Abstract
T-cell receptor diversity enables the cellular immune response to recognize a broad range of viral and other pathogenic agents. An increasingly common method of characterizing T-cell receptor diversity and usage in response to antigenic challenges involves the identification of clonal expansions by PCR amplification of the CDR3 region of distinct TCRVβ families. Though clonal expansions often appear evident upon visual inspection of the results, a systematic method is needed for the valid enumeration of these expansions. Here, we describe a novel analysis method, termed the MaGiK method, for systematically identifying and enumerating clonal T-cell expansions and for applying the results to investigations of the T-cell receptor repertoire.
The antigen-specific cellular immune response of humans is mediated through T-cell receptor (TCR) recognition of foreign peptides presented by human leukocyte antigens (HLA) (12). The TCR expressed on the vast majority of T cells is a heterodimer consisting of an α- and a β-polypeptide chain (11). The TCRβ chain consists of variable (V), joining, diversity, and constant (C) regions encoded by gene segments spanning 685 kilobases of chromosome 7 (10). Diversity and antigen specificity of T-cell antigen recognition is primarily attributed to the hypervariable or third complementarity-determining region (CDR3) of the TCR (2). The extensive diversity of the CDR3 results from the occurrence of random nucleotide additions or deletions during TCR gene rearrangements (8). This variation is observable as multiple lengths of PCR amplicons, each set separated by 3 base pairs, arising from single TCRVβ and Cβ primers. X-ray crystallography has revealed that the CDR3 of the TCR physically contacts HLA-presented peptides (4). Thus, the CDR3 plays an important role in determining the antigen specificity of the T cell.
Antigen-specific responses give rise to oligoclonal populations within the peripheral T-cell compartment (1). These responses are observable as clonal expansions of T-cell subsets with distinct TCRVβ CDR3 lengths. Thus, differences in the frequency of T-cell clones' pre- and postantigenic stimulation can be measured by analyzing CDR3 distributions, giving the investigator a measure of the diversity of the T-cell response.
Measuring the TCR repertoire has been achieved by two methods, flow cytometry and spectratyping by PCR. Each method possesses its own advantages and disadvantages. Absolute quantitation of the frequency of individual members of the T-cell repertoire is easily achieved with the use of flow cytometry. Flow cytometry also allows quantitation of the abundance of specific TCRs expressed on the cell surface. The greatest limitation of flow cytometry for the use of T-cell repertoire comparisons is the lack of sensitivity of the monoclonal antibodies that identify TCR families. For example, biologically significant clonal expansions may comprise a relatively small percentage of a TCR family and may not measurably increase the percentage of the entire family. Other limitations include the incomplete repertoire coverage (∼60% [unpublished data]) of commercially available monoclonal antibodies, as well as a requirement for a relatively large number of cells (approximately 4 million peripheral blood mononuclear cells [PBMC]). Our observations indicate that gross differences in the extent of clonality between individuals are not revealed by flow cytometric analysis of the T-cell repertoire (unpublished data). Alternatively, spectratyping by PCR provides the ability for greater resolution given a fewer number of cells. However, absolute quantitation following reverse transcription (RT)- PCR is not currently achievable. At best, RT-PCR-based methods are semiquantitative despite the intricate statistical methods that must be applied to the data analysis.
The identification, quantitation, and comparison of CDR3 profiles provides valuable information relating to the T-cell immune response. For example, cross-sectional analyses can be used to compare the T-cell repertoires of groups with differing risks of disease or longitudinal analyses can be applied to studies of vaccine effectiveness. Regardless of the study design, each investigation must identify and enumerate clonal expansions in a manner that can be applied to cross-sectional or longitudinal comparisons. Ideally, absolute values of the quantity of each CDR3 length would be used for comparison. However, even with the use of internal quantitation controls, uncontrollable RT-PCR variation negates the use of absolute CDR3 length magnitudes. While accounting for this variation greatly complicates the methods, analysis of CDR3 profiles remains approachable.
Here, we present a novel statistical analysis method, the MaGiK (Matud, Giorgi, Killian) method, and describe its use in the analysis of the CD4 TCR repertoire of healthy individuals. As both the diversity and the magnitude of the T-cell response to many diseases factor into prognoses, this method offers both clinical and diagnostic value.
