAX amino acids are removed, and the C-terminal cysteine undergoes methyl esterification (16, 17). Although both B-type lamins and LA are farnesylated and carboxymethylated, unique to LA is a second cleavage inside the nucleus causing the removal of an additional 15 C-terminal amino acids from the mature protein, including the farnesylated cysteine. This final cleavage step and the resulting loss of the farnesyl anchor presumably releases prelamin A from the nuclear membrane and allows it to be inserted into the nuclear lamina. In HGPS, although preprogerin can be farnesylated, its internal deletion of amino acids 606-656 removes the endoprotease recognition site necessary for executing the final cleavage step (Fig. 1). The importance of this cleavage is evident by the fact that mutations in ZMPSTE24 cause a severe form of mandibuloacral dysplasia, one of the laminopathies that is phenotypically similar to HGPS (18). ZMPSTE24 is the human homolog of yeast STE24 and is responsible for this final cleavage of LA (19).
Fig. 1.
Translation of the LMNA gene yields the prelamin A protein, which requires posttranslational processing for incorporation into the nuclear lamina. The prelamin A protein contains a CAAX box at the C terminus that signals isoprenylation (in this case, the addition of a farnesyl group to the cysteine by the enzyme FTase). After farnesylation, the terminal three amino acids (SIM) are cleaved, and the terminal farnesylated cysteine undergoes methyl esterification. A second cleavage step by the ZMPSTE24 endoprotease then removes the terminal 15 aa, including the farnesyl group. This final cleavage step is blocked in progeria.
We hypothesized that retention of the farnesylated C terminus causes progerin to become permanently anchored in the nuclear membrane and unable to be released. The central rod domain of progerin then allows dimerization with mature nonfarnesylated LA and assembly into a multiprotein complex, resulting in dominant-negative disruption of the nuclear scaffolding and underlying heterochromatin and leading to the characteristic nuclear blebbing seen in HGPS (11). Also, we hypothesized that farnesyltransferase (FTase) inhibitors (FTIs) would inhibit the formation of progerin and that decreasing the amount of this aberrant protein could potentially improve disease status in HGPS and other laminopathies.
In this study, we have examined the ability of both genetic mutation and pharmacological treatment to prevent the dysmorphic nuclear phenotype seen in HGPS. The results support our hypothesis that it is the permanently farnesylated state of progerin that allows it to exert its dominant-negative effects and cause the devastating effect on nuclear structure and function. Also, we have demonstrated the ability of FTIs to reverse this nuclear phenotype. Because FTIs are currently under evaluation in phase III clinical trials as anticancer drugs (20), we have identified FTIs as a potential therapy for HGPS.
Materials and Methods
Constructs and Mutagenesis. The pEGFP-myc-LA vector (referred to here as WT LA-CSIM) and the LAΔ50 vector (referred to here as progerin-CSIM) were created as described in ref. 11; these vectors encode GFP-tagged LA fusion proteins. The WT LA-CSIL, progerin-CSIL, WT LA-SSIM, and progerin-SSIM mutations were created by site-directed mutagenesis using the following oligonucleotides: CSIL, 5′-CCCCAGAACTGCAGCATCTTATAATCTAGAGTCGACGGTA-3′; and SSIM, 5′-CCAGAGCCCCCAGAACTCAAGCATCATGTAATCTAGAG-3′ (QuikChange II XL site-directed mutagenesis kit, Stratagene).
Cell Culture. The cell lines used were the normal human fibroblast line AG06299 and the HGPS fibroblast lines AG06917, AG11513, and AG11498 (NIA Aging Cell Culture Repository, Coriell Cell Repositories, Camden, NJ). Fibroblasts were cultured in MEM (Invitrogen/GIBCO) supplemented with 15% FBS (HyClone)/2 mM l-glutamine/50 units/ml penicillin/50 mg/ml streptomycin. HeLa and HEK 293 cell lines were cultured in DMEM (Invitrogen/GIBCO) supplemented with 10% FBS and antibiotics.
