Abstract
Facultatively symbiotic corals provide important experimental models to explore the establishment, maintenance, and breakdown of the mutualism between corals and members of the algal family Symbiodiniaceae. Here, we report the de novo chromosome-scale genome assembly and annotation of the facultatively symbiotic, temperate coral Astrangia poculata. Though widespread segmental/tandem duplications of genomic regions were detected, we did not find strong evidence of a whole-genome duplication event. Comparison of the gene arrangement between As. poculata and the tropical coral Acropora millepora revealed considerable conserved colinearity despite ∼415 million years of divergence. Gene families related to sperm hyperactivation and innate immunity, including lectins, were found to contain more genes in Ac. millepora relative to As. poculata. Sperm hyperactivation in Ac. millepora is expected given the extreme requirements of gamete competition during mass spawning events in tropical corals, while lectins are important in the establishment of coral–algal symbiosis. By contrast, gene families involved in sleep promotion, feeding suppression, and circadian sleep/wake cycle processes were expanded in As. poculata. These expanded gene families may play a role in As. poculata's ability to enter a dormancy-like state (winter quiescence) to survive freezing temperatures at the northern edges of the species' range.
Keywords: chromosome-level genome assembly, corals, scleractinian, facultative symbiosis, evolution, gene family expansion, whole-genome duplication, dormancy, mass spawning
Introduction
Anthozoa, the largest class within the phylum Cnidaria, includes some of the most ecologically important and oldest clades of marine metazoans, estimated to have evolved as early as 771 million years ago (Mya) (McFadden et al. 2021). Among these are corals, a diverse group of solitary and colonial organisms that can form a symbiotic association with algae of the family Symbiodiniaceae in the shallow water of the tropic and temperate zones (LaJeunesse et al. 2018). Stony corals of the order Scleractinia contain the engineers of reef ecosystems and are generally divided into 2 major clades, Complexa and Robusta, which diverged ∼415 Mya (Romano and Palumbi 1996; Kitahara et al. 2010; Stolarski et al. 2011). Over the past several decades, tropical coral species have undergone mass mortality due to their sensitivity to bleaching in the face of anthropogenic climate change (Bellwood et al. 2004; DeCarlo et al. 2017; Hughes et al. 2017). Understanding differences between species of variable temperature tolerance and adaptive strategies has become a focus for conservation efforts. In the “robust” clade, the temperate coral Astrangia poculata (the Northern Star Coral) has recently emerged as a model system for these comparisons due to its facultative symbiosis and temperature tolerance ranging from near freezing to 24°C in Narragansett Bay, the northern part of its distribution (Jacques et al. 1983).
Astrangia poculata, like many other shallow water corals, hosts a photosynthesizing algal symbiont, Breviolum psygmophilum (Lajeunesse et al. 2012). The symbiosis is facultative, with a gradient of symbiont density existing among individual polyps within a single colony of As. poculata and between sympatric colonies (Dimond and Carrington 2007) as revealed by an aposymbiotic “white” appearance, nearly or entirely devoid of symbionts, or a symbiotic “brown” appearance (Fig. 1a). Unlike many other shallow water coral species, As. poculata occurs across a broad temperature and latitudinal gradient from southern Massachusetts to the Gulf of Mexico (Cummings 1983; Peters et al. 1988; Dimond and Carrington 2007; Dimond et al 2013). At the northernmost parts of its range, during winter months high in the intertidal zone, As. poculata experiences a quiescence characterized by a dormancy state of reduced feeding and growth (Supplementary Fig. 1; Jacques et al. 1983; Grace 2017). This appears to be an adaptation of As. poculata to the intertidal zone, where it is vulnerable to desiccation, predation, and extreme shifts in salinity and temperature. Astrangia poculata's hardiness combined with the ability of researchers to experimentally isolate the contributions of the host and the symbiont in aposymbiotic and symbiotic colonies has made this species particularly interesting for the study of coral symbiosis in the face of climate change.
Fig. 1.
Photograph of As. poculata and phylogenetic tree. a) Underwater photograph of As. poculata colony with extended tentacles exhibiting aposymbiotic (white appearance, left) and symbiotic (brown appearance, right) states. Photograph credit to Sean P. Grace. b) Phylogeny of cnidarians included in the comparative genomic analyses of this study. Scleractinian coral clades are highlighted (top box, yellow = Complexa; bottom box, blue = Robusta). Astrangia poculata and Ac. millepora are highlighted in red. The species tree was inferred using the STAG algorithm and rooted using the STRIDE algorithm. Branch lengths represent the number of substitutions per site.
Previous work found signatures of adaptation in the thermal tolerance of As. poculata across the species' range, with a cold-adapted population exhibiting higher metabolic rates than warm-adapted populations across several temperature treatments (Aichelman et al. 2019). Symbiotic and aposymbiotic As. poculata respond differently to warm and cold temperatures at both the organismal and transcriptomic levels. Symbiotic colonies significantly outperformed aposymbiotic colonies at 9, 18, and 24°C with respect to wound healing (Burmester et al. 2017), while aposymbiotic colonies responded more strongly transcriptionally to a cold exposure than to a heat exposure treatment, particularly through upregulation of genes involved in the myosin complex, proteasome core, translation regulator activity, nucleic acid binding, extracellular matrix structural constituent, muscle system process, and proteolysis (Wuitchik et al. 2021). In a separate thermal stress experiment, the algal endosymbiont B. psygmophilum responded more strongly than the As. poculata host to chronic heat stress (Chan et al. 2021). Further, season, rather than symbiont state, was shown to drive the structure of the microbiome in As. poculata (Sharp et al. 2017). Thus, across multiple scales of measurement, thermal variation, particularly exposure to cold temperatures, appears to play an important role in the biology and ecology of this coral. However, the evolutionary roots and the genomic mechanisms driving the response to environmental change remain unclear for As. poculata, and for many other corals.
The use of comparative genomics has shed light on the evolution of basal metazoans. For example, studies have suggested the potential roles of whole-genome duplication (WGD) (Mao and Satoh 2019), horizontal gene transfer (Bhattacharya et al. 2016), and de novo biosynthesis pathways (Ying et al. 2018) in coral evolutionary trajectories. Many of these comparisons have focused on complex vs robust lineages, for which there exists a deep evolutionary split based on molecular and phylogenetic evidence (Fig. 1b; Romano and Palumbi 1996; Kitahara et al. 2010; Stolarski et al. 2011). However, it is not well understood how phylogenetically widespread these genomic traits may be, and the inclusion of other corals representing a wider array of ecological niches and adaptive strategies is warranted, which requires additional genomic resources.
While over the past several years there has been an increase in the availability of genomic resources for cnidarians, currently few of them are at chromosome-scale (however, see Fuller et al. 2020; Hu et al. 2020; Stephens et al. 2022; Locatelli et al. 2024) and none represent a facultatively symbiotic, temperate coral. To fill this gap, we present here the chromosome-scale assembly of a male As. poculata colony. The assembly is among the most contiguous and complete of available coral genome assemblies to date. We characterized the structure and content of the As. poculata genome to investigate potential genomic drivers underlying this species' unique temperature tolerance and flexible symbiotic state. To determine the sex of the sequenced colony, we examined histological sections from the colony. Our aims were to (1) produce a high-quality reference genome for As. poculata, (2) use this genome assembly to explore several potential genomic mechanisms that may contribute to the unique plasticity of the species, and (3) characterize the degree of similarity of the gene repertoire and genome organization among As. poculata and other corals.
Methods
Genome sequencing
An aposymbiotic colony was collected from Fort Wetherill State Park in Jamestown, Rhode Island, USA in October of 2017. To avoid sequencing the symbiont, a colony with a minimal density of B. psygmophilum in its tissue was selected based on the colony's white appearance. A subsample of this colony was collected, frozen, and sent for DNA extraction, library preparation, and sequencing at Dovetail Genomics (Scotts Valley, CA). Two subsamples from this colony were maintained in aquaria at the Pennsylvania State University, 2 at Boston University, and 2 at the University of Rhode Island for future use.