MATERIALS AND METHODS
Donors.
Thirty-five healthy individuals were selected from University of California—Los Angeles (UCLA) Blood Bank donors, human immunodeficiency virus type 1 (HIV-1)-seronegative participants in the Multicenter AIDS Cohort Study (a study of men who have sex with men), and UCLA nonlaboratory workers (3, 7). Thirty-one of 35 individuals studied were men, approximately half of whom were homosexual men. The group's mean age and CD4/CD8 ratio were 39 years (range, 30 to 50 years) and 1.7 (range, 0.9 to 4.9), respectively. Note that this group of donors is presented here for demonstration of the MaGiK method only and is not necessarily representative of broader populations of healthy individuals.
CDR3 length analyses using RT-PCR.
PBMC were isolated by gradient separation and then cryopreserved. Positive selection for CD4+ T lymphocytes was performed by incubating thawed PBMC with anti-CD4 immunomagnetic beads (Dynal, Great Neck, N.Y.) for 30 min at 4°C on a rotating shaker, as recommended by the manufacturer. The average purity of separated CD4+ cells was >99%. CDR3 length assays were performed as previously described (6). Briefly, RNA was extracted from cell suspensions in TriReagent (Sigma, St. Louis, Mo.) and reverse transcribed using Moloney murine leukemia virus reverse transcriptase and a TCRβ chain C region primer (CTCAGCTCCAGTG). Specific combinations of one, two, or three TCRVβ-specific forward primers (0.1 μM each) were then used for each multiplex PCR on the resulting cDNA. Each reaction also contained a fluorescently labeled (with 6-Fam, Tet, or Hex) reverse primer (0.1 μM) specific for the TCRβ C region. PCR primers and reaction tubes are described in Table 1. The nucleotide lengths of the fluorescently labeled CDR3 amplicons were measured by a model 310 Genetic Analyzer (Applied Biosystems, Foster City, Calif.). The model 310 Genetic Analyzer is a capillary-based sequencer and genetic fragment analyzer that can simultaneously detect the fluorescence intensity of four separate colors (three PCR product colors and one size-standard color). Genetic fragment quantity, e.g., discrete CDR3 lengths, is determined from the fluorescence intensity of the fragment and the nucleotide length of the fragment. Fluorescence data was collected and processed using Genescan and Genotyper software packages (Applied Biosystems). PCRs and CDR3 measurements were performed in duplicate.
TABLE 1.
Description of primer combinations used for the TCRβ multiplex PCRsa
| PCR tube(s) | Primer(s) | Sequence(s) | Cβ fluorescent label |
|---|---|---|---|
| 1-12 | Cβ | CTTCTGATGGCTCAAACAC | 6-Fam, Tet, or Hex |
| 1 | Vβ01 | CAACAGTTCCCTGACTTGCAC | 6-Fam |
| Vβ18 | GAGTCAGGAATGCCAAAGGAA | ||
| 2 | Vβ02 | TCAACCATGCAAGCCTGACCT | Tet |
| Vβ04 | CATATGAGAGTGGATTTGTCATT | ||
| Vβ08 | TACTTTAACAACAACGTTCCG | ||
| 3 | Vβ03 | TCTAGAGAGAAGAAGGAGCGC | Hex |
| Vβ13.1 | GACCAAGGAGAAGTCCCCAAT | ||
| 4 | Vβ05.1 | TTCAGTGAGACACAGAGAAAC | 6-Fam |
| Vβ05.2 | CCTAACTATAGCTCTGAGCTG | ||
| 5 | Vβ06 | AGGCCTGAGGGATCCGTCTC | Tet |
| Vβ24 | AAAGATTTTAACAATGAAGCAGAC | ||
| 6 | Vβ07 | CTGAATGCCCCAACAGCTCTC | Hex |
| Vβ22 | CAGAGAAGTCTGAAATATTCGA | ||
| 7 | Vβ09 | AAATCTCCAGACAAAGCTCAC | 6-Fam |
| Vβ16 | GAGTCTAAACAGGATGAGTCC | ||
| 8 | Vβ11 | ACAGTCTCCAGAATAAGGACG | Tet |
| Vβ12 | GACAAAGGAGAAGTCTCAGAT | ||
| 9 | Vβ13.2 | GTTGGTGAGGGTACAACTGCC | Hex |
| Vβ15 | GTCTCTCGACAGGCACAGGCT | ||
| 10 | Vβ14 | TCTCGAAAAGAGAAGAGGAAT | 6-Fam |
| Vβ17 | CACAGATAGTAAATGACTTTCAG | ||
| Vβ20 | TCTGAGGTGCCCCAGAATCTC | ||
| 11 | Vβ21 | GATATGAGAATGAGGAAGCAG | Tet |
| 12 | Vβ23 | TCATTTCGTTTTATGAAAAGATGC | Hex |
Twelve PCR reaction tubes, containing varying TCRVβ-specific primers, were created for each sample. Each PCR tube contained one of three fluorescently labeled TCRCβ reverse primers. This feature reduced the analysis time by allowing the PCR products to be combined into four tubes prior to spectratyping.