Transient Transfections. Cells were plated at ≈25,000 cells per chamber of four-chamber slides (Nunc no. 154526, Lab-Tek, Nalge). After 24 h, HeLa and HEK 293 cell lines were transiently transfected with 0.8 μg of each construct by using Lipofectamine 2000 (Invitrogen) under standard conditions.
FTI Treatment. At the time of transient transfection, HeLa cells were treated with one dose of 0, 0.5, 1.0, or 2.0 μM selective FTI lonafarnib (Sarasar SCH66336, Schering-Plough). HEK 293 cells were treated with one dose of 0, 1, 2, 5, or 10 μM selective FTI L-744832 (Biomol, Plymouth Meeting, PA). Also, NIH 3T3 cells were treated with 5 μM FTI-2153, which is highly selective for FTase, or GGTase inhibitor (GGTI)-2166, which is highly selective for GGTase I (kind gifts of S. M. Sebti and A. D. Hamilton, Yale University, New Haven, CT). These FTIs have the same mechanism of action and can be used interchangeably to inhibit protein farnesylation in vitro. However, only lonafarnib is a clinical candidate. Normal and HGPS fibroblasts were treated with one daily dose of 0, 0.5, 1.0, or 2.0 μM lonafarnib for 3 days.
GFP Localization and Fluorescence Microscopy. At 48 h after transient transfection, the HeLa and HEK 293 cells were visualized for GFP localization by using an Axioplan fluorescence microscope (Zeiss). After a 3-day treatment with lonafarnib, HGPS fibroblasts were washed two times with PBS (pH 7.2) and fixed for 10 min at room temperature with 1% paraformaldehyde in PBS. After three washes with PBS, the cells were permeabilized for 5 min at room temperature with 0.5% Triton X-100 in PBS and blocked with 5% horse serum for 30 min at room temperature. Cells were then incubated for 1 h at room temperature with the primary Ab diluted in blocking solution, a polyclonal mouse anti-human LA/C at 1:10 (mAb 3211; Chemicon). Three more washes with PBS were then followed by incubation with the secondary Ab (Alexa Fluor 488-conjugated donkey anti-mouse IgG; Molecular Probes) for 45 min and three additional PBS washes. Slides were then mounted with mounting medium containing Vectastain DAPI (Vector Laboratories).
Mobility-Shift Experiments. NIH 3T3 cells, grown in high-glucose DMEM (DMEM-H; Invitrogen/GIBCO) supplemented with 10% FCS at 37°C in 10% CO2, were transiently transfected with empty vector, progerin-CSIM, or progerin-SSIM by using Lipofectamine Plus transfection reagent (Invitrogen) according to the manufacturer's instructions. Transfected cells were treated for 48 h with vehicle (DMSO) or inhibitors of prenylation (5 μM FTI-2153, 5 μM GGTI-2166, or both). Cells were lysed directly in 2× Laemmli loading buffer, and total cell lysates were resolved on SDS/8% PAGE. Proteins were transferred to Immobilon poly(vinylidene difluoride) (Millipore), blotted with anti-GFP mAb (clone B34, Covance, Berkeley, CA), and visualized by using SuperSignal enhanced chemiluminescence (Pierce).
Morphometric Analysis. To examine the overall percentage of blebbed cells in cells with different amounts and lengths of FTI treatment, 200 nuclei were classified as either blebbed (if they contained two or more lobulations) or not blebbed, as described (11). These classifications were done by three independent observers, all without knowledge of the specifics of the cells that were being examined, and the three independent data sets were then averaged. Armitage's trend test (21) was used to test for a dose-dependent response for each cell type.