Sequencing involved a multistrategy approach and included 2 Illumina (San Diego, CA) datasets of paired-end 150 base-pair (bp) reads: one with 414 million reads and an estimated average insert size of 395 bp, and the second with 235 million reads and an estimated average insert size of 484 bp. Three Hi-C libraries were produced with 198 million, 266 million, and 257 million of 150 bp paired-end reads each. DNA extraction, library preparation, and sequencing of the Illumina and Hi-C data were carried out by Dovetail Genomics (https://dovetailgenomics.com/). All Illumina and Hi-C reads were trimmed using Cutadapt v2.9 with default settings (Martin 2011).
In preparation for Oxford Nanopore (Oxford, UK) sequencing, extracted genomic DNA was purified with AMPure XP Beads to improve the purity. Short fragments were discarded using Circulomics Short Reads Eliminator XS (Circulomics, Pacific Biosciences). The library was prepared using the Nanopore Ligation Sequencing Kit LSK109, starting with 2.1 μg of DNA, and yielded 1.4 μg of library. The final library was sequenced with a MinION on an R9.4 flow cell with fast base calling (Jain et al. 2016). The flow cell was washed and reloaded 3 times (281 ng of DNA for the first load, 187 ng for subsequent loads). A total output of 6.79 Gigabases (Gb) was obtained with an N50 of 18 kilobases (kb) and an N90 of 5 kb. The reads were trimmed of the adaptors with Porechop (available at https://github.com/rrwick/Porechop), using default parameters. After trimming, the dataset reached 6.77 Gb.
De novo genome assembly
Several assembly strategies were compared to select the most robust approach (Supplementary Table 1). Assemblers that were investigated include Raven (Vaser and Šikić 2019), Flye (Kolmogorov et al. 2019), Canu (Koren et al. 2017), and wtdbg2 (Ruan and Li 2020). Ultimately, the genome was assembled using wtdbg2 v2.5 under default settings. Haplotigs were purged using purge_haplotigs v1.1.1 (Roach et al. 2018) with default settings [following the recommendations in Guiglielmoni et al. (2021)] with both Illumina datasets mapped using bowtie2 v2.3.5.1 (Langmead and Salzberg 2012). Polishing was done using HyPo v1.0.3 (Kundu et al. 2019). Hi-C reads were mapped to the assembly and processed using bowtie2 v2.3.5.1 and hicstuff v2.3.0 (Matthey-Doret et al. 2020) with the parameters --enzyme DpnII --iterative --aligner bowtie2. The draft assembly was then scaffolded using instaGRAAL v0.1.6 no-opengl branch (Baudry et al. 2020), with default parameters (--levels 4, --cycles 100, --coverage-std 1, --neighborhood 5). The output was then refined using the module instaGRAAL-polish. BUSCO v5.4.7 was used to assess the completeness of the assembly (Simão et al. 2015) against the Metazoa odb10 lineage. The genome size was estimated with the Illumina dataset of 235 million reads and the module kmercount.sh from BBmap v38.79 (Bushnell 2014). The circularized mitochondrial genome was assembled from the Illumina reads using NOVOPlasty v2.7.2 (Dierckxsens et al. 2016) with the publicly available Acropora digitifera mitochondrial genome as a reference (GenBank: KF448535.1). Our 14 chromosome-scale scaffolds were named Ap1, Ap2, …, Ap14 on the basis of their homologies with Acropora millepora chromosome-scale scaffolds bearing the same number (in the absence of a published karyotype for As. poculata).
Transcriptome assembly
RNA sequencing data from Chan et al. (2021) were used to construct a de novo transcriptome. First, to limit the potential for symbiont contamination, only reads for aposymbiotic individuals were used for the host transcriptome assembly. Reads were trimmed using Cutadapt v3.4 (Martin 2011) with a quality cutoff of 15 and a minimum read length of 50 nucleotides. The trimmed reads were then assembled into transcripts using rnaSPades v3.12.0 (Bushmanova et al. 2019). To further reduce the possibility of contamination from the algal endosymbiont, a custom database of Symbiodiniaceae protein sequences was assembled that included the following species: Cladocopium goreaui (Liu et al. 2018b), B. psygmophilum (Parkinson et al. 2016), Breviolum minutum (Shoguchi et al. 2013), Fugacium kawaguti (Liu et al. 2018a, 2018b), Symbiodinium fitti (Reich et al. 2021), Symbiodinium microadriaticum (Aranda et al. 2016), and Symbiodinium tridacnidorum (González-Pech et al. 2021). A BLAST nucleotide-to-protein alignment (blastx) of the assembled As. poculata transcriptome was conducted against this database using BLAST v2.6.0 (Altschul et al. 1990; Camacho et al. 2009). Transcripts with at least 80% identity and with at least 100 bp mapped length were filtered from the transcriptome.
Genome annotation
To annotate repetitive content, de novo transposable element family identification and modeling were conducted using RepeatModeler v1.0.1 (Smit and Hubley 2008). RepeatMasker v4.0.7 (Smit et al. 2015) was then used to soft-mask repetitive regions prior to gene modeling. Subsequent gene prediction included a multitool approach. First, ab initio gene prediction was done using GeneMark-ES v3.51 (Lukashin and Borodovsky 1998). Gene prediction using protein-based evidence was conducted with Exonerate v2.4.0 (Slater and Birney 2005) using the UniProt eukaryote database (downloaded 2017 December 28). RNA-seq reads from Chan et al. (2021) were aligned to the genome using STAR v020201 (Dobin et al. 2013) and incorporated into the automated training of gene prediction using Braker2 v2.1.2 (Brůna et al. 2021). From the resulting predictions, high-quality gene models (HiQ), here defined as those having ≥90% RNA-seq coverage support, were extracted. Gene prediction informed by transcriptomic evidence was carried out using PASA v2.3.3 (Haas et al. 2003) with the flag --TRANSDECODER to keep only the longest open reading frame per group of transcript isoforms (see section “Transcriptome assembly” above for transcriptome assembly details). Consensus gene predictions were acquired using EvidenceModeler v1.1.1 (Haas et al. 2008) weighting each line of evidence as follows: ab initio predictions: 1; RNA-seq based evidence: 1; protein-based evidence: 1; HiQ: 5; and transcriptomic-based evidence: 10. PASA v2.3.3 was run a second time to update gene models and add annotations of untranslated regions (UTRs) using the consensus gene prediction from EVidenceModeler. Finally, tRNAscan-Se v1.3.1 (Chan and Lowe 2019) was used to annotate transfer RNAs. Genome Annotation Generator (GAG) v2.0.1 (Geib et al. 2018) was used to extract coding sequence (CDS), protein, and mRNA sequences.
Functional annotation of predicted genes was conducted based on sequence similarity using BLAST v2.6.0 (Altschul et al. 1990; Camacho et al. 2009) searches via blastp with the settings of eval = 1e−5, max target seqs = 5, and max hsp = 1. Blast queries were conducted against 3 databases: NCBI nr, UniProt Swiss-Prot, and TrEMBL. Hits to each database were combined and annotated with Gene Ontology (GO) terms using the UniProt-GOA mapping.
Investigating the possibility of whole-genome duplication
Given the recent suggestion of a possible WGD event in the genus Acropora (Mao and Satoh 2019), we set out to determine whether a similar event may have occurred in As. poculata. Detection and classification of duplication in the genome was carried out in several ways. First, estimates were conducted using the tool MCScanX under default settings (Wang et al. 2012). Results were then compared to a run under more relaxed settings (max gap size increased to 50). In both cases, duplication origins were classified using the duplicate_gene_classifier module.
A second method of WGD detection employed was the tool wgd v1.1.2 (Zwaenepoel and Van de Peer 2018), which relies on the distributions of synonymous substitutions per synonymous site (Ks). To conduct the analysis, genome-wide CDSs were filtered for the longest translatable isoform of each gene. wgd v1.1.2 was then run using the diamond aligner to compute the whole-paranome (the collection of all duplicate genes in a genome). A Ks distribution was constructed in pairwise mode and kernel density estimates were subsequently fit to the distribution and visualized. A Ks distribution for anchor pairs, defined as paralogs located on colinear duplicated segments, was similarly constructed and visualized. The shape of the Ks distribution was inspected for detection of ancient WGD with the expectation of an exponential decay shape in the absence of a WGD event (Zwaenepoel and Van de Peer 2018).
