Abstract
Many proteins can adopt self-propagating β-sheet-rich structures, termed amyloid fibrils. The [URE3] and [PSI+] prions of Saccharomyces cerevisiae are infectious amyloid forms of the proteins Ure2p and Sup35p, respectively. Ure2p forms prions primarily as a result of its sequence composition, as versions of Ure2p with the prion domain amino acids shuffled are still able to form prions. Here we show that prion induction by both Ure2p and Ure2-21p, one of the scrambled versions of Ure2p, is clearly dependent on the length of the inducing fragment. For Ure2-21p, no single sequence is found in all of the inducing fragments, highlighting the sequence independence of prion formation. Furthermore, the sequence of the Sup35p prion domain can also be randomized without blocking prion formation. Indeed, a single shuffled sequence could give rise to several prion variants. These results suggest that [PSI+] formation is driven primarily by the amino acid composition of the Sup35p prion domain, and that the Sup35p oligopeptide repeats are not required for prion maintenance.
Keywords: [PSI+], amyloid, Sup35
Numerous human diseases are associated with the formation of amyloid fibrils, including Alzheimer's disease, Parkinson's disease, and the transmissible spongiform encephalopathies (TSEs). Of these, only the TSEs are infectious. [URE3] and [PSI+] are prion (infectious protein) forms of the Saccharomyces cerevisiae proteins Ure2p and Sup35p, respectively (1). In both cases, prion formation results from conversion of the active native protein into an inactive, self-propagating, infectious amyloid form (2-4). Therefore, yeast prions provide a useful model for examining the forces that drive amyloid fibril formation and that cause certain amyloids to be infectious.
Ure2p is involved in nitrogen catabolite repression (reviewed in ref. 5). Loss of Ure2p activity, either because of deletion of the URE2 gene or because of the presence of the [URE3] prion, allows cells to take up ureidosuccinate (USA), an intermediate in uracil biosynthesis (5). Sup35p is an essential protein involved in translation termination; depressed Sup35p activity results in suppression of certain nonsense mutations (6).
Ure2p and Sup35p are each composed of an N-terminal prion domain (PD) that is dispensable for function and a C-terminal functional domain (7-11). The Sup35p PD is composed of residues 1-114, whereas that of Ure2p is generally described as amino acids 1-89 or 1-65. For Ure2p, residues 90-354 constitute the functional domain. The Sup35p functional domain is composed of amino acids 254-685, whereas amino acids 115-253 constitute a highly charged region termed M. M is not required for prion formation or the translation termination activity of Sup35p, but its deletion destabilizes [PSI+] (12).
For Ure2p, randomizing the order of the amino acids in the PD does not prevent prion formation, suggesting that the amino acid composition of the PD, not the primary sequence, is the predominant feature that drives prion formation (13). The PDs of both Ure2p and Sup35p have unusually high glutamine (Q) and asparagine (N) content (48% for Ure2p-1-89 and 46% for Sup35p), and mutational studies of Ure2p and Sup35p have implicated these Q/N residues in prion propagation (11, 14).
Here we examined the minimum fragments required to induce prion formation by Ure2p and by Ure2-21p, a version of Ure2p in which the prion domain amino acids were scrambled (13). Overexpression of the PD of Ure2p increases the frequency of prion formation, presumably because the misfolding event that initiates prion formation is a random occurrence, and therefore increasing the pool of PDs increases the chances of this initial misfolding (1, 9). For wild-type Ure2p, we found that while length was an important factor in prion induction, sequence composition also played a significant role. For Ure2-21, length was the primary determinant of prion-inducing ability. Furthermore, there was no amino acid sequence of Ure2-21p that was necessary to induce prion formation.
In Sup35, the Q/N residues are particularly concentrated in the first 40 amino acids of the PD, whereas the remainder of the PD contains a series of imperfect oligopeptide repeats (PQG-GYQQYN) (15). It was proposed that while the first 40 amino acids of the PD drive Sup35p aggregation, the oligopeptide repeats are required for chaperone-dependent replication of the aggregates (16).
We constructed seven versions of the Sup35 PD in which the order of the amino acids in the PD was randomized while keeping the amino acid content constant. Two of these expressed poorly and were therefore not well suited for study. Surprisingly, four of the remaining five formed stable prions, and the fifth formed unstable prions. These results indicate that prion formation by Sup35p shows primary sequence independence similar to that of Ure2p, and that prion maintenance does not absolutely require the oligopeptide repeats.
Materials and Methods
Strains and Media. A complete strain list can be found in Table 2, which is published as supporting information on the PNAS web site. Standard yeast media and methods were as previously described (17), except that YPD contained 0.5% yeast extract instead of the standard 1%. Galactose/raffinose dropout medium contained 2% galactose and 1% raffinose. In all experiments, yeast were grown at 30°C.
Building URE2 and URE2-21 Induction Plasmids. For URE2, mutagenic oligonucleotides were used to amplify the desired URE2 fragments, installing start and stop codons in the desired locations (see Tables 3 and 4, which are published as supporting information on the PNAS web site, for oligonucleotides and plasmids, respectively). PCR products were digested with BamHI and XhoI and ligated into BamHI/XhoI-cut pH317 (18).