Results
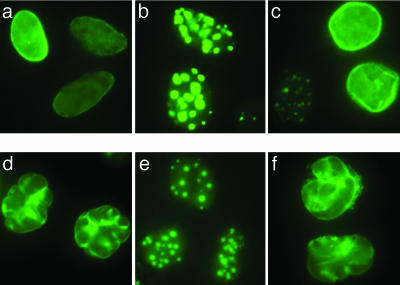
We sought to examine the ability of CAAX box mutants or FTIs to block the nuclear blebbing that is the cellular hallmark of the HGPS phenotype. To test the effects of inhibiting all prenylation, missense mutations of the cysteine residue of the CSIM motifs of WT LA and progerin were generated (designated SSIM). These constructs, marked for visualization by an N-terminal GFP fusion, were transfected into HeLa cells and imaged after 48 h. Without prenylation, all of the prelamin A was relocated from its normal location at the nuclear periphery into nucleoplasmic aggregates, and there was no progerin-induced nuclear blebbing (Fig. 2). Observations by confocal microscopy suggest that these nucleoplasmic aggregates are distributed throughout the nucleoplasm (data not shown). Mutating the CSIM terminal sequence to CSIL is predicted to favor geranylgeranylation rather than farnesylation (14) and, therefore, generates an FTI-resistant form of the protein. In this case, progerin-induced nuclear blebbing persisted (Fig. 2), indicating that the persistent modification of any prenoid group promotes the progerin phenotype. Similar results for all of these constructs were obtained with HEK 293 cells and NIH 3T3 cells (data not shown).
Fig. 2.
Subcellular localization and induction of nuclear blebbing by prenylation mutants of WT and progerin LA. Transient transfection of HeLa cells with expression vectors encoding the indicated WT or progerin LA protein. (a) Normal WT LA-CSIM CAAX motif. (b) WT LA with the CSIM sequence mutated to SSIM, relocalizing all LA into nucleoplasmic aggregates. (c) WT LA with the CSIM sequence mutated to CSIL, predicted to be geranylgeranylated, resulting in a mix of nucleoplasmic and nuclear membrane localization. (d) The progerin-CSIM construct recreates the hallmark nuclear blebbing seen in HGPS. (e) When the CSIM of the progerin-CSIM is mutated to SSIM (progerin-SSIM), blebbing is prevented and progerin accumulates in nucleoplasmic aggregates. (f) Progerin-CSIL causes blebbing that is indistinguishable from that of authentic progerin-CSIM. Cells were visualized 48 h after transfection, and photomicrographs were taken at ×60 magnification.
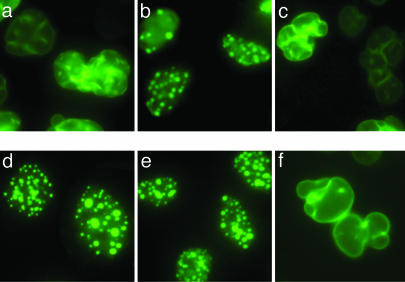
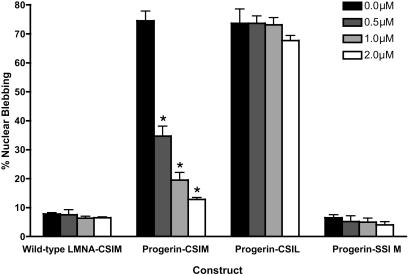
We then explored the effect of treating HeLa cells transiently transfected with WT LA-CSIM, progerin-CSIM, progerin-SSIM, and progerin-CSIL with the clinical candidate FTI lonafarnib (22). After a single dose of 0, 0.5, 1.0, or 2.0 μM at the time of transfection, cells were visualized 48 h later (Figs. 3 and 4). In the WT LA-CSIM-transfected cells, cell counting demonstrated that the percentage of nuclear blebbing was unchanged despite treatment, remaining at ≈5%, which is the typical blebbed percentage for normal WT cells (11). In contrast, the cells that were transfected with the progerin-CSIM constructs showed a dramatic dose-dependent response (P < 0.0001), with the nuclei approaching normal blebbing percentages with a single dose of 2.0 μM lonafarnib. The progerin-CSIL mutant, predicted to be geranylgeranylated, was completely resistant to treatment with the FTI, which is evidence that inhibition of the farnesylation of progerin, and not of any other endogenous farnesylated protein, is responsible for the improvement in nuclear phenotype seen with FTI treatment. As expected, there was no difference seen with FTI treatment of the progerin-SSIM mutant, because prenylation had already been completely inhibited. Similar results for all of these constructs were obtained with HEK 293 and NIH 3T3 cells when the FTIs L-744832 or FTI-2153 were used in doses of 1.0-10 μM (data not shown).