Lastly, the whole-genome synteny aligner Satsuma2 (available at https://github.com/bioinfologics/satsuma2) was used to detect microhomologous regions between As. poculata chromosomes. Syntenic blocks of homologous sequences arranged in a colinear fashion between chromosomes were then plotted using the tool Orthodotter (https://github.com/institut-de-genomique/orthodotter) to produce an Oxford grid (Edwards 1991), an approach used previously to detect WGD in arthropods (Schwager et al. 2017).
Comparative genomics: gene family and conserved synteny analysis
To characterize the genome organization and content of As. poculata relative to other cnidarians, we completed several comparative genomic analyses. Phylogenetic orthology inferences were carried out using OrthoFinder2 v2.4.0 (Emms and Kelly 2019) on As. poculata and 23 other available cnidarian proteomes (Fig. 1b; Supplementary Table 2) using default parameters with the species tree inferred using the STAG algorithm (Emms and Kelly 2018) and rooted using the STRIDE algorithm (Emms and Kelly 2017). The phylogenetic species tree was visualized using the R package “ggtree” v3.6.2 (Xu et al. 2022 ). GO enrichment of gene families unique to As. poculata was conducted using the clusterProfiler package (Yu et al. 2012) implemented in R v4.0.5 with a P-value cutoff of 0.05, a multiple testing correction method of Benjamini–Hochberg procedure (Benjamini and Hochberg 1995), and a q-value cutoff of 0.2.
Acropora millepora, an obligately symbiotic coral of the complex clade, was selected for more detailed comparison with As. poculata as its assembly was similarly complete. The sizes of gene families common to the 2 species were compared following the methods of González-Pech et al. (2021). Using the orthogroups previously identified by Orthofinder2, size differences of gene families shared between As. poculata and Ac. millepora were evaluated using Fisher's exact test with the multiple testing correction method of Benjamini–Hochberg (Benjamini and Hochberg 1995) and a significance threshold of adjusted P ≤ 0.05. GO enrichment of gene families significantly larger in As. poculata and in Ac. millepora was conducted separately using the same methods as described above.
Conserved synteny between As. poculata and Ac. millepora was assessed with MCScanX_h (Wang et al. 2012) using the homologous genes between the species identified by OrthoFinder2. Default parameters were used to identify colinear blocks (gap size of 25 genes allowable, minimum of 5 genes per colinear block).
Histological examination
Three small fragments from one of the 2 still-living fragments of the sequenced As. poculata colony that were being maintained at Boston University were placed in Z-Fix concentrate:seawater (1:4) fixative in November 2017 and processed in the Histology Laboratory at George Mason University (GMU). Samples were decalcified in 10% ethylenediaminetetraacetic acid (EDTA) pH 7, trimmed into 5 mm strips and placed in labeled cassettes, dehydrated in a series of ethanol solutions (70–100%), cleared using Clearify (StatLab), and embedded in Paraplast Regular (Leica Microsystems Surgipath). Tissue sections, 4 µm thick, were mounted on clean glass microscope slides and stained with Harris's hematoxylin (Statlab) and eosin–phloxine and Giemsa (StatLab) procedures (Price and Peters 2018), then covered with Permount (Fisher Scientific) and a glass cover slip and examined using light microscopy.
Results
Genome statistics and assembly quality
Of the 4 assemblers tried, wtdbg2 (Ruan and Li, 2020) produced the highest quality assembly (Fig. 2; Supplementary Table 1). After purging, polishing, and scaffolding, the assembly using wtdbg2 had an N50 of 31 Mb and a BUSCO score of 95.5% (93.8% single copies, 1.7% duplicate copies, 2.1% fragmented, and 2.4% missing; Table 1). The assembly size of 458 Mb is in line with BBMap predicted haploid size of 453 Mb (estimated ploidy of 2 and 40.95% repetitive). The 14 chromosome-level scaffolds match the known 2n = 28 formula typical of most scleractinian corals (Fig. 2a; Supplementary Table 3; Flot et al. 2006). Further, the k-mer completeness was 52.24%, close to the expected 50% for a haploid assembly (Fig. 2b). Gene prediction of the As. poculata genome assembly yielded 44,839 gene models, of which 1,613 had functional annotations to transposons, leaving 38,998 protein-coding genes excluding repetitive elements (Table 1). RepeatMasker predicted 39.29% of the genomic bases as repetitive elements, with an estimated GC content of 38.50%. Additionally, tRNAscan-Se identified 6,951 transfer RNAs. The assembled transcriptome (used to assist in gene modeling), included 516,681 assembled transcripts and a BUSCO score of 90.8% (Single: 32.3%, Duplicated: 58.5%, Fragmented: 6.4%, Missing: 2.8%). The high level of duplicates in the transcriptome results from retaining all isoforms for every gene.
Fig. 2.
Astrangia poculata genome assembly. a) Proximity ligation sequencing data (Hi-C) contact map displaying the 14 chromosome-level scaffolds of the As. poculata assembly. Interaction points between chromosomes are represented by red dots with binning = 100 kb. Chromosomes are ordered by size from smallest to largest. b) K-mer Analysis Toolkit (KAT) plot (Mapleson et al. 2017) of distinct k-mer duplicity and the number of times these k-mers are represented in the final As. poculata genome assembly, with k = 27.
Table 1.
Assembly summary statistics for the As. poculata genome.
| Metric | Value |
|---|---|
| Assembly size (Mb) | 458 |
| Number of contigs | 488 |
| N50 (Mb) | 31 |
| Genome BUSCO (%) [singles; duplicates; missing; fragmented] | 95.5 (93.8; 1.7; 2.1; 2.4) |
| k-mer completeness (%) | 52.24 |
| Number of genes | 44,839 |
| Gene density (genes/Mb) | 97.9 |
| Average gene length (bp) | 5,204 |
| Average exon length (bp) | 244 |
| Average intron length (bp) | 1,071 |
| Average CDS length (bp) | 1,268 |
| Gene model BUSCO (%) [singles; duplicates; missing; fragmented] | 92.9 (87.1; 5.8; 2.7; 4.4) |
Whole-genome duplication
Duplication analysis via MCScanX (Wang et al. 2012) revealed 88 syntenic blocks with 853 duplicated genes classified as putatively originating from WGD or segmental duplication (SD; Supplementary Table 4). When relaxing MCScanX parameters by allowing a max gap size of 50 genes within colinear blocks (default gap size is 25), the number of duplications classified as whole genome or segmental increased to 1,005. Detected colinear blocks often involved more than 2 colinear regions, with some involving 3 or even 4 colinear regions. However, a Ks-based approach using the tool wgd (Zwaenepoel and Van de Peer 2018) resulted in an exponential decay shape of the distribution of synonymous substitutions per synonymous site, suggesting no signature of a WGD event in As. poculata (Fig. 3). While we did detect many anchor pairs (colinear paralogs), the anchor Ks distribution also declined exponentially. This was consistent with the failure of Orthodotter to detect large colinear regions in the genome of As. poculata (Fig. 4), suggesting that WGD, if it occurred, was too ancient and the genome had subsequently undergone too much rearrangement to leave an obvious colinearity signature. Rather, these results suggest widespread duplications (tandem, proximal, and dispersed) within the As. poculata genome, which may have resulted in novel functional gene copies through the process of neofunctionalization (Teshima and Innan 2008).
Fig. 3.
Distribution of synonymous substitutions per synonymous site (Ks) for all inferred duplications in the As. poculata genome with light gray representing all paralogous gene pairs and black representing anchor gene pairs.
Fig. 4.
Oxford grid representing pairs of homologous regions detected by SatsumaSynteny across the genome of As. poculata. On this grid (not drawn to scale), each point represents a pair of identical or nearly identical 4,096 bp regions. Ap1, Ap2, …, Ap14 represent the 14 chromosome-scale scaffolds in the assembly of the genome of As. poculata.