For N- and C-terminal deletions of the Ure2-21 PD, mutagenic oligonucleotides were used to amplify the desired fragments from pER98, installing start and stop codons in the desired locations. PCR products were digested with BamHI and PstI and ligated into BamHI/PstI-cut pER98 (13). For internal deletions, N- and C-terminal fragments were amplified in separate PCRs. Products of these reactions were then combined and reamplified with the outer oligonucleotides. PCR fragments were then cloned as above. All URE2- and URE2-21-inducing plasmids were confirmed by DNA sequencing.
SUP35 Prion Domain Randomization. Randomizations of Sup35p amino acids 3-114 were designed as previously described for URE2 (13), except that five overlapping oligonucleotides were used to code for each scrambled Sup35p (Table 5, which is published as supporting information on the PNAS web site). Overlapping oligonucleotides were combined and amplified by PCR, generating the scrambled PDs (see Table 5 for details). PCR products were cotransformed with AatII/HindIII-cut pJ526 (from Dan Masison, National Institutes of Health) into yeast strain 780-1D/pJ533 (ref. 19; from Dan Masison). Transformants were selected on SD-leu and replica plated to 5-fluoroorotic acid-containing medium to select for loss of pJ533. Plasmids expressing scrambled SUP35s were confirmed by DNA sequencing.
Plasmids for [PSI+] Induction Experiments. Scrambled Sup35 PDs were amplified by PCR (see Table 6, which is published as supporting information on the PNAS web site, for oligonucleotides) from yeast strains containing the full-length scrambled SUP35s, installing a stop codon. PCR products were digested with BamHI and PstI and inserted into BamHI/PstI-cut pKT24 (from Kim Taylor, NABI, Rockville, MD). Ligation products were transformed into Escherichia coli and analyzed by DNA sequencing.
Western Blots. Cells were grown overnight on YPAD, then diluted to OD600 = 0.1 and grown to OD600 = 0.5-0.6. Cells from 50 ml of culture were collected by centrifugation and resuspended in 600 μl of PBS (pH 7.4) supplemented with Complete Protease Inhibitor Tablets (Roche; 1 tablet per 10 ml). Cells were lysed in a mini bead beater (three times, 1 min each). Protein concentrations were determined using a Pierce BCA Protein Assay Kit. Extracts were fractionated by centrifugation at 16,000 × g for 30 min. Pellets were resuspended in a volume of buffer equivalent to the volume centrifuged. Ten micrograms of total lysate, along with an equal volume of supernatant and resuspended pellet, was separated electrophoretically on SDS/10% PAGE gels in Nu-PAGE Mops SDS Running Buffer (Invitrogen). Proteins were transferred to poly(vinylidene difluoride) membranes (Invitrogen) according to the supplier's instructions with an XCell II Blot Module (Invitrogen). Anti-Sup35p antiserum was a gift from Dan Masison. Alkaline phosphatase-conjugated anti-rabbit IgG (Promega) was used as secondary antibody. Bands were detected by CDP-Star Chemiluminescence Reagent (Perkin-Elmer) and exposure to x-ray film.
Results
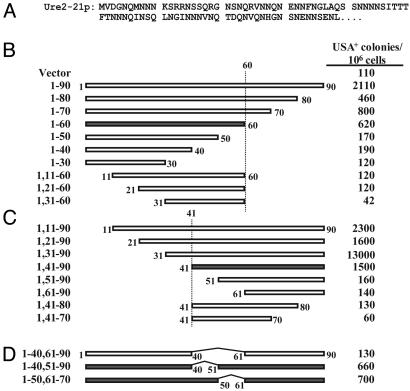
Mapping the URE2 Prion-Inducing Domain. [ure-o] cells (lacking the [URE3] prion) become USA+ (able to take up USA) at a frequency of about 2/105 cells (Fig. 1). Transiently overexpressing the Ure2 PD increases the frequency of prion formation (1). We transiently overexpressed a series of N- and C-terminal truncations of the Ure2 PD (Fig. 1) in cells bearing a full-length chromosomal copy of URE2, and determined the frequency of USA+ colony formation. The minimal N-terminal fragment that was sufficient to induce prion formation at levels similar to the full-length PD was Ure21-59 (Fig. 1B). For C-terminal fragments, Ure21, 18-65 induced very well (Fig. 1C). Although Ure21, 20-65 and Ure21, 25-65 both induced reasonably well, the intervening sequences Ure21, 21-65 and Ure21, 24-65 did not, suggesting that the specific residues found at the N terminus can affect induction (Fig. 1C). This could be either because the N-terminal residues have a direct effect on inducing ability or because they affect the expression level of the fragment. None of a series of C-terminal truncations of Ure21, 18-65 induced well, suggesting that the exact boundaries of the minimal prion domain are not fixed but are dependent on the length of the inducing fragment (Fig. 1D).
Fig. 1.
Induction of prion formation for wild-type Ure2p. (A) Amino acid sequence of Ure2 PD. (B-D) Plasmids containing a GAL1 promoter (vector; pH317) or expressing various fragments of Ure2p from a GAL1 promoter were introduced into strain YHE711, which contains a chromosomal copy of URE2. Strains were grown in galactose/raffinose dropout medium, and then serial dilutions were plated on USA medium to select for prion-containing cells. Colonies were counted after 5 days.