Fig. 3.
FTI treatment prevents nuclear blebbing caused by farnesylated, but not geranylgeranylated, progerin. HeLa cells were transiently transfected with expression vectors encoding the progerin-CSIM, progerin-SSIM, or progerin-CSIL constructs and treated with either 0 (Upper) or 2 (Lower) μM FTI lonafarnib/SCH66336. (a and d) The progerin-CSIM construct recreates the hallmark nuclear blebbing seen in HGPS, whereas FTI treatment almost abolishes it and leads to relocalization of progerin into nucleoplasmic aggregates. (b and e) Unprocessed progerin-SSIM mimics the phenotype of FTI-treated cells expressing authentic progerin-CSIM, with all progerin relocalized into nucleoplasmic aggregates, and does not change significantly with treatment. (c and f) Progerin-CSIL, predicted to be geranylgeranylated, recreates nuclear blebbing but is resistant to FTI treatment. Cells were visualized 48 h after transfection and photomicrographs were taken at ×60 magnification.
Fig. 4.
The percentage of blebbed HeLa nuclei is shown when transiently transfected with one of the indicated four constructs and treated with a single dose of lonafarnib/SCH66336 at the time of transfection. Percentages were read by three independent observers, who scored 200 cells for each experiment without knowledge of the construct being used. Error bars show SEM, and the P values are for the comparison of the treated cells with the untreated control. WT LA-CSIM does not change significantly from the typical 5% nuclear blebbing of normal cells. The progerin-CSIM shows a dramatic dose-response. The progerin-CSIL, expected to be geranylgeranylated, is resistant to the inhibition of farnesylation. The progerin-SSIM, which cannot be farnesylated or geranylgeranylated, demonstrates virtually no blebbing. *, P < 0.001.
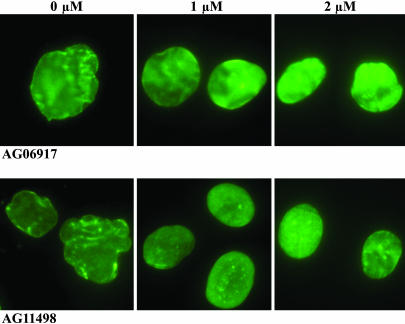
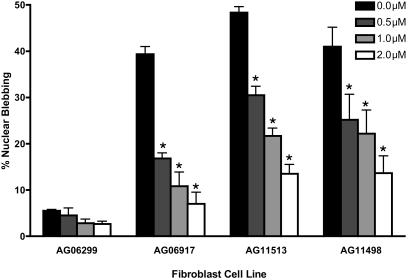
The next step was to examine the effects of FTIs on nuclear blebbing in cells from individuals with HGPS (in this case, dermal fibroblasts). Cultured fibroblasts from three HGPS patients and one unaffected mother were treated for 3 days with one daily dose of lonafarnib. To observe nuclear morphology at the end of treatment, nuclei were stained with an Ab for LA and LC (Figs. 5 and 6). On day 3, all three HGPS fibroblast lines demonstrated a significant dose-dependent reduction in nuclear blebbing that was unaffected by patient age or cell passage number (P < 0.0001).
Fig. 5.
FTI treatment causes reversion of the nuclear blebbing in two different progerin-expressing HGPS human fibroblasts. Cells were stained with anti-LA Ab after being treated for 3 days with 0, 1, or 2 μM FTI lonafarnib/SCH66336. (Original magnification ×60.)
Fig. 6.
The percentage of blebbed nuclei when HGPS skin fibroblasts were treated once daily for 3 days with the shown doses of lonafarnib. AG06299 is from the 34-year-old unaffected mother (passage 31) of an affected 6-year-old patient (AG06917; passage 14). AG11513 is from an affected 8-year-old patient (passage 7), and AG11498 is from an affected 14-year-old patient (passage 11). Percentages were read by three independent observers, who scored 200 cells for each experiment without knowledge of the genotype of the fibroblasts and the dose of the FTI being used. Error bars show SEM, and the P values are for the comparison of the treated cells with the untreated control. *, P < 0.001.