Statistical analysis.
Clonal T-cell expansions were identified by using a novel statistical approach to analyzing CDR3 distributions that we termed MaGiK analysis. The analysis is demonstrated in Fig. 1 and Table 2. Expansions were defined as adjusted proportions whose values exceeded the mean control CD4+ CDR3 length value by at least 3 standard deviations. The mean CD4+ CDR3 value for each length in each TCRVβ family is referred to as the control value in this text. The control values were derived from the CDR3 fragment distributions of 35 donors. For each donor and T-cell subset, a ratio of the number of expansions per number of CDR3 lengths measured (the expansion ratio [ER]) was computed. The ER is the summary measure derived by the MaGiK method of CDR3 analysis. The ER is a useful summary measure of the relative proportion of the number of expansions present in the T-cell repertoire, especially when the total number of CDR3 lengths measured varies across samples. The ER provides an easily interpreted measure of the percentage of expanded CDR3 lengths in the T-cell repertoire.
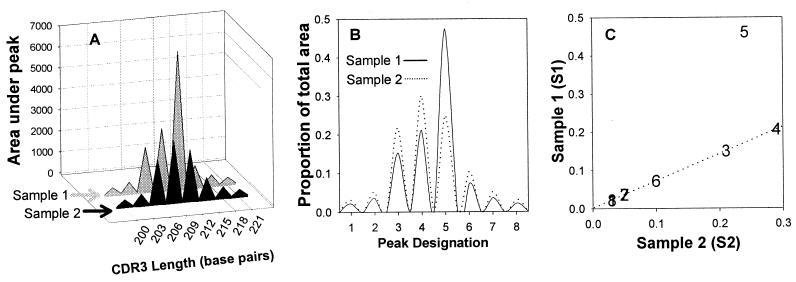
FIG. 1.
The MaGiK method of TCR repertoire comparison. CDR3 lengths for a TCRVβ family between hypothetical samples 1 and 2 (S1 and S2, respectively) are compared. S1 represents a TCRβ family with a clonal expansion. S2 represents an idealized TCRβ family with CDR3 length magnitudes averaged from several controls with no expansions. (A) The area (ai) under each peak (i) was calculated. (B) Each peak's proportional representation (pi = ai/Σai) was computed. For each peak, a sample proportion ratio [PRi = S1(pi)/S2(pi)] was computed. The median of the ratios (Rm) from all peaks in a family was then calculated. S1 proportions were then multiplied by the median PR [aS1(pi) = S1(pi) × medianPR] to standardize their distributions. (C) Expansions were identified among the S1 peaks whose adjusted proportions aS1(pi) were greater than 3 standard deviations above the corresponding S2 peak's proportion.
TABLE 2.