Mapping the URE2-21-Inducing Domains. URE2-21 encodes one of the five Ure2ps with a scrambled PD (Fig. 2A), all of which could form prions and amyloid (13). The minimal N-terminal fragment that was sufficient to induce prion formation by full-length URE2-21 was Ure2-211-60, and the minimal C-terminal fragment was Ure2-211, 41-90 (Fig. 2 B and C).
Fig. 2.
Induction of prion formation for Ure2-21p. (A) Amino acid sequence of Ure2-21 PD. (B-D) Plasmids containing a GAL1 promoter (vector; pKT24) or expressing various fragments of Ure2-21p from a GAL1 promoter were introduced into strain YER139, which contains a chromosomal copy of URE2-21. USA selection was conducted as in Fig. 1. The four shaded fragments share no sequence that is common to all of them.
Versions of URE2-21 with both C-terminal and N-terminal truncations showed that the N- and C-terminal boundaries of the induction domain were not fixed, but instead were dependent on the length of the fragment (Fig. 2 B and C). We started with Ure2-211-60, the minimal N-terminal fragment, and made a series of N-terminal deletions (Fig. 2B). Whereas the first 40 amino acids were dispensable for induction in the context of the full-length PD (Fig. 2C), not even 10 amino acids can be deleted from the N terminus of Ure2-211-60 without completely eliminating induction (Fig. 2B). Similarly, although deletion of the final 30 amino acids from the full-length PD reduces induction only ≈3-fold, not even 10 amino acids could be deleted from the C terminus of Ure2-211, 41-90 without eliminating induction. These results suggest that length may be the primary determinant of inducing ability, with fragments longer than ≈50-60 amino acids consistently inducing well and shorter fragments failing to induce.
Of all of the N- and C-terminal truncation mutants of Ure2-21p that were able to induce prion formation, the only region that is common to all of them is amino acids 40-60. When we deleted amino acids 40-60, the resulting fragment was unable to induce prion formation (Fig. 2D). However, we found that we could delete either amino acids 40-50 or 50-60 and still get induction. Therefore, the four fragments Ure2-211-60, Ure2-211, 41-90, Ure2-211-50, 61-90, and Ure2-211-41, 50-90 (shaded in Fig. 2) are all able to induce prion formation even though there is no portion of prion domain sequence common to all four. Thus no single segment of the Ure2-21 PD is absolutely required for induction.
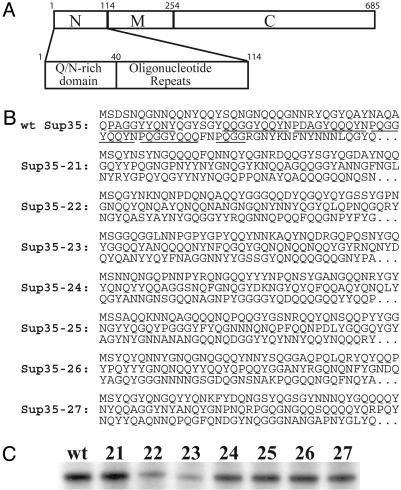
Randomizing the Order of the Amino Acids in the Sup35 PD. Amino acids 1-40 of the Sup35 PD are very Q/N-rich (55%) and are thought to drive aggregation of the protein, whereas amino acids 41-114 contain five and a half imperfect oligopeptide repeats that are thought to be important for prion maintenance (Fig. 3A and refs. 16 and 20-22). Similar repeats are found in the human prion protein (23), although not in Ure2p.
Fig. 3.
Scrambled Sup35 proteins. (A) Schematic of Sup35p, showing the PD, the M domain, and the functional C-terminal domain (C). The PD is enlarged below. (B) Amino acid sequences of scrambled and wild-type PDs. For each protein, amino acids 1-114 are shown, and the remainder of the protein is the same as wild-type Sup35p. Oligopeptide repeats are underlined in the wild-type Sup35p. (C) Western blots of expression levels of scrambled SUP35s in derivatives of 780-1D/pJ533 (wt) in which the plasmid expressing the wild-type SUP35 was replaced by plasmids expressing the scrambled versions.
We constructed seven versions of Sup35p, named Sup35-21, -22, -23, -24, -25, -26, and -27, in which the order of amino acids 3-114 was completely randomized while keeping the rest of the protein and the overall amino acid composition of the PD unchanged (Fig. 3B). Each of the scrambled Sup35ps could substitute for wild-type Sup35p. SUP35-22 and SUP35-23 were poorly expressed (Fig. 3C), allowing partial read-through of stop codons and making prion formation difficult to assay. The remaining five were all expressed at levels similar to the wild-type protein (Fig. 3C) and were studied further.
All Scrambled Sup35ps Form Ade+ Colonies. The active soluble [psi-] form of Sup35p and the inactive aggregated [PSI+] state can be differentiated by monitoring nonsense suppression of the mutant ade2-1 allele (24). ade2-1 mutants are unable to grow without adenine and form red colonies in the presence of limiting adenine because of accumulation of a pigment derived from the substrate of Ade2p. In cells containing the weak nonsense suppressor tRNA SUQ5 (SUP16), [PSI+] cells are able to grow in the absence of adenine and form white or pink colonies when grown on media containing limiting adenine.