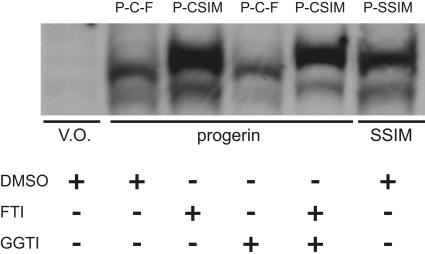
If treatment with FTIs is to be considered a viable option for HGPS, it is important to determine whether authentic progerin, like K-Ras and some other farnesylated proteins (23), could still be geranylgeranylated in the presence of FTIs and, therefore, resistant to FTI treatment. To explore this possibility, we performed a series of experiments examining the migration of progerin on a highly resolving gel when treated with an FTI, a GGTI, or a combination of both drugs. The appearance of a shift in mobility to a slower-migrating form is a standard method of demonstrating the ability of FTIs to prevent the processing of farnesylated proteins. As expected (Fig. 7), treatment with GGTI alone had no effect on the mobility of progerin, demonstrating that progerin is not normally geranylgeranylated. However, GGTI completely shifted the mobility of progerin-CSIL, whereas FTI had no effect (data not shown), confirming that this mutant was processed as expected. Importantly, progerin treated with either an FTI or FTI plus GGTI migrated identically with vehicle-treated, completely unprocessed progerin-SSIM mutant, demonstrating that the processing of progerin can be completely inhibited by FTI treatment alone and that progerin is not alternatively geranylgeranylated when farnesylation is inhibited.
Fig. 7.
FTI treatment alone is sufficient to impair progerin processing. NIH 3T3 cells transiently expressing empty vector (V.O.), GFP-tagged progerin-CSIM (progerin), or GFP-tagged progerin-SSIM (SSIM) were treated for 48 h with vehicle (DMSO) or inhibitors of prenylation (5 μM FTI-2153 or GGTI-2166). Total cell lysates were resolved by SDS/PAGE and immunoblotted with anti-GFP to detect progerin-CSIM or the fully unprocessed mutant progerin-SSIM. Treatment with FTI, but not GGTI, induces a mobility shift of progerin-CSIM to that of the unprocessed progerin-SSIM, demonstrating that progerin is an FTI target that does not undergo alternative prenylation. The labels at the top of each lane indicate the molecular entity that constitutes the major species in each experiment. P, progerin sequence up to CAAX motif; C, cysteine; F, farnesyl group. The lower bands in the progerin and SSIM lanes are presumed to be breakdown products.
Discussion
Since its description by Jonathan Hutchinson in 1886 (24), HGPS has received little attention from scientific researchers because of its extreme rarity (1 in 4 million live births). However, after the discovery of the genetic mutation behind the disease in 2003 (4, 5), there has been a resurgence of interest in the study of this dramatic premature aging disease and its potential links to both normal aging and atherosclerotic disease. Here, we present evidence that FTIs may serve as a potential therapy for this devastating condition.
Farnesylation, a posttranslational modification involving the addition of a 15-carbon isoprene moiety, was implicated as a potential anticancer target when it was discovered that the oncoprotein Ras, which has been estimated to be involved in up to 30% of all human cancers (25), required farnesylation for its function (26). Since then, a vigorous research effort in the pharmaceutical industry has identified and developed a number of small-molecule compounds that potently and selectively inhibit FTase (20). Two of these drugs (lonafarnib/SCH66336 from Schering-Plough, Kenilworth, NJ, and tipifarnib/R115777 from Johnson & Johnson, New Brunswick, NJ) have entered phase III trials and have been well tolerated, including in trials involving children (27). Lonafarnib competes with protein substrates for binding to the FTase enzyme (28).