Data used to generate Fig. 1
| Amplicon length (base pairs) | Peak (i) | Area
|
Proportion
|
Proportion ratio (PRi) | Adjusted proportions
|
|||
|---|---|---|---|---|---|---|---|---|
| Sample 2 S2(ai) | Sample 1 S1(ai) | Sample 2 C(pi) | Sample 1 S(pi) | Sample 2 C(pi) | Sample 1 aS(pi) | |||
| 200 | 1 | 300 | 300 | 0.030 | 0.021 | 0.71 | 0.030 | 0.030 |
| 203 | 2 | 500 | 500 | 0.050 | 0.035 | 0.71 | 0.050 | 0.050 |
| 206 | 3 | 2,100 | 2,100 | 0.210 | 0.148 | 0.71 | 0.210 | 0.210 |
| 209 | 4 | 2,900 | 2,900 | 0.290 | 0.205 | 0.71 | 0.290 | 0.290 |
| 212 | 5 | 2,400 | 6,500 | 0.240 | 0.461 | 1.92 | 0.240 | 0.649 |
| 215 | 6 | 1,000 | 1,000 | 0.100 | 0.070 | 0.71 | 0.100 | 0.100 |
| 218 | 7 | 500 | 500 | 0.050 | 0.035 | 0.71 | 0.050 | 0.050 |
| 221 | 8 | 300 | 300 | 0.030 | 0.021 | 0.71 | 0.030 | 0.030 |
RESULTS AND DISCUSSION
TCR repertoire in CD4+ T cells.
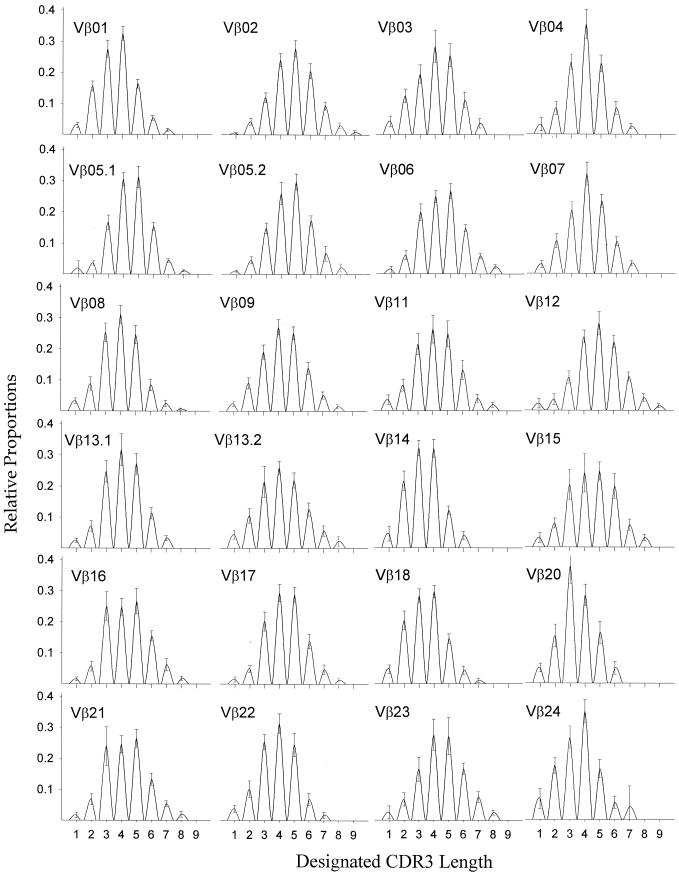
We investigated CD4+ TCR diversity in 35 healthy individuals. Means and standard deviations for the CDR3 distributions of each of the 24 TCRVβ families among the 35 individuals are shown in Fig. 2. The average ratio of expansions per number of CDR3 lengths measured (ER) was 0.01 for CD4+ T cells. Consistent with previously reported findings (5), an approximately Gaussian distribution of CDR3 lengths was observed for each individual TCRVβ family.
FIG. 2.
Spectratypes for 24 TCRVβ families, showing the mean values derived from the CD4+ T cells of 35 healthy subjects. Relative proportions (y axis) are plotted for each discrete CDR3 length (x axis). Standard deviations for each CDR3 length are indicated by bars.
Several methods have been used to quantitate and compare CDR3 profiles (5, 9). The simplest method is to visually identify expansions as CDR3 lengths whose magnitudes appear increased in comparison to an expected Gaussian approximation. Naturally, visual identification introduces subjective variation and bias and therefore is not a truly quantitative analysis. Also, visual identification of expansions when more than one peak is expanded becomes increasingly difficult and nonintuitive. Another applied method is the comparison of proportions of the magnitudes of each CDR3 length. Typically, differences are computed between the proportions. However, this approach is flawed as it ignores the interdependence of the proportions, i.e., an expansion in one peak increases its proportion and simultaneously decreases the proportions of all other peaks within a given TCR family.