Cells expressing each scrambled Sup35p and deleted for wild-type Sup35p were red when grown on limiting adenine (Fig. 4A). When these cells were plated on medium lacking adenine, only rare colonies were formed (Table 1). One hallmark of a prion is that increasing protein expression increases the frequency of prion formation (1). For each scrambled SUP35, transient overexpression significantly increased the frequency of Ade+ colony formation (Table 1), suggesting that they are forming prions.
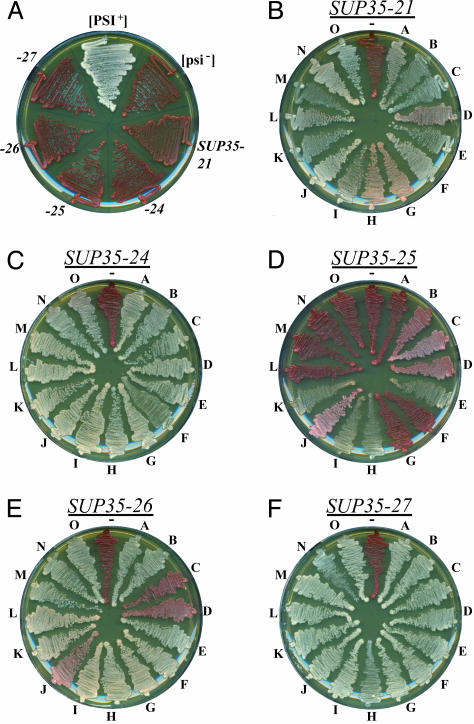
Fig. 4.
Stability of Ade+ phenotype. (A) Wild-type [PSI+] and [psi-] strains, and [psi-] strains expressing SUP35-21, -24, -25, -26, and -27 were streaked onto YPD. (B-F) For each scrambled SUP35, 15 Ade+ isolates (A-O) were streaked onto YPD, along with their respective parent Ade- strain.
Table 1. Ade+ induction and curing.
| Ade+ colonies/106 cells*
|
|||
|---|---|---|---|
| SUP35 allele | Uninduced | Induced | Curing† |
| SUP35-21 | 1.3 | 4.7 | 14/15 |
| SUP35-24 | 0.3 | 7.2 | 15/15 |
| SUP35-25 | 0.2 | 2.8 | 0/15 |
| SUP35-26 | 3.3 | 26 | 12/15 |
| SUP35-27 | 4.5 | 370 | 15/15 |
Strains expressing each full-length scrambled SUP35 from the SUP35 promoter were transformed with either plasmid pKT24 containing the GAL1 promoter (Uninduced) or with a derivative of pKT24 in which the PD of the same scrambled SUP35 was under control of the GAL1 promoter (Induced). Strains were grown for 4 days in galactose/raffinose-dropout medium. Serial dilutions were plated on medium lacking adenine, and colonies were counted after 6 days. Frequencies of Ade+ colony formation are the average of at least three independent experiments.
For each scrambled SUP35, 15 Ade+ colonies were streaked onto YPD medium with and without the addition of guanidine·HCI, grown to single colonies at 30°C, and then tested for loss of the Ade+ phenotype. Numbers represent the fraction of Ade+ isolates that maintained the Ade+ phenotype in the absence of guanidine and lost it in the presence of guanidine.
Sup35-21p, -24p, -26p, and -27p Form Stable Prions. For each scrambled SUP35, 15 Ade+ colonies were streaked onto medium containing limiting adenine. When this is done with spontaneous Ade+ isolates of cells expressing wild-type Sup35p, a range of colors from white to dark pink is observed, resulting from different variants of the prion (25). Each variant is thought to represent a different amyloid conformation (26, 27). For Sup35-21p, a similar range of white to pink colors was observed (Fig. 4B). Any color differences for Sup35-24p and -27p were much more subtle, with all of the cells either white or light pink (Fig. 4 C and F). Most of the Sup35-26p isolates were white or light pink, with three isolates a much darker pink (Fig. 4E). By contrast, the majority of the Ade+ colonies formed by cells expressing Sup35-25p were red or dark pink, indicating that the Ade+ phenotype was either weak or lost (Fig. 4D).
Growth of cells on media containing low concentrations of guanidine cures both [URE3] and [PSI+] (ref. 28; M. Aigle, cited in ref. 1) as a result of inactivation of Hsp104p (29-31), a chaperone required to break up prion aggregates and form new prion seeds (32-34). The Ade+ isolates from Fig. 4 B-F were grown on YPD media with and without the addition of guanidine. Single colonies were then restreaked on YPD and the color phenotype was observed. For all scrambled SUP35s except SUP35-25 (see below for discussion of SUP35-25), the majority of the Ade+ isolates became red after guanidine treatment, indicating loss of the Ade+ phenotype (Table 1 and Fig. 5A).
Fig. 5.