Similar to Ras, the LA precursor is also farnesylated, with farnesylation serving as a required step to insert prelamin A into the nuclear membrane as well as to allow for the two downstream cleavage steps which complete the processing of LA (13). With the knowledge that the single C-to-T base change seen in nearly all cases of HGPS created a cryptic splice site and, thus, deleted the normal second endoproteolytic cleavage site in the LA-processing pathway, it was hypothesized that progerin was forced to retain its farnesyl group and, therefore, could not dissociate itself from the nuclear membrane. With other members of the nuclear lamina also potentially becoming trapped in complexes with the mislocalized progerin, a mechanistic connection between this permanently farnesylated state and the striking nuclear blebbing and disrupted nuclear architecture seen in HGPS cells was proposed, and the possibility of preventing or reversing this phenotype through FTIs was raised (4).
We hypothesized that FTIs would reduce the dominant-negative effect of mature progerin on nuclear membrane structure. However, this pharmacologic effect would not be specific for progerin; FTIs would also decrease levels of WT LA, increase the levels of unfarnesylated prelamin A, and no doubt affect the posttranslational processing of dozens of other farnesylated proteins. Although total cellular LA would be expected to decrease, we hypothesized that the net effect could still be beneficial, because the abnormal splice that leads to progerin is produced rather inefficiently by the mutant allele, and thus, progerin is present in much lower quantities than WT LA. Also, only a small amount of mature LA is necessary for proper nuclear-envelope assembly (29). In support of this idea, clinical trials using FTIs demonstrate little toxicity, even when levels of unfarnesylated prelamin A are raised significantly (30). However, the absence of LA leads to serious cellular consequences and disease (31).
Also, we explored the possibility that progerin would be geranylgeranylated in the presence of FTIs, which might limit the usefulness of this approach. In the presence of FTIs, oncogenic K-Ras and N-Ras serve as alternative substrates for the related enzyme GGTase I, thus remaining biologically active and fully capable of signal transduction and malignant transformation (22, 32-34). Thus, although H-Ras does not undergo alternative prenylation when FTase activity is blocked, FTIs are not effective inhibitors of the two Ras isoforms most commonly mutationally activated in human tumors, compromising the effectiveness of FTIs in the treatment of cancer. Fortunately, the data reported here confirm that such alternative isoprenylation does not occur with progerin, the processing of which is inhibited fully by FTI alone.
Our results indicate that farnesylated progerin is responsible for the dysmorphic nuclear morphology seen in HGPS, because inhibition of farnesylation by both genetic mutation and pharmacological intervention prevents nuclear blebbing and redistributes all of the mutant progerin protein from the nuclear membrane into nucleoplasmic aggregates. Beyond the prevention of this nuclear phenotype in transfected cells caused by ectopically expressed progerin, the ability of lonafarnib to cause a significant reduction in the percentage of blebbed nuclei seen in HGPS fibroblasts, even at high passage numbers, is encouraging evidence of its therapeutic potential against endogenous progerin. Perhaps the effects of the removal of farnesylated progerin, whether they be through gene expression changes or through the actual mechanical stabilization of the nuclear scaffolding, would rescue the cells that are most prone to damage in HGPS. Recent work supports this idea by showing that by simply reducing levels of farnesylated prelamin A by 50%, the nuclear blebbing and progeria-like phenotype seen in Zmpste24-deficient mice could be eliminated (35).
However, it cannot be presumed that this improvement in the nuclear deformity seen in vitro in HGPS cells would necessarily translate into phenotypic improvement in HGPS patients. Indeed, nuclear blebbing might not be the best gauge of disease phenotype at the whole-body level in humans, because other LMNA mutations cause human disease without any effect on nuclear shape (36). Similarly, in tissues in which cells have already been lost or irreversibly damaged, FTI treatment might be unable to induce cellular repopulation. Nevertheless, the results reported here support aggressive pursuit of the potential of FTI treatment for HGPS and perhaps also for other laminopathies in which there is evidence of nuclear deformation.
Acknowledgments
We thank Schering-Plough for providing the drug lonafarnib/SCH66336; Saïd Sebti and Andy Hamilton for providing FTI-2153 and GGTI-2166; Thomas Glover and Michael Glynn for sharing their early data on the possible use of FTIs in the treatment of HGPS; Dina Faddah for help in analyzing the nuclear shape changes seen with FTI treatment; and Darryl Leja for his assistance in preparing the figures. This work was supported in part by National Cancer Institute National Cooperative Drug Discovery Grant U19 CA67771 (to C.J.D. and A.D.C.).