Comparative genomics: gene family size and conserved synteny
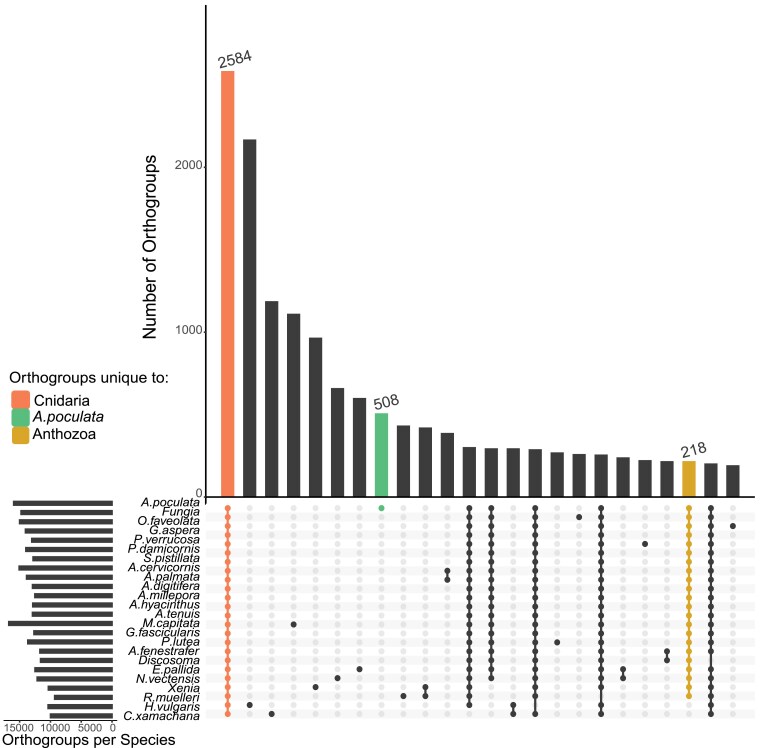
Using a gene family analysis involving a total of 24 species, we defined a “core” cnidarian genome that consisted of 2,584 gene families shared among all cnidarians present in the analysis (Fig. 5). We found 508 gene families unique to As. poculata. Interestingly, only 218 orthogroups were present in all anthozoans included in the analysis. GO enrichment in clusterProfiler (Yu et al. 2012) of the gene families unique to As. poculata resulted in 143 enriched GO terms (significance threshold of P < 0.05 after adjusting for multiple testing; Supplementary Table 5).
Fig. 5.
An UpSet plot representing the number of orthogroups containing each species included in the analysis. Each dot represents the presence of a given species in orthogroups, with orthogroups unique to As. poculata (green), shared amongst cnidarians (orange), and shared among anthozoans (gold) highlighted.
Gene families that were common to As. poculata and Ac. millepora were assessed for differences in gene numbers using Fisher's exact test following the methods of González-Pech et al. (2021). In total, 170 gene families were identified as significantly different in gene numbers between the 2 species (Fig. 6a; Supplementary Table 6; adjusted P ≤ 0.05). This included 73 gene families that were significantly larger in Ac. millepora and 97 gene families that were significantly larger in As. poculata. Gene families larger in Ac. millepora that were most different in size compared to As. poculata (according to log2 fold change) included gene families that putatively encoded for zinc finger CCHC domain-containing proteins, cation channel sperm-associated proteins, RING-box proteins, lectins, and serine/threonine-protein kinases. In contrast, gene families larger in As. poculata that were most different in size compared to Ac. millepora by log2 fold change included orthogroups that putatively encoded for transposable elements, G protein-coupled receptors, zinc finger MYM-type proteins, RNA-directed DNA polymerases, orexin receptors, ATP-dependent DNA helicases, E3 ubiquitin-protein ligases, and histone H3 (Fig. 6b; Supplementary Table 6). GO enrichment analysis resulted in 723 and 635 enriched terms for expanded families in As. poculata and Ac. millepora, respectively. Enriched terms in Ac. millepora expanded gene families included CatSper complex, sperm principal piece, zinc ion binding, and viral latency (Supplementary Table 7), while enriched terms in expanded As. poculata families included transposition, G protein-coupled receptor activity, and peptide receptor activity (Supplementary Table 8).
Fig. 6.
Comparison of As. poculata and Ac. millepora genomes. a) Volcano plot of gene family size comparison using Fisher's exact test between As. poculata and Ac. millepora. Points are colored according to whether they are significantly larger in As. poculata (blue), significantly larger in Ac. millepora (gold), or not significantly different (gray) with adjusted P > 0.05. The top 20 [note: equal rank (i.e. equal log2 fold change values) orthogroups are all depicted, leading to 23 gene families for Ac. millepora] significant gene families for each species are labeled with orthogroup IDs in red. For readability, orthogroups are labeled with leading “OG” and zeros removed from their IDs. b) The number of genes for these top gene families that are significantly larger in As. poculata (panel “A. poculata”) and Ac. millepora (panel “A. millepora”). Each bar represents the number of genes in the gene family for As. poculata (blue) and Ac. millepora (gold). Gene families with putative functions in reproduction, symbiosis, innate immunity, transposition, and quiescence are highlighted in red. c) A bar plot representing the conserved synteny between the genomes of As. poculata (AP) and Ac. millepora (AM). Each chromosome of As. poculata is painted with the color of the Ac. millepora chromosome with which there is conserved synteny. White spaces indicate regions where colinear blocks were not detected.
In addition to gene family size analysis, orthologues common to As. poculata and Ac. millepora were evaluated for conserved synteny. MCScanX analysis of colinearity revealed a high level of conserved gene synteny between As. poculata and Ac. millepora with 3,719 syntenic blocks identified of at least 5 colinear genes. In total, 56.63% of orthologous genes were present in the colinear blocks between the 2 divergent coral species (Fig. 6c).
Histology
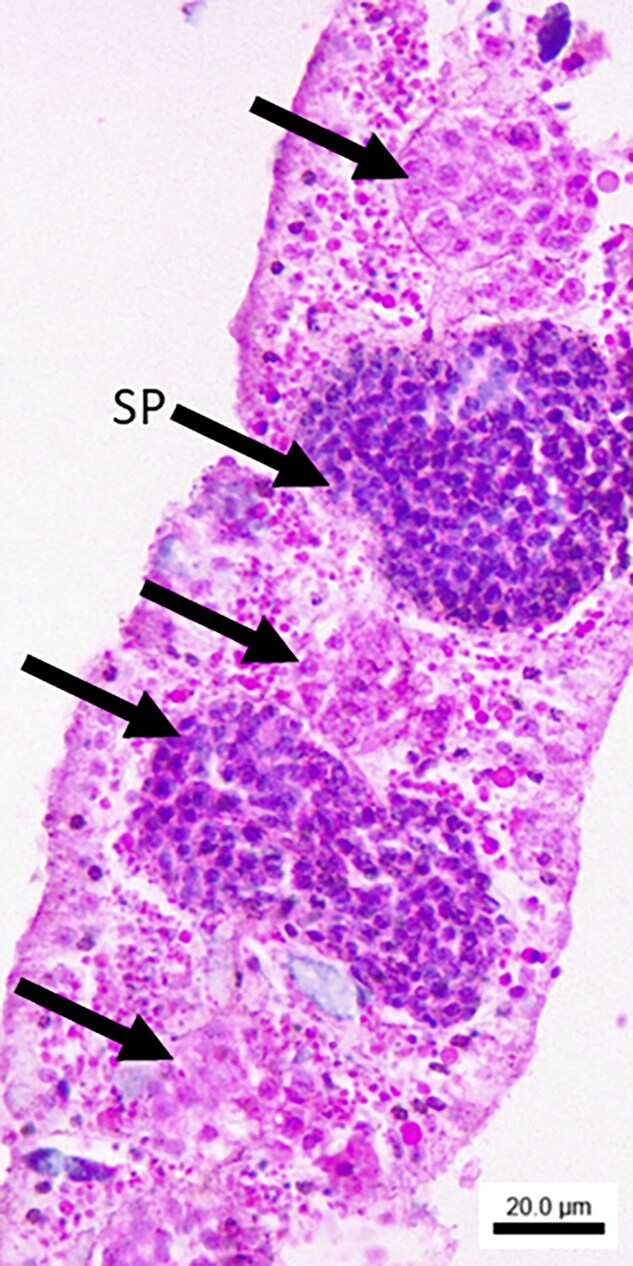
Examination of the histology slides revealed that the sequenced As. poculata colony was male, with developing spermaries in stages II–IV (Szmant et al. 1980) in 2 of the tissue sections (Fig. 7).