Curing, aggregation, and cytoduction of Ade+ isolates. (A) Ade+ isolates of strains expressing SUP35-21, -24, -26, and -27 were grown on YPD and YPD with 4 mM guanidine·HCl. Single colonies were taken from the YPD (+) and YPD plus guanidine (Gd) plates and restreaked onto YPD, along with the respective Ade- parent strain (-). (B) Cell lysates from Ade+ isolates and from their Ade- parent strain were separated by centrifugation into soluble (S) and pellet (P) fractions. Proteins in each fraction were separated by polyacrylamide gel electrophoresis and immunoblotted with antiserum specific for Sup35p. (C and D) Ade+ isolates were used as donors for cytoduction reactions. Cytoductions were performed as previously described (13). Recipient cells expressed the same scrambled Sup35p. Cytoductants (+), donors, and the precytoduction Ade- recipient (-) were then streaked onto YPD. For SUP35-21, four donors containing different [PSI+] strains were used to test whether strain variations were maintained through cytoduction.
The Sup35-21, -24, -26, and -27 protein in the curable Ade+ isolates was in all cases less soluble than in the respective isogenic Ade- strains (Fig. 5B), consistent with the Ade+ phenotype resulting from a prion. We tested these isolates for infectivity by using cytoduction (or cytoplasmic mixing). Cytoductions transfer cytoplasmic elements, but not nuclear genes (35). For each scrambled SUP35 except SUP35-25, four curable Ade+ isolates were used as cytoduction donors to recipient cells expressing the same scrambled SUP35. In all cases, recipient cells became Ade+ (Fig. 5 C and D), demonstrating that prion formation is responsible for the Ade+ phenotype. Furthermore, for SUP35-21, the only scrambled SUP35 where a clear range of Ade+ phenotypes was observed, recipient cells showed color phenotypes similar to those of their respective donors (Fig. 5C).
Sup35-25p Forms Unstable Prions. Sup35-25p had the lowest rate of Ade+ colony formation (Table 1), and only a few were stably Ade+ (Fig. 4D, sectors E, H, I, and K). None of the stable Ade+ colonies were able to transmit the Ade+ phenotype by cytoduction, indicating that the phenotype is not the result of a prion (data not shown).
Many of the other Sup35-25p clones had an unstable Ade+ phenotype, as indicated by a mixture of red and pink cells when streaked on nonselective media (Fig. 4D, sectors B, C, D, and J; see Fig. 6A for enlargement). When maintained on media lacking adenine, these cells retain the ability to grow without adenine, but prolonged growth on nonselective media led to complete loss of the Ade+ phenotype. Furthermore, when these cells were grown without adenine, the distribution of Sup35p shifted such that less was soluble (Fig. 6B). This shift was subtle, as most of the Sup35-25p is insoluble even under nonselective conditions.
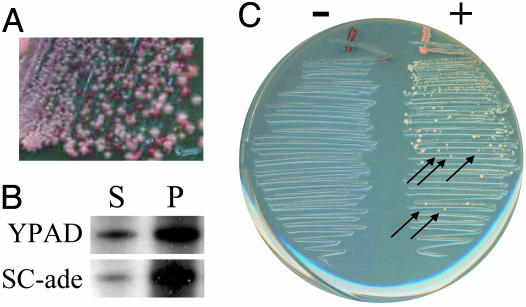
Fig. 6.
Analysis of Sup35-25p. (A) SUP35-25 Ade+ isolate J was streaked from SC-ade onto YPD. (B) Ade+ isolates of cells expressing SUP35-25 were pregrown for 48 h on YPAD or SC-ade. Cells were then grown for two to three generations in YPAD. Cell lysates were separated by centrifugation into soluble (S) and pellet (P) fractions and analyzed by Western blotting. (C) Ade+ isolates of cells expressing SUP35-25 pregrown for 48 h on either YPAD (-) or SC-ade (+) were used as cytoduction donors to recipient cells expressing SUP35-25. Cytoductions were streaked on medium selecting for Ade+ recipient cells. Colonies were tested by replica plating to distinguish cytoductants (indicated with arrows) from diploids.
These results suggested that Sup35-25p may form unstable prions, as has been observed previously with the Sup35-Δ22/69 mutant of Sup35p (36) and with the Ure2-22p scrambled version of Ure2p (13). We thus performed cytoduction experiments under conditions that select for the prion, in a manner similar to ref. 36. Ade+ cells were grown for 48 h on either YPAD or SC-ade and then used as cytoduction donors. Matings were performed as in normal cytoductions, but mixtures were then streaked directly onto medium selecting for recipient cells that had gained the Ade+ phenotype. Ade+ colonies were observed when donor cells had been constantly maintained on medium lacking adenine, but not for cells maintained without selection (Fig. 6C). Many of the colonies were cytoductants (arrows in Fig. 6C), demonstrating that the Ade+ phenotype is the result of an unstable prion.
Discussion
We previously demonstrated that the order of the amino acids in the Ure2 PD can be randomized without abolishing the prion-forming ability of the protein, highlighting the importance of sequence composition in driving prion formation. Here we examined the minimal sequence requirements for prion induction for both wild-type Ure2p and the scrambled version Ure2-21p.