Author contributions: B.C.C., M.R.E., C.J.D., A.D.C., and F.S.C. designed research; B.C.C., M.R.E., J.P.M., and J.J.F. performed research; A.D.C. contributed new reagents/analytic tools; B.C.C., M.R.E., J.P.M., J.J.F., R.V., K.N.C., L.B.G., C.J.D., A.D.C., and F.S.C. analyzed data; and B.C.C., M.R.E., C.J.D., A.D.C., and F.S.C. wrote the paper.
Abbreviations: HGPS, Hutchinson-Gilford progeria syndrome; LA/C, lamin A/C; FTase, farnesyltransferase; FTI, FTase inhibitor; GGTase, geranylgeranyltransferase; GGTI, GGTase inhibitor.
References
- 1.DeBusk, F. L. (1972) J. Pediatr. 80, 697-724. [DOI] [PubMed] [Google Scholar]
- 2.Baker, P. B., Baba, N. & Boesel, C. P. (1981) Arch. Pathol. Lab. Med. 105, 384-386. [PubMed] [Google Scholar]
- 3.Sarkar, P. K. & Shinton, R. A. (2001) Postgrad. Med. J. 77, 312-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eriksson, M., Brown, W. T., Gordon, L. B., Glynn, M. W., Singer, J., Scott, L., Erdos, M. R., Robbins, C. M., Moses, T. Y., Berglund, P., et al. (2003) Nature 423, 293-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Sandre-Giovannoli, A., Bernard, R., Cau, P., Navarro, C., Amiel, J., Boccaccio, I., Lyonnet, S., Stewart, C. L., Munnich, A., Le Merrer, M., et al. (2003) Science 300, 2055. [DOI] [PubMed] [Google Scholar]
- 6.Goldman, R. D., Gruenbaum, Y., Moir, R. D., Shumaker, D. K. & Spann, T. P. (2002) Genes Dev. 16, 533-547. [DOI] [PubMed] [Google Scholar]
- 7.Gruenbaum, Y., Margalit, A., Goldman, R. D., Shumaker, D. K. & Wilson, K. L. (2005) Nat. Rev. Mol. Cell Biol. 6, 21-31. [DOI] [PubMed] [Google Scholar]
- 8.Mounkes, L., Kozlov, S., Burke, B. & Stewart, C. L. (2003) Curr. Opin. Genet. Dev. 13, 223-230. [DOI] [PubMed] [Google Scholar]
- 9.Navarro, C. L., De Sandre-Giovannoli, A., Bernard, R., Boccaccio, I., Boyer, A., Genevieve, D., Hadj-Rabia. S., Gaudy-Marqueste, C., Smitt, H. S., Vabres, P., et al. (2004) Hum. Mol. Genet. 13, 2493-2503. [DOI] [PubMed] [Google Scholar]
- 10.Rober, R. A., Weber, K. & Osborn, M. (1989) Development (Cambridge, U.K.) 105, 365-378. [DOI] [PubMed] [Google Scholar]
- 11.Goldman, R. D., Shumaker, D. K., Erdos, M. R., Eriksson, M., Goldman, A. E., Gordon, L. B., Gruenbaum, Y., Khuon, S., Mendez, M., Varga, R., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 8963-8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scaffidi, P. & Misteli, T. (2005) Nat. Med. 11, 440-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beck, L. A., Hosick, T. J. & Sinensky, M. (1990) J. Cell Biol. 110, 1489-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox, A. D. & Der, C. J. (2002) Curr. Opin. Pharmacol. 2, 388-393. [DOI] [PubMed] [Google Scholar]
- 15.Hennekes, H. & Nigg, E. A. (1994) J. Cell Sci. 107, 1019-1029. [DOI] [PubMed] [Google Scholar]