Fig. 7.
Representative section from the sequenced As. poculata colony stained with hematoxylin and eosin. The scale bar = 20 µm. Black arrows point to mesenteries showing developing spermaries (SP).
Discussion
Astrangia poculata has increasingly been used as a model coral system due to its temperature tolerance and flexibility in symbiont state (Jacques et al. 1983; Peters et al. 1988; Dimond and Carrington 2007; Burmester et al 2017; 2018; Sharp et al. 2017; Aichelman et al. 2019; Chan et al. 2021; DiRoberts et al. 2021; Wuitchik et al. 2021). Because As. poculata is facultatively symbiotic, the host and the algal symbiont response to manipulation can be distinguished—a study design that is often impossible in adult tropical corals, many of whom do not occur naturally in an aposymbiotic state. In addition to associating with an algal endosymbiont of the family Symbiodiniaceae, As. poculata creates a calcium carbonate skeleton similar to reef-building corals (Hayes and Goreau 1977; Peters et al. 1988). However, corals are a diverse group of organisms with respect to their biology and ecology (e.g. habitat, morphology, and response to environmental change) (Chappell 1980; Fabricius et al. 2011; Muir et al. 2018; Kusumoto et al. 2020). Thus, it is important to recognize the differences, as well as the similarities, between As. poculata and other corals. Here, we have developed a chromosome-scale genome assembly for As. poculata, which has allowed us to characterize some of these differences and similarities between As. poculata and other cnidarians. These results provided insight into potential genomic drivers of cnidarian biology, such as As. poculata's astounding ability to enter a dormancy state when exposed to extreme cold temperatures, and elucidated the demands placed on sperm during mass spawning events of tropical corals.
No evidence of ancient whole-genome duplication, but recent duplications are abundant
WGD can generate new genetic material upon which selection or genetic drift may act (Ohno 1970; Holland and Ocampo Daza 2018). A possible WGD event in the most recent common ancestor of the genus Acropora was suggested using phylogenomic and comparative genomic techniques (Mao and Satoh 2019). However, it is unknown if such an event may have occurred at other points in the scleractinian lineage where polyploidism is common. Our conservative results from MCScanX indicated that 853 duplicated genes (1.8%) were classified as possibly originating from large-scale duplication events, such as segmental or WGD (Supplementary Table 4). To further identify whether this may indeed be indicative of an ancient WGD event, we examined the distribution of synonymous substitutions per synonymous site (Ks). Under a model of a constant rate of duplication and loss, there should be an exponential decay shape to a Ks distribution, which is the shape we find in the distribution for As. poculata (Fig. 3). In contrast, when a WGD event has occurred, it leaves a signature peak in the Ks distribution (Zwaenepoel and Van de Peer 2018). Additionally, we examined the Ks distribution of the anchor pairs (paralogs located on colinear duplicated segments). However, many anchors detected represented small Ks values (0–0.1) and also followed an exponentially decaying shape. Similarly, a search for microsynteny using SatsumaSynteny followed with colinearity detection using Orthodotter did not reveal abundant pairs of colinear regions (Fig. 4). These results suggest that while large-scale SDs may be present in the As. poculata genome, we are unable to detect a strong signature of an ancient WGD event. Because these approaches may not be able to detect very ancient duplications, a phylogenomic approach (Zwaenepoel and Van de Peer 2018) will be required to further test for an ancient WGD event in Scleractinia as additional high-quality genome assemblies representative of each taxonomic group across the cnidarian phylogeny become available.
While there was no evidence of an ancient WGD event in As. poculata, we found a high level of large-scale SDs, tandem duplications, and putatively transposon-derived gene models (Table 1). This may explain the higher gene density of As. poculata (97.9 genes/Mb; Table 1) relative to the complex coral, Ac. millepora (63.4 genes/Mb). The gene density in As. poculata is in-line with those of other Robusta corals (Pocillipora cf effusa and Pocillopora damicornis) where pervasive tandem duplications were also detected (Noel et al. 2023).
Genome content of temperate As. poculata vs tropical Ac. millepora
Orthologue analysis resulted in 508 orthogroups unique to As. poculata (Fig. 5). These gene families were found to be enriched in 143 GO terms, including terms involving ubiquitin E3 ligases, regulation of transcription and gene expression, G protein-coupled receptor activity, and transposition (Supplementary Table 5). To further investigate the unique genomic content of As. poculata, we selected one tropical coral to include in a more detailed comparison. The As. poculata genome assembly is among the most complete and contiguous coral genomes to date. Many of the other currently available cnidarian genome assemblies remain considerably more fragmented. For this reason, we limited our subsequent genomic analyses to comparisons between As. poculata and Ac. millepora. Acropora millepora has a chromosome-scale genome assembly (Fuller et al. 2020), and contrasts with As. poculata in its ecology and evolutionary history. While As. poculata is a facultatively symbiotic temperate coral of the robust clade, Ac. millepora is an obligately symbiotic tropical coral of the complex clade.
Conserved micro- and macrosynteny between complex and robust clades
Synteny analysis between As. poculata and Ac. millepora revealed considerable conserved colinearity (56.63%; Fig. 6c) despite ∼415 Mya of divergence of the 2 clades, Robusta (As. poculata) and Complexa (Ac. millepora) (Stolarski et al. 2011). This surprisingly high level of colinearity is in line with previous work comparing other complex and robust coral species. Ying et al. (2018) found that the extent of conserved gene order within Scleractinia, regardless of clade, was relatively high compared to the level of conserved synteny between sea anemones Exaiptasia and Nematostella in the order Actinaria. Our results further lend support to this conclusion of consistently high conserved gene order across scleractinians.
Differential gene family expansions related to innate immunity and symbiosis
While gene order analysis highlighted similarities between As. poculata and Ac. millepora, gene family size comparisons revealed differences (Fig. 6a and b; Supplementary Table 6). Gene family expansions are often observed during adaptation in corals (van Oppen and Medina 2020). Notably, of the gene families expanded in Ac. millepora relative to As. poculata, the gene family with the largest log2 fold change contained several copies of zinc finger CCHC domain-containing protein 3 (orthogroup OG0000470; Fig. 6a and b; Supplementary Table 6), which plays a role in the innate immune response to viruses (Lian et al. 2018). Defense against viruses is likely important in Ac. millepora, as previous work has identified massive viral outbreaks in this species (Correa et al. 2016). Further, obligately symbiotic cnidarians have a more advanced innate immunity repertoire relative to nonsymbiotic relatives, possibly driven by the dynamic and constant interaction with the algal endosymbiont (Shinzato et al. 2011; Voolstra et al. 2017; Cunning et al. 2018; Shumaker et al. 2019; van Oppen and Medina 2020). These previous findings relied on comparisons to more distantly related, nonsymbiotic anthozoans. However, here we determine that this holds with a comparison to a more closely related facultatively symbiotic coral, as opposed to a noncoral anthozoan relative.
The molecular elements governing the uptake, maintenance, and breakdown of symbiosis in corals still remain largely unclear. Previous work does indicate that features of the innate immune system of the host, notably lectins, play an important role (Kvennefors et al. 2008; Fransolet et al. 2012; Zhou et al. 2018; Hu et al. 2020; Takeuchi et al. 2021). Lectins are pattern-recognition proteins that bind to carbohydrates (Goldstein et al. 1980). Within the top 15 orthogroups significantly larger in Ac. millepora relative to As. poculata were gene families related to innate immunity that have previously been implicated in establishing symbiotic associations in corals, including C-type lectins and macrophage mannose receptors that mediate endocytosis of glycoproteins (orthogroups OG0000360, OG0000958, and OG0001199; Fig. 6a and b; Supplementary Table 6). Kvennefors et al. (2008) isolated a mannose-binding lectin in Ac. millepora and demonstrated its affinity to binding to both pathogens and algal dinoflagellates of the family Symbiodiniaceae. Since then, comparative genomics and single-cell RNA sequencing have further emphasized the role of lectins in the cnidarian-algal symbiosis in additional species (Cunning et al. 2018; Hu et al. 2020). Here, we were able to compare 2 scleractinian corals, one facultatively symbiotic and the other obligately symbiotic. We found that pattern-recognition proteins (e.g. lectins) are expanded in Ac. millepora relative to As. poculata. Future study characterizing the potential differences in the establishment and maintenance of symbiosis between obligately vs facultatively symbiotic corals is warranted, and the results here highlight lectin-related gene families as excellent targets.