If the amino acid composition of Ure2p were the primary factor driving prion formation, the ability of a fragment of the PD to induce prion formation should be primarily dependent on the length and composition of the fragment. For the wild-type PD, the amino acids that drive prion formation might be concentrated into some portion of the PD; therefore, fragments in this region should induce particularly well. By contrast, after randomization of amino acid order, these amino acids would presumably be spread out roughly uniformly throughout the PD; hence, for Ure2-21p, we would expect that induction should depend primarily on the length of the inducing fragment.
For wild-type Ure2p, a combination of length and sequence composition determines inducing ability. Amino acids 59-65, which are expendable in the context of the full-length PD, are necessary for induction in the context of the N-terminally truncated fragment Ure21, 18-65. This observation clearly suggests some sort of length requirement. Whether the requirement is some minimum absolute length or is something more specific such as a minimum number of asparagines is unclear. However, length is not the only factor determining inducing ability, because significantly more can be removed from the N terminus than the C terminus of the PD without eliminating induction. This difference is perhaps not surprising because glutamines and asparagines have previously been shown to be important for prion formation by both Ure2p and Sup35p (11, 14), and amino acids 44-65 of Ure2p are very asparagine rich.
As for wild-type Ure2p, a clear length dependence was observed for Ure2-21p, with specific regions being dispensable for induction in the context of the full PD but required in the context of truncated PDs. Again, the length dependence was not absolute, because 40 amino acids could be removed from the N terminus of the PD without preventing induction, whereas only 30 could be removed from the C terminus.
Negative results in induction experiments are difficult to interpret for two reasons. First, the likely variable expression of different fragments could result in poor induction by poorly expressed fragments. Second, an inducing fragment may form a prion whose structure is incompatible with the full-length protein, a sort of species barrier. Such a problem is particularly likely for internal deletions such as Ure2-211-40, 60-90. Consequently, it is more informative to focus on the fragments that were able to induce. Specifically, the four fragments Ure2-211-60, Ure2-211, 41-90, Ure2-211-50, 60-90, and Ure2-211-40, 50-90 were all able to induce prion formation despite having no portion of the PD sequence common to all four, proving that there is no single region of the PD that is absolutely required for induction.
To examine whether other amyloid-forming proteins might show a degree of primary sequence independence similar to that of Ure2p, we randomized the order of amino acids in the Sup35 PD and tested the ability of the resulting proteins to form prions. That all of the expressed scrambled Sup35ps could still form prions is quite surprising. A number of single point mutations have been found to block [PSI+] induction or propagation (14, 37). Such mutations should have only a modest impact on the overall PD composition, and therefore would suggest that specific primary features are important.
How can single point mutations block prion propagation whereas complete sequence scrambling has such modest effects? Part of the answer may lie in the experimental details. Previous experiments looking for mutations that block [PSI+] generally start with a [PSI+] strain expressing wild-type Sup35p, then put in a plasmid expressing mutant Sup35p and look for loss of the Ade+ phenotype. Such experiments are really testing for a species barrier: the inability of a given mutant to propagate a specific variant of [PSI+]. Such mutants may be capable of forming prions de novo, but simply unable to propagate the specific prion present in the cell, as has been shown for one such mutant (38). By contrast, our experiments are examining the ability of each sequence to form prions de novo. When plasmids expressing the scrambled Sup35ps were put into strains with wild-type [PSI+], the Ade+ phenotype was lost (data not shown).
A second, more subtle, explanation is also possible. Presumably there is some set of criteria that an amyloid structure must meet to be stable, such as the need to place charged residues and prolines either on turns or outside the core amyloid structure. If the requirement for a stable prion were rather general such as, for example, a Q/N-rich domain containing at least one stretch of >20 residues without any prolines or charged residues, then most scrambled versions of the Sup35 PD (with 9.6% charged/proline residues) would be able to form prions. However, in most cases a single properly placed charged residue could disrupt this ability.
Nevertheless, the ability of scrambled Sup35ps to form prions is surprising based on the considerable evidence suggesting that the oligopeptide repeats of Sup35p are important for [PSI+] stability. Deletion of the repeats impairs [PSI+] propagation (8), and insertion of additional repeats increases the frequency of [PSI+] (21). It has been proposed that the oligopeptide repeats may act as chaperone binding sites, allowing aggregates to be broken up to make new seeds (16). This proposal is based on the observations that deletion of one or more of the repeats prevents stable [PSI+] propagation without preventing incorporation into preexisting Sup35p aggregates (16, 20) and that addition of oligopeptide repeats to expanded polyQ tracts allows them to behave as prions (16).
Our results suggest that the oligopeptide repeats per se are not required for prion maintenance. It is appealing to think that the repeats are important because similar repeats are found in mammalian PrP (prion protein). However, none of the experiments discussed above ever replaced one or more of the repeats with a sequence of similar composition and length that lacked repeats. Therefore, it is possible that the presence of the repeats was coincidental, and that this region is important solely for its length and composition.