- 16.Bergo, M. O., Gavino, B., Ross, J., Schmidt, W. K., Hong, C., Kendall, L. V., Mohr, A., Meta, M., Genant, H., Jiang, Y., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 13049-13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai, Q., Choy, E., Chiu, V., Romano, J., Slivka, S. R., Stietz, S. A., Michaelis, S. & Phillips, M. R. (1998) J. Biol. Chem. 273, 15030-15034. [DOI] [PubMed] [Google Scholar]
- 18.Agarwal, A. K., Fryns, J. P., Auchus, R. J. & Garg, A. (2003) Hum. Mol. Genet. 12, 1995-2001. [DOI] [PubMed] [Google Scholar]
- 19.Pendas, A., M., Zhou, Z., Cadinanos, J., Freije, J. M., Wang, J., Hultenby, K., Astudillo, A., Wernerson, A., Rodriguez, F., Tryggvason, K., et al. (2002) Nat. Genet. 31, 94-99. [DOI] [PubMed] [Google Scholar]
- 20.Sebti, S. M. & Der, C. J. (2003) Nat. Rev. Cancer 3, 945-951. [DOI] [PubMed] [Google Scholar]
- 21.Armitage, P. (1955) Biometrics 11, 375-386. [Google Scholar]
- 22.Ganguly, A. K., Doll, R. J. & Girijavallabhan, V. M. (2001) Curr. Med. Chem. 8, 1419-1436. [DOI] [PubMed] [Google Scholar]
- 23.Zhang, F. L., Kirschmeier, P., Carr, D., James, L., Bond, R. W., Wang, L., Patton, R., Windsor, W. T., Syto, R., Zhang, R., et al. (1997) J. Biol. Chem. 272, 10232-10239. [DOI] [PubMed] [Google Scholar]
- 24.Hutchinson, J. (1886) Medicochir. Trans. 69, 473-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bos, J. L. (1989) Cancer Res. 50, 1352. [Google Scholar]
- 26.Reiss, Y., Goldstein, J. L., Seabra, M. C., Casey, P. J. & Brown, M. S. (1990) Cell 62, 81-88. [DOI] [PubMed] [Google Scholar]
- 27.Doll, R. J., Kirschmeier, P. & Bishop, W. R. (2004) Curr. Opin. Drug Discovery Dev. 7, 478-486. [PubMed] [Google Scholar]
- 28.Bishop, W. R., Bond, R., Petrin, J., Wang, L., Patton, R., Doll, R., Njoroge, G., Catino, J., Schwartz, J. & Windsor, W. (1995) J. Biol. Chem. 270, 30611-30618. [DOI] [PubMed] [Google Scholar]
- 29.Lourim, D. & Krohne, G. (1993) J. Cell Biol. 123, 501-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taveras, A. G., Kirschmeier, P. & Baum, C. M. (2003) Curr. Top. Med. Chem. 3, 1103-1114. [DOI] [PubMed] [Google Scholar]
- 31.Sullivan, T., Escalante-Alcade, D., Bhatt, H., Anver, M., Bhat, N., Nagashima, K., Stewart, C. L. & Burke, B. (1999) J. Cell Biol. 147, 913-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whyte, D. B., Kirschmeier, P., Hockenberry, T. N., Nunez-Oliva, I., James, L., Catino, J. J., Bishop, W. R. & Pai, J. K. (1997) J. Biol. Chem. 272, 14459-14464. [DOI] [PubMed] [Google Scholar]
- 33.Rowell, C. A., Kowalczyk, J. J., Lewis, M. D. & Garcia, A. M. (1997) J. Biol. Chem. 272, 14093-14097. [DOI] [PubMed] [Google Scholar]
- 34.James, G., Goldstein, J. L. & Brown, M. S. (1996) Proc. Natl. Acad. Sci. USA 93, 4454-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fong, L. G., Ng, J. K., Meta, M., Cote, N., Yang, S. H., Stewart, C. L., Sullivan, T., Burghardt, A., Majumdar, S., Reue, K., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 18111-18116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broers, J. L., Hutchison, C. J. & Ramaekers, F. C. (2004) J. Pathol. 204, 478-488. [DOI] [PubMed] [Google Scholar]