Figure 1A presents hypothetical data for two samples for which the areas under each peak were identical except for the peak centered at 212 base pairs, or peak 5. Visually, an expansion is evident at peak 5. Notice that even though only one peak was expanded in this example, the individual CDR3 length proportions of the total TCRVβ family for the two samples were all different. In this example, it is readily apparent that directly computing differences between the proportions of the two samples would lead to the incorrect conclusion that all of the peaks were different in magnitude. Therefore, to correctly compare the samples, we calculated each peak's proportional representation in a given family, as shown in Fig. 1B.
An outline of the MaGiK T-cell repertoire analysis method is provided in Table 3. MaGiK analysis begins with the assumption that major expansions are present in fewer than 50% of the peaks within a given family. This assumption is required when absolute magnitude values cannot be ascertained, since expansions must then be determined based on the relative distribution of CDR3 lengths. The large number of differing antigens required for greater than 50% of the peaks to be expanded lends credibility to this assumption. Another assumption is that deletions do not significantly affect CDR3 length distributions. Deletions of entire CDR3 lengths are improbable events, since each CDR3 length consists of numerous T-cell variants. Genetic heterogeneity of α- and β-chain gene segments gives rise to T-cell variants, as does other genetic and phenotypic heterogeneity among cells sharing a common CDR3 length and TCRβ variable segment.
TABLE 3.
Outline of the MaGiK method of TCR repertoire analysis
| T-cell source | Spectratyping steps |
|---|---|
| Controls | 1. Perform CDR3 spectratyping on the CD4+-T-cell subsets of ≥10 healthy controls. Fetal thymocytes or cord blood also make excellent controls but frequently are more difficult to obtain. Spectratyping on a capillary-based instrument such as the ABI model 310 Genetic Analyzer yields reproducible fluorescence intensity and fragment length data. |
| 2. Import the spectratyping data into a database application such as Microsoft Access. | |
| 3. For each Vβ family, identify the discrete nucleotide ranges that comprise each CDR3 length. | |
| 4. Compute the mean and standard deviation for each CDR3 length. | |
| Test samples | 1. Perform CDR3 spectratyping on the samples. |
| 2. Import the spectratyping data into a database application such as Microsoft Access. | |
| 3. For each CDR3 length in a sample, compute a sample-to-control ratio by dividing the sample value by the control mean. | |
| 4. Next, for each Vβ family, calculate the median sample-to-control ratio. | |
| 5. Multiply each CDR3 length proportion in each Vβ family by its respective median sample-to-control ratio. This achieves a standardization of sample values based on a common control set. | |
| 6. Establish and apply a criterion that defines clonal expansions. Values that exceed the control means by more than 3 standard deviations are statistically significant (P < 0.05). Biological significance may vary depending on the context, thus necessitating adjustment of the expansion definition in some instances. |
CDR3 fragment analysis must address an issue common to quantitative PCR: varying template amounts. MaGiK analysis, like other reported methods, deals with this issue by comparing CDR3 lengths as proportions of the total area for a given TCRVβ family. Importantly, unlike other methods, MaGiK analysis deals with the interdependence of proportions and allows for the objective identification of individual expansions while accounting for the net effect of expansions in a TCR family and simultaneously provides an estimate of the overall degree of TCR repertoire perturbation based on the total number of observed expansions. Also, the ER is a biologically intuitive value as opposed to other derived statistics. The ER, a measure of the observed number of expansions per total number of CDR3 lengths measured, is useful when results are not obtained for equal numbers of CDR3 lengths and/or TCR subfamilies among all individuals. This is frequently the case when the number of cells available is limited or when the repertoire is extremely skewed. The MaGiK analysis method is also applicable to CDR3 assays using PCR primers differing from ours. It may be extended as well to investigation of CD8+-T-cell and B-cell repertoires.
The MaGiK method of analysis offers a systematic means of assessing the degree of clonality, and thus antigen-specific responses, within the T-cell repertoire. By definition, an expanded population of T cells is one that is increased in frequency above the expected value. We have elected to use the CD4+-T-cell repertoire, averaged from the data from 35 healthy donors, as a reference that has been observed to contain very few expansions. Adjusted sample CDR3 lengths that exceed the control value are by definition expanded.