Sexual reproduction in mass spawning tropical corals
In addition to innate immunity, top gene families larger in Ac. millepora compared to As. poculata included those related to sexual reproduction (Fig. 6a and b; Supplementary Table 6), with functions involved in sperm cell hyperactivation (cation channel sperm-associated protein subunit epsilon; orthogroup OG0000900) and meiosis (RING-box protein 1; orthogroup OG0000130). Further, significantly enriched GO terms in the Ac. millepora expanded gene families included CatSper complex (GO:0036128), sperm principal piece (GO:0097228), and sperm flagellum (GO:0036126; Supplementary Table 7). These findings are interesting because of the difference in reproductive modes between Ac. millepora and As. poculata. Astrangia poculata is a gonochoric species that reproduces via broadcast spawning wherein gametes are released into the water column prior to fertilization (Szmant et al. 1980). Spawning occurs annually from August to September based on the seasonal maximum temperature, with a second cycle sometimes observed in October or November (Peters et al. 1988). Astrangia poculata sperm are unlikely to have to compete with many other corals for the fertilization of eggs because there are only a few other temperate coral species. We were able to confirm that the sequenced As. poculata colony was male as indicated by the presence of spermaries (Fig. 7). In contrast, Ac. millepora is a hermaphroditic broadcast spawning species that reproduces during “mass spawning” events synchronized with the occurrence of the full moon (Kaniewska et al. 2015). During these annual spawning events, Ac. millepora releases gametes simultaneously with over 100 other coral species, as well as hundreds of other invertebrates over the course of only a few nights (Babcock et al. 1986; Harrison 2011). This creates competition between gametes, as well as the opportunity for interspecific hybridization. Expansion of these reproduction-related gene families has the potential to soften or maintain species boundaries and would be great targets for future studies examining adaptation to mass spawning in corals.
Genome plasticity in As. poculata: transposition and epigenetics
Gene families that were larger in As. poculata relative to Ac. millepora included families of transposable elements (Fig. 6a and b; Supplementary Table 6; orthogroups OG0000015 and OG0000418). Further, significantly enriched GO terms in the As. poculata expanded gene families included transposition (GO:0032196) and DNA-mediated transposition (GO:0006313; Supplementary Table 8). Retrotransposition contributed to gene family expansion in Symbiodiniaceae (Lin et al. 2015; González-Pech et al. 2021) and, based on our results, may also play a role in the host As. poculata. Further, transposable elements promote adaptation and drive genome plasticity in many species, including bacteria, fungi, plants, and animals (Bennett 2004; Feschotte and Pritham 2007; Leitch and Leitch 2008; Mat Razali et al. 2019; Yuan et al. 2021). Epigenetic modification is involved in host genome regulation of transposable elements (Matzke et al. 2000). Interestingly, we also found that a family of histone H3 proteins was expanded in As. poculata relative to Ac. millepora (orthogroup OG0000255; Fig. 6a and b; Supplementary Table 6). In eukaryotes, histone H3 is one of the core histone proteins involved in structuring chromatin (Kornberg 1977; McGhee and Felsenfeld 1980). The sequence variants, as well as different modification states of histone H3, are thought to influence gene regulation (Sarma and Reinberg 2005; Hake et al. 2006; Kouzarides 2007; Loyola and Almouzni 2007; Maehara et al. 2015; Jiang and Berger 2017; Klemm et al. 2019). In plants, histone H3 plays a role in development and abiotic stress (Yuan et al. 2013; Otero et al. 2014). These results suggest that transposable elements and epigenetic modification may play an important role in the plasticity of As. poculata.
Winter quiescence in As. poculata
Among the top gene families expanded in As. poculata was a family of G-coupled protein receptors, including RYamide receptors, orexin receptors, and neuropeptide SIFamide receptors (orthogroup OG0000211; Fig. 6a and b; Supplementary Table 6). Expanded As. poculata gene families were also enriched for G protein-coupled receptor activity (GO:0004930; Supplementary Table 8). In Drosophila, RYamide receptors are possibly associated with feeding suppression (Ida et al. 2011), while neuropeptide SIFamide receptors have been associated with the promotion of sleep (Park et al. 2014). Similarly, orexin receptors are known to regulate circadian sleep/wake cycles in mammals (Chemelli et al. 1999). The expansion of this gene family in As. poculata relative to Ac. millepora may explain As. poculata's ability to enter a dormant state when exposed to near-freezing temperatures. During winter months in the intertidal and subtidal regions at the northernmost edges of the species' range, As. poculata enters this dormancy, referred to as “winter quiescence,” when water temperatures plummet to below 6°C (Grace 2017). During this state, polyps are retracted and oral discs are puffed out, while feeding is reduced or ceased entirely (Jacques et al. 1983; Grace 2017; Supplementary Fig. 1). This finding has relevance to warmer waters, e.g. some Mediterranean anthozoans enter a similar “summer dormancy” state, including corals (Coma et al. 2000; Caroselli et al. 2015). Overall, these results indicate that gene family expansions may have contributed to the adaption of As. poculata to the high variance in environmental conditions that this species experiences temporally and spatially across its range.
Conclusion
In this study, we present the first chromosome-scale assembly of the facultatively symbiotic, temperate coral As. poculata. Our contribution of a high-quality genome resource for As. poculata advances the use of this species as an experimental model and lays the groundwork for numerous future studies, as As. poculata is an important emerging model for coral health (Neff 2020). Further, comparison of the As. poculata genome to the tropical, obligately symbiotic coral Ac. millepora uncovered potential genomic drivers of unique features of not only As. poculata, but of Ac. millepora, as well. Taken together, these results have generated genomic targets for future study of adaptation in these species and emphasize the power of comparative genomics to reveal novel insights into the biology of corals.
Supplementary Material
Acknowledgments
We thank all authors who made their coral genome assemblies available for the comparative genomic analyses in this work. Special thanks to Sam Piorkowski and Claire Klippel for their computational and aquaria support; and to Amanda Drill for histology assistance.
Contributor Information
Kathryn H Stankiewicz, Department of Biology, The Pennsylvania State University, University Park, PA 16802, USA; Institute for Systems Biology, Seattle, WA 98109, USA.
Nadège Guiglielmoni, Department of Marine Biology, Université libre de Bruxelles (ULB), Brussels 1050, Belgium.
Sheila A Kitchen, Department of Biology, The Pennsylvania State University, University Park, PA 16802, USA; Department of Marine Biology, Texas A&M University at Galveston, Galveston, TX 77554, USA.
Jean-François Flot, Department of Marine Biology, Université libre de Bruxelles (ULB), Brussels 1050, Belgium; Interuniversity Institute of Bioinformatics in Brussels —(IB), Brussels 1050, Belgium.
Katie L Barott, Department of Biology, University of Pennsylvania, Philadelphia, PA 19104, USA.
Sarah W Davies, Department of Biology, Boston University, Boston, MA 02215, USA.
John R Finnerty, Department of Biology, Boston University, Boston, MA 02215, USA.
Sean P Grace, Department of Biology & Werth Center for Coastal and Marine Studies, Southern Connecticut State University, New Haven, CT 06515, USA.
Leslie S Kaufman, Department of Biology, Boston University, Boston, MA 02215, USA.
Hollie M Putnam, Department of Biological Sciences, University of Rhode Island, Kingston, RI 02881, USA.
Randi D Rotjan, Department of Biology, Boston University, Boston, MA 02215, USA.
Koty H Sharp, Department of Biology, Marine Biology, and Environmental Science, Roger Williams University, Bristol, RI 02809, USA.
Esther C Peters, Department of Environmental Science and Policy, George Mason University, Fairfax, VA 22030, USA.