Numerous other Q/N-rich proteins exist in yeast (39). If amino acid composition is the primary determinant of prion-forming ability, why have more of these not been shown to be prions? The most obvious answer is that although considerable evidence points to the importance of glutamines and asparagines, little is known about the contributions of other amino acids. This may explain why the oligopeptide repeat region is not sufficient for aggregation despite its length and high Q/N content (16); other sequence composition features such as the high number of prolines may preclude aggregation. It is also striking that for both Ure2p and Sup35p, the PDs are at one extreme end of the protein sequence and relatively unstructured (40, 41), making the PDs easily accessible for prion formation. Some Q/N-rich proteins may also have domains that would be capable of forming prions in isolation but are prevented from doing so when in the context of a stably folded protein. Finally, some of the Q/N-rich proteins may have the intrinsic ability to form prions, but issues such as cellular location or expression timing or levels may prevent prion formation.
Although all of the expressed scrambled Sup35ps formed prions, they displayed different characteristics. They showed differing frequencies of [PSI+] formation (Table 1), [PSI+] stabilities, ranges of [PSI+] variants (Fig. 4), and solubility (Figs. 5B and 6B), in both the prion and nonprion state. This observation clearly suggests that while sequence composition is an important factor in determining prion-forming ability, the details of primary sequence also play an important role.
Explaining the minor impact of scrambling on prion formation is difficult because the molecular structure of Sup35p amyloid is not known. Recent papers have suggested either a parallel in-register β-sheet (42, 47) or a head-to-head and tail-to-tail β-helix (43), whereas peptides from certain other amyloid-forming proteins adopt an antiparallel β-sheet structure (44, 45). In an antiparallel β-sheet or β-helix, most contacts along the fibril axis are between nonidentical (presumably complementary) residues. Therefore, scrambling should disrupt most such interactions. By contrast, in a parallel in-register β-sheet, each residue in a peptide interacts along the fiber axis with the identical residue in the next peptide; such interactions should be maintained after sequence scrambling, thereby lessening the impact of scrambling on fibril stability (46). Thus, maintaining the ability to be a prion and form amyloid after scrambling is suggestive of parallel β-sheet structure.
Supplementary Material
Acknowledgments
M.J.T. was a member of the Howard Hughes Medical Institute-National Institutes of Health Research Scholar Program. This research was supported by the intramural program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
Author contributions: E.D.R., H.K.E., M.J.T., and R.B.W. designed research; E.D.R., H.K.E., and M.J.T. performed research; E.D.R., H.K.E., M.J.T., and R.B.W. analyzed data; and E.D.R., H.K.E., and R.B.W. wrote the paper.
Abbreviations: PD, prion domain; USA, ureidosuccinate.
References
- 1.Wickner, R. B. (1994) Science 264, 566-569. [DOI] [PubMed] [Google Scholar]
- 2.King, C. Y., Tittmann, P., Gross, H., Gebert, R., Aebi, M. & Wüthrich, K. (1997) Proc. Natl. Acad. Sci. USA 94, 6618-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glover, J. R., Kowal, A. S., Schirmer, E. C., Patino, M. M., Liu, J. J. & Lindquist, S. (1997) Cell 89, 811-819. [DOI] [PubMed] [Google Scholar]
- 4.Taylor, K. L., Cheng, N., Williams, R. W., Steven, A. C. & Wickner, R. B. (1999) Science 283, 1339-1343. [DOI] [PubMed] [Google Scholar]
- 5.Cooper, T. G. (2002) FEMS Microbiol. Rev. 26, 223-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kushnirov, V. V. & Ter-Avanesyan, M. D. (1998) Cell 94, 13-16. [DOI] [PubMed] [Google Scholar]
- 7.Liebman, S. W. & Derkatch, I. L. (1999) J. Biol. Chem. 274, 1181-1184. [DOI] [PubMed] [Google Scholar]
- 8.Ter-Avanesyan, M. D., Dagkesamanskaya, A. R., Kushnirov, V. V. & Smirnov, V. N. (1994) Genetics 137, 671-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masison, D. C. & Wickner, R. B. (1995) Science 270, 93-95. [DOI] [PubMed] [Google Scholar]
- 10.Ter-Avanesyan, M. D., Kushnirov, V. V., Dagkesamanskaya, A. R., Didichenko, S. A., Chernoff, Y. O., Inge-Vechtomov, S. G. & Smirnov, V. N. (1993) Mol. Microbiol. 7, 683-692. [DOI] [PubMed] [Google Scholar]