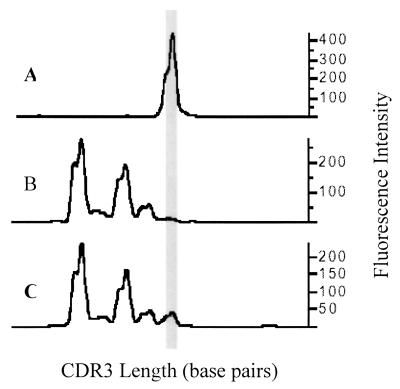
Direct sequencing of CDR3 PCR fragments has been used to demonstrate the presence of clonal expansions. For instance, if a given CDR3 fragment comprises 50% or more of the total PCR product for a given TCRVβ family, direct sequencing of the total PCR product for that TCRVβ family reveals a cleanly readable sequence 95% of the time (6). However, the ability to obtain a cleanly readable sequence might, in some cases, be sufficient to indicate the presence of clonal expansions though it is not a necessary requirement. Direct sequencing only reveals very large clonal expansions or clonal dominance. As depicted in Fig. 3, clonal expansions of T cells may or may not be of sufficient magnitude to result in clonal dominance.
FIG. 3.
Identifying diminutive clonal expansions. We identified a clonal expansion of HIV-specific CD8+ T cells (TCRVβ21+) by using HLA class I (A0201) tetramers complexed with a Gag peptide (SLYNTVATL). The CDR3 length distributions of three fluorescence-activated cell-sorted T-cell subsets are shown. (A) CDR3 length distribution for the tetramer positive subset; (B and C) tetramer-negative and total CD8+-T-cell subsets, respectively. The gray vertical bar aligns the expanded CDR3 length across the panels.
Furthermore, we have observed the occurrence of a biologically significant clonal expansion directed against an HLA-A201-restricted HIV-1 epitope (SLYNTVATL) that does not comprise 50% of a TCRVβ family. Using flow cytometry and HLA class I tetramer/HIV-1 peptide complexes, we have observed multiple T-cell clones that comprise the response to a dominant epitope presented by the tetramer. None of the nine different TCRVβ clones detected in three individuals comprised 50% of the corresponding Vβ family (unpublished data). These HIV-specific clones (those that were clearly expansions because we did not detect them in HIV-seronegative patients) would likely not be identified by direct sequencing. However, these expansions were revealed by MaGiK analysis.
Analysis of the diversity in the CDR3 of TCR families is a valuable method for investigating the cellular immune response. Comparing TCR diversity between persons with opposed infection risks and prognoses may provide valuable immunologic insight. However, it is imperative that such comparisons are not flawed by invalid analysis methods.
We have presented MaGiK analysis as an analytical method for comparing TCR repertoires. The method functions through the computation of basic statistics used to compare CDR3 distributions of T-cell repertoires. Analysis of the T-cell repertoire is clinically useful for identifying infections and determining prognoses. In addition, this method has important application in vaccine development and efficacy assessment, as it may be applied to quantitation of the magnitude and diversity of the antigen-specific T-cell response.
Acknowledgments
This work was supported by NIH grants AI-35040, AI-36704, and AI-37613.
The research presented here included data collected on participants in the Multicenter AIDS Cohort Study (MACS), which includes the following sites and investigators: from the Johns Hopkins University School of Hygiene and Public Health, Baltimore, Md., J. B. Margolick (principal investigator), H. Armenian, B. Crain, A. Dobs, H. Farzadegan, N. Kass, S. Lai, Justin McArthur, S. Strathdee, and E. Taylor; from the Howard Brown Health Center and Northwestern University Medical School, Chicago, Ill., J. P. Phair (principal investigator), J. S. Chmiel, B. Cohen, M. O'Gorman, D. Variakojis, and S. M. Wolinsky; from the University of California UCLA Schools of Public Health and Medicine, Los Angeles, R. Detels and B. Jamieson (principal investigators), B. R. Visscher (coprincipal investigator), A. Butch, J. Fahey, O. Martínez-Maza, E. N. Miller, J. Oishi, P. Satz, E. Singer, H. Vinters, O. Yang, and S. Young; from the University of Pittsburgh Graduate School of Public Health, Pittsburgh, Pa., C. R. Rinaldo (principal investigator), L. Kingsley (coprincipal investigator), J. T. Becker, P. Gupta, J. Mellors, S. Riddler, and A. Silvestre; from the Data Coordinating Center at the Johns Hopkins University School of Hygiene and Public Health, Baltimore, Md., A. Muñoz (principal investigator), L. P. Jacobson (coprincipal investigator), L. Ahdieh, S. Cole, S. Gange, C. Kleeberger, S. Piantadosi, E. Smit, S. Su, and P. Tarwater; and from NIH, Bethesda, Md., the National Institute of Allergy and Infectious Diseases, C. Williams and P. Miotti, and the National Cancer Institute, S. Melnick.