Iliana B Baums, Department of Biology, The Pennsylvania State University, University Park, PA 16802, USA; Helmholtz Institute for Functional Marine Biodiversity at the University of Oldenburg (HIFMB), Carl von Ossietzky Universität Oldenburg, Oldenburg 26129, Germany; Alfred Wegener Institute, Helmholtz-Centre for Polar and Marine Research, Bremerhaven 27570, Germany; Institute for Chemistry and Biology of the Marine Environment (ICBM), School of Mathematics and Science, Carl von Ossietzky Universität Oldenburg, Oldenburg 26129, Germany.
Data availability
All raw sequence data and the genome assembly have been submitted to NCBI under BioProject PRJNA1123198. The genome assembly and annotation files are also available at https://zenodo.org/records/14110456. Supplemental files are available at https://zenodo.org/records/14226509. The scripts associated with the data analysis presented here are available at https://github.com/kate-stankiewicz/apoculata_genome_assembly.
Supplemental material available at G3 online.
Funding
This work was made possible by the NSF grant OCE-1537959 to I.B.B., NIH T32 Kirschstein-NRSA: Computation, Bioinformatics, and Statistics (CBIOS) support to KHS. I.B.B. acknowledges support by the Open Access publication fund of Alfred-Wegener-Institut Helmholtz-Zentrum für Polar- und Meeresforschung.
Literature cited
- Aichelman HE, Zimmerman RC, Barshis DJ. 2019. Adaptive signatures in thermal performance of the temperate coral Astrangia poculata. J Exp Biol. 222(5):jeb189225. doi: 10.1242/jeb.189225. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Aranda M, Li Y, Liew YJ, Baumgarten S, Simakov O, Wilson MC, Piel J, Ashoor H, Bougouffa S, Bajic VB, et al. 2016. Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci Rep. 6(1):39734. doi: 10.1038/srep39734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock RC, Bull GD, Harrison PL, Heyward AJ, Oliver JK, Wallace CC, Willis BL. 1986. Synchronous spawnings of 105 scleractinian coral species on the Great Barrier Reef. Mar Biol. 90(3):379–394. doi: 10.1007/BF00428562. [DOI] [Google Scholar]
- Baudry L, Guiglielmoni N, Marie-Nelly H, Cormier A, Marbouty M, Avia K, Mie YL, Godfroy O, Sterck L, Cock JM, et al. 2020. instaGRAAL: chromosome-level quality scaffolding of genomes using a proximity ligation-based scaffolder. Genome Biol. 21(1):148. doi: 10.1186/s13059-020-02041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellwood DR, Hughes TP, Folke C, Nyström M. 2004. Confronting the coral reef crisis. Nature. 429(6994):827–833. doi: 10.1038/nature02691. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodological). 57(1):289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- Bennett PM. 2004. Genome plasticity. In: Woodford N, Johnson AP, editors. Genomics, Proteomics, and Clinical Bacteriology: Methods and Reviews. Totowa (NJ): Humana Press. p. 71–113. [Google Scholar]
- Bhattacharya D, Agrawal S, Aranda M, Baumgarten S, Belcaid M, Drake JL, Erwin D, Foret S, Gates RD, Gruber DF, et al. 2016. Comparative genomics explains the evolutionary success of reef-forming corals. eLife. 5:e13288. doi: 10.7554/eLife.13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruna T, Hoff KJ, Lomsadze A, Stanke M, Borodovsky M. 2021. BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genom Bioinform. 3(1):lqaa108. doi: 10.1093/nargab/lqaa108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmester EM, Breef-Pilz A, Lawrence NF, Kaufman L, Finnerty JR, Rotjan RD. 2018. The impact of autotrophic versus heterotrophic nutritional pathways on colony health and wound recovery in corals. Ecol Evol. 8(22):10805–10816. doi: 10.1002/ece3.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmester EM, Finnerty JR, Kaufman L, Rotjan RD. 2017. Temperature and symbiotic state impact healing in experimentally wounded corals. Mar Ecol Prog Ser. 570:87–99. doi: 10.3354/meps12114. [DOI] [Google Scholar]
- Bushmanova E, Antipov D, Lapidus A, Prjibelski AD. 2019. rnaSPAdes: a de novo transcriptome assembler and its application to RNA-Seq data. GigaScience. 8(9):giz100. doi: 10.1093/gigascience/giz100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell B. 2014. BBMap: a fast, accurate, splice-aware aligner. 9th Annual Genomics of Energy & Environment Meeting, March 17–20, 2014, Walnut Creek (CA).
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics. 10(1):421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caroselli E, Falini G, Goffredo S, Dubinsky Z, Levy O. 2015. Negative response of photosynthesis to natural and projected high seawater temperatures estimated by pulse amplitude modulation fluorometry in a temperate coral. Front Physiol. 6:317. doi: 10.3389/fphys.2015.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AN, González-Guerrero LA, Iglesias-Prieto R, Burmester EM, Rotjan RD, Finnerty JR, Baums IB. 2021. An algal symbiont Breviolum psygmophilum responds more strongly to chronic high temperatures than its facultatively symbiotic coral host Astrangia poculata. bioRxiv 430325. doi: 10.1101/2021.02.08.430325, preprint: not peer reviewed. [DOI] [Google Scholar]
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14. doi: 10.1007/978-1-4939-9173-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J. 1980. Coral morphology, diversity and reef growth. Nature. 286(5770):249–252. doi: 10.1038/286249a0. [DOI] [Google Scholar]
- Chemelli RM, Willie JT, Sinton CM, Elmquist JK, Scammell T, Lee C, Richardson JA, Williams SC, Xiong Y, Kisanuki Y, et al. 1999. Narcolepsy in orexin knockout mice: molecular genetics of sleep regulation. Cell. 98(4):437–451. doi: 10.1016/S0092-8674(00)81973-X. [DOI] [PubMed] [Google Scholar]
- Coma R, Ribes M, Gili J-M, Zabala M. 2000. Seasonality in coastal benthic ecosystems. Trends Ecol Evol. 15(11):448–453. doi: 10.1016/S0169-5347(00)01970-4. [DOI] [PubMed] [Google Scholar]
- Correa AMS, Ainsworth TD, Rosales SM, Thurber AR, Butler CR, Vega Thurber RL. 2016. Viral outbreak in corals associated with an in situ bleaching event: atypical herpes-like viruses and a new megavirus infecting Symbiodinium. Front Microbiol. 7:127 doi: 10.3389/fmicb.2016.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings CE. 1983. The biology of Astrangia danae [Ph.D. Dissertation]. [Kingston (RI)]: University of Rhode Island.
- Cunning R, Bay RA, Gillette P, Baker AC, Traylor-Knowles N. 2018. Comparative analysis of the Pocillopora damicornis genome highlights role of immune system in coral evolution. Sci Rep. 8(1):16134. doi: 10.1038/s41598-018-34459-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCarlo TM, Cohen AL, Wong GTF, Davis KA, Lohmann P, Soong K. 2017. Mass coral mortality under local amplification of 2°C ocean warming. Sci Rep. 7(1):44586. doi: 10.1038/srep44586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi: 10.1093/nar/gkw955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimond J, Carrington E. 2007. Temporal variation in the symbiosis and growth of the temperate scleractinian coral Astrangia poculata. Mar Ecol Prog Ser. 348:161–172. doi: 10.3354/meps07050. [DOI] [Google Scholar]
- Dimond JL, Kerwin AH, Rotjan R, Sharp K, Stewart FJ, Thornhill DJ. 2013. A simple temperature-based model predicts the upper latitudinal limit of the temperate coral Astrangia poculata. Coral Reefs. 32(2):401–409. doi: 10.1007/s00338-012-0983-z. [DOI] [Google Scholar]
- DiRoberts LE, Dudek A, Ray NE, Fulweiler RW, Rotjan RD. 2021. Testing assumptions of nitrogen cycling between a temperate, model coral host and its facultative symbiont: symbiotic contributions to dissolved inorganic nitrogen assimilation. Mar Ecol Prog Ser. 670:61–74. doi: 10.3354/meps13731. [DOI] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England). 29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JH. 1991. The Oxford Grid. Ann Hum Genet. 55(1):17–31. doi:1111/j.1469-1809.1991.tb00394.x. [DOI] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2017. STRIDE: species tree root inference from gene duplication events. Mol Biol Evol. 34(12):3267–3278. doi: 10.1093/molbev/msx259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2018. STAG: species tree inference from all genes [preprint]. bioRxiv. https://www.biorxiv.org/content/10.1101/267914v1. [Google Scholar]
- Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20(1):238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabricius KE, Langdon C, Uthicke S, Humphrey C, Noonan S, De’ath G, Okazaki R, Muehllehner N, Glas MS, Lough JM. 2011. Losers and winners in coral reefs acclimatized to elevated carbon dioxide concentrations. Nat Clim Chang. 1(3):165–169. doi: 10.1038/nclimate1122. [DOI] [Google Scholar]
- Feschotte C, Pritham EJ. 2007. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 41(1):331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flot J-F, Ozouf-Costaz C, Tsuchiya M, Van Woesik R. 2006. Comparative coral cytogenetics. Proceedings of the 10th International Coral Reef Symposium:1: 4–8.