- 11.Maddelein, M. L. & Wickner, R. B. (1999) Mol. Cell. Biol. 19, 4516-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu, J. J., Sondheimer, N. & Lindquist, S. L. (2002) Proc. Natl. Acad. Sci. USA 99, Suppl. 4, 16446-16453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross, E. D., Baxa, U. & Wickner, R. B. (2004) Mol. Cell. Biol. 24, 7206-7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DePace, A. H., Santoso, A., Hillner, P. & Weissman, J. S. (1998) Cell 93, 1241-1252. [DOI] [PubMed] [Google Scholar]
- 15.Kushnirov, V. V., Ter-Avanesyan, M. D., Telckov, M. V., Surguchov, A. P., Smirnov, V. N. & Inge-Vechtomov, S. G. (1988) Gene 66, 45-54. [DOI] [PubMed] [Google Scholar]
- 16.Osherovich, L. Z., Cox, B. S., Tuite, M. F. & Weissman, J. S. (2004) PLoS Biol. 2, E86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherman, F. (1991) Methods Enzymol. 194, 3-21. [DOI] [PubMed] [Google Scholar]
- 18.Edskes, H. K. & Wickner, R. B. (2000) Proc. Natl. Acad. Sci. USA 97, 6625-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song, Y., Wu, Y. X., Jung, G., Tutar, Y., Eisenberg, E., Greene, L. E. & Masison, D. C. (2005) Eukaryot. Cell 4, 289-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parham, S. N., Resende, C. G. & Tuite, M. F. (2001) EMBO J. 20, 2111-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, J. J. & Lindquist, S. (1999) Nature 400, 573-576. [DOI] [PubMed] [Google Scholar]
- 22.Crist, C. G., Nakayashiki, T., Kurahashi, H. & Nakamura, Y. (2003) Genes Cells 8, 603-618. [DOI] [PubMed] [Google Scholar]
- 23.Kretzschmar, H. A., Stowring, L. E., Westaway, D., Stubblebine, W. H., Prusiner, S. B. & Dearmond, S. J. (1986) DNA 5, 315-324. [DOI] [PubMed] [Google Scholar]
- 24.Cox, B. S. (1965) Heredity 26, 211-232. [DOI] [PubMed] [Google Scholar]
- 25.Derkatch, I. L., Chernoff, Y. O., Kushnirov, V. V., Inge-Vechtomov, S. G. & Liebman, S. W. (1996) Genetics 144, 1375-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bessen, R. A. & Marsh, R. F. (1994) J. Virol. 68, 7859-7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chien, P., Weissman, J. S. & DePace, A. H. (2004) Annu. Rev. Biochem. 73, 617-656. [DOI] [PubMed] [Google Scholar]
- 28.Tuite, M. F., Mundy, C. R. & Cox, B. S. (1981) Genetics 98, 691-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferreira, P. C., Ness, F., Edwards, S. R., Cox, B. S. & Tuite, M. F. (2001) Mol. Microbiol. 40, 1357-1369. [DOI] [PubMed] [Google Scholar]
- 30.Jung, G. & Masison, D. C. (2001) Curr. Microbiol. 43, 7-10. [DOI] [PubMed] [Google Scholar]
- 31.Jung, G., Jones, G. & Masison, D. C. (2002) Proc. Natl. Acad. Sci. USA 99, 9936-9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paushkin, S. V., Kushnirov, V. V., Smirnov, V. N. & Ter-Avanesyan, M. D. (1996) EMBO J. 15, 3127-3134. [PMC free article] [PubMed] [Google Scholar]
- 33.Ness, F., Ferreira, P., Cox, B. S. & Tuite, M. F. (2002) Mol. Cell. Biol. 22, 5593-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wegrzyn, R. D., Bapat, K., Newnam, G. P., Zink, A. D. & Chernoff, Y. O. (2001) Mol. Cell. Biol. 21, 4656-4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conde, J. & Fink, G. R. (1976) Proc. Natl. Acad. Sci. USA 73, 3651-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borchsenius, A. S., Wegrzyn, R. D., Newnam, G. P., Inge-Vechtomov, S. G. & Chernoff, Y. O. (2001) EMBO J. 20, 6683-6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doel, S. M., McCready, S. J., Nierras, C. R. & Cox, B. S. (1994) Genetics 137, 659-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kochneva-Pervukhova, N. V., Poznyakovski, A. I., Smirnov, V. N. & Ter-Avanesyan, M. D. (1998) Curr. Genet. 34, 146-151. [DOI] [PubMed] [Google Scholar]
- 39.Michelitsch, M. D. & Weissman, J. S. (2000) Proc. Natl. Acad. Sci. USA 97, 11910-11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pierce, M. M., Baxa, U., Steven, A. C., Bax, A. & Wickner, R. B. (2005) Biochemistry 44, 321-328. [DOI] [PubMed] [Google Scholar]
- 41.Scheibel, T. & Lindquist, S. L. (2001) Nat. Struct. Biol. 8, 958-962. [DOI] [PubMed] [Google Scholar]
- 42.Nelson, R., Sawaya, M. R., Balbirnie, M., Madsen, A. O., Riekel, C., Grothe, R. & Eisenberg, D. (2005) Nature 435, 773-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krishnan, R. & Lindquist, S. L. (2005) Nature 435, 765-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Makin, O. S., Atkins, E., Sikorski, P., Johansson, J. & Serpell, L. C. (2005) Proc. Natl. Acad. Sci. USA 102, 315-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balbach, J. J., Ishii, Y., Antzutkin, O. N., Leapman, R. D., Rizzo, N. W., Dyda, F., Reed, J. & Tycko, R. (2000) Biochemistry 39, 13748-13759. [DOI] [PubMed] [Google Scholar]
- 46.Ross, E. D., Minton, A. & Wickner, R. B. (2005) Nat. Cell Biol., in press. [DOI] [PubMed]
- 47.Chan, J. C., Oyler, N. A., Yau. W. M. & Tycko, R. (2005) Biochemistry 44, 10669-10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.