REFERENCES
- 1.Currier, J. R., H. Deulofeut, K. S. Barron, P. J. Kehn, and M. A. Robinson. 1996. Mitogens, superantigens, and nominal antigens elicit distinctive patterns of TCRB CDR3 diversity. Hum. Immunol. 48:39-51. [DOI] [PubMed] [Google Scholar]
- 2.Davis, M. M., and P. J. Bjorkman. 1988. T-cell antigen receptor genes and T-cell recognition. Nature 334:395-402. [DOI] [PubMed] [Google Scholar]
- 3.Detels, R., P. English, B. R. Visscher, L. Jacobson, L. A. Kingsley, J. S. Chmiel, J. P. Dudley, L. J. Eldred, and H. M. Ginzburg. 1989. Seroconversion, sexual activity, and condom use among 2915 HIV seronegative men followed for up to 2 years. J. Acquir. Immune Defic. Syndr. 2:77-83. [PubMed] [Google Scholar]
- 4.Garcia, K. C., M. Degano, R. L. Stanfield, A. Brunmark, M. R. Jackson, P. A. Peterson, L. Teyton, and I. A. Wilson. 1996. An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science 274:209-219. [DOI] [PubMed] [Google Scholar]
- 5.Gorochov, G., A. U. Neumann, A. Kereveur, C. Parizot, T. Li, C. Katlama, M. Karmochkine, G. Raguin, B. Autran, and P. Debre. 1998. Perturbation of CD4+ and CD8+ T-cell repertoires during progression to AIDS and regulation of the CD4+ repertoire during antiviral therapy. Nat. Med. 4:215-221. [DOI] [PubMed] [Google Scholar]
- 6.Gregersen, P. K., R. Hingorani, and J. Monteiro. 1995. Oligoclonality in the CD8+ T-cell population: analysis using a multiplex PCR assay for CDR3 length. Ann. New York Acad. Sci. 756:19-27. [DOI] [PubMed] [Google Scholar]
- 7.Kaslow, R. A., D. G. Ostrow, R. Detels, J. P. Phair, B. F. Polk, and C. R. Rinaldo, Jr. 1987. The multicenter AIDS cohort study: rationale, organization, and selected characteristics of the participants. Am. J. Epidemiol. 126:310-318. [DOI] [PubMed] [Google Scholar]
- 8.Komori, T., A. Okada, V. Stewart, and F. W. Alt. 1993. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science 261:1171-1175. [DOI] [PubMed] [Google Scholar]
- 9.Oksenberg, J. R. 1997. The antigen T cell receptor: selected protocols and applications. R.G. Landes, New York, N.Y.
- 10.Rowen, L., B. F. Koop, and L. Hood. 1996. The complete 685-kilobase DNA sequence of the human beta T cell receptor locus. Science 272:1755-1762. [DOI] [PubMed] [Google Scholar]
- 11.Wilson, R. K., E. Lai, P. Concannon, R. K. Barth, and L. E. Hood. 1988. Structure, organization and polymorphism of murine and human T-cell receptor alpha and beta chain gene families. Immunol. Rev. 101:149-172. [DOI] [PubMed] [Google Scholar]
- 12.Zinkernagel, R. M., and P. C. Doherty. 1974. Restriction of in vitro T cell-mediated cytotoxicity in lymphocytic choriomeningitis within a syngeneic or semiallogeneic system. Nature 248:701-702. [DOI] [PubMed] [Google Scholar]