- Fransolet D, Roberty S, Plumier J-C. 2012. Establishment of endosymbiosis: the case of cnidarians and Symbiodinium. J Exp Mar Biol Ecol. 420–421:1–7. doi: 10.1016/j.jembe.2012.03.015. [DOI] [Google Scholar]
- Fuller ZL, Mocellin VJL, Morris LA, Cantin N, Shepherd J, Sarre L, Peng J, Liao Y, Pickrell J, Andolfatto P, et al. 2020. Population genetics of the coral Acropora millepora: toward genomic prediction of bleaching. Science. 369(6501):eaba4674. doi: 10.1126/science.aba4674. [DOI] [PubMed] [Google Scholar]
- Geib SM, Hall B, Derego T, Bremer FT, Cannoles K, Sim SB. 2018. Genome annotation generator: a simple tool for generating and correcting WGS annotation tables for NCBI submission. GigaScience. 7(4):1–5. doi: 10.1093/gigascience/giy018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein IJ, Hughes RC, Monsigny M, Osawa T, Sharon N. 1980. What should be called a lectin? Nature. 285(5760):66. doi: 10.1038/285066b0. [DOI] [Google Scholar]
- González-Pech RA, Stephens TG, Chen Y, Mohamed AR, Cheng Y, Shah S, Dougan KE, Fortuin MDA, Lagorce R, Burt DW, et al. 2021. Comparison of 15 dinoflagellate genomes reveals extensive sequence and structural divergence in family Symbiodiniaceae and genus Symbiodinium. BMC Biol. 19(1):73. doi: 10.1186/s12915-021-00994-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grace S. 2017. Winter quiescence, growth rate, and the release from competition in the temperate scleractinian coral Astrangia poculata (Ellis & Solander 1786). Northeast Nat (Steuben). 24(sp7):B119–B134. doi: 10.1656/045.024.s715. [DOI] [Google Scholar]
- Guiglielmoni N, Houtain A, Derzelle A, Van Doninck K, Flot J-F. 2021. Overcoming uncollapsed haplotypes in long-read assemblies of non-model organisms. BMC Bioinformatics. 22(1):303. doi: 10.1186/s12859-021-04118-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK Jr, Hannick LI, Maiti R, Ronning CM, Rusch DB, Town CD, et al. 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31(19):5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, White O, Buell CR, Wortman JR. 2008. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 9(1):R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, Bazett-Jones DP, Allis CD, Hunt DF. 2006. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J Biol Chem. 281(1):559–568. doi: 10.1074/jbc.M509266200. [DOI] [PubMed] [Google Scholar]
- Harrison PL. 2011. Sexual reproduction of scleractinian corals. In: Dubinsky Z, Stambler N, editors. Coral Reefs: An Ecosystem in Transition. Dordrecht: Springer. p. 59–85. [Google Scholar]
- Hayes RL, Goreau NI. 1977. Intracellular crystal-bearing vesicles in the epidermis of scleractinian corals, Astrangia danae (Agassiz) and Porites porites (Pallas). Biol Bull. 152(1):26–40. doi: 10.2307/1540724. [DOI] [PubMed] [Google Scholar]
- Holland LZ, Ocampo Daza D. 2018. A new look at an old question: when did the second whole genome duplication occur in vertebrate evolution? Genome Biol. 19(1):209. doi: 10.1186/s13059-018-1592-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Zheng X, Fan C-M, Zheng Y. 2020. Lineage dynamics of the endosymbiotic cell type in the soft coral Xenia. Nature. 582(7813):534–538. doi: 10.1038/s41586-020-2385-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TP, Kerry JT, Álvarez-Noriega M, Álvarez-Romero JG, Anderson KD, Baird AH, Babcock RC, Beger M, Bellwood DR, Berkelmans R, et al. 2017. Global warming and recurrent mass bleaching of corals. Nature. 543(7645):373. doi: 10.1038/nature21707. [DOI] [PubMed] [Google Scholar]
- Ida T, Takahashi T, Tominaga H, Sato T, Kume K, Ozaki M, Hiraguchi T, Maeda T, Shiotani H, Terajima S, et al. 2011. Identification of the novel bioactive peptides dRYamide-1 and dRYamide-2, ligands for a neuropeptide Y-like receptor in Drosophila. Biochem Biophys Res Commun. 410(4):872–877. doi: 10.1016/j.bbrc.2011.06.081. [DOI] [PubMed] [Google Scholar]
- Jacques TG, Marshall N, Pilson MEQ. 1983. Experimental ecology of the temperate scleractinian coral Astrangia danae. Mar Biol. 76(2):135–148. doi: 10.1007/BF00392730. [DOI] [Google Scholar]
- Jain M, Olsen HE, Paten B, Akeson M. 2016. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 17(1):239. doi: 10.1186/s13059-016-1103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Berger F. 2017. Histone variants in plant transcriptional regulation. Biochim Biophys Acta Gene Regul Mech. 1860(1):123–130. doi: 10.1016/j.bbagrm.2016.07.002. [DOI] [PubMed] [Google Scholar]
- Kaniewska P, Alon S, Karako-Lampert S, Hoegh-Guldberg O, Levy O. 2015. Signaling cascades and the importance of moonlight in coral broadcast mass spawning. eLife. 4:e09991. doi: 10.7554/eLife.09991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitahara MV, Cairns SD, Stolarski J, Blair D, Miller DJ. 2010. A comprehensive phylogenetic analysis of the Scleractinia (Cnidaria, Anthozoa) based on mitochondrial CO1 sequence data. PLoS One. 5(7):e11490. doi: 10.1371/journal.pone.0011490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm SL, Shipony Z, Greenleaf WJ. 2019. Chromatin accessibility and the regulatory epigenome. Nat Rev Genet. 20(4):207–220. doi: 10.1038/s41576-018-0089-8. [DOI] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 37(5):540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27(5):722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD. 1977. Structure of chromatin. Annu Rev Biochem. 46(1):931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell. 128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kundu R, Casey J, Sung W.-K. 2019. HyPo: super fast & accurate polisher for long read genome assemblies. bioRxiv 882506. doi: 10.1101/2019.12.19.882506, preprint: not peer reviewed. [DOI] [Google Scholar]
- Kusumoto B, Costello MJ, Kubota Y, Shiono T, Wei C-L, Yasuhara M, Chao A. 2020. Global distribution of coral diversity: biodiversity knowledge gradients related to spatial resolution. Ecol Res. 35(2):315–326. doi: 10.1111/1440-1703.12096. [DOI] [Google Scholar]
- Kvennefors ECE, Leggat W, Hoegh-Guldberg O, Degnan BM, Barnes AC. 2008. An ancient and variable mannose-binding lectin from the coral Acropora millepora binds both pathogens and symbionts. Dev Comp Immunol. 32(12):1582–1592. doi: 10.1016/j.dci.2008.05.010. [DOI] [PubMed] [Google Scholar]
- LaJeunesse TC, Parkinson JE, Gabrielson PW, Jeong HJ, Reimer JD, Voolstra CR, Santos SR. 2018. Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr Biol. 28(16):2570–2580.e6. doi: 10.1016/j.cub.2018.07.008. [DOI] [PubMed] [Google Scholar]
- Lajeunesse TC, Parkinson JE, Reimer JD. 2012. A genetics-based description of Symbiodinium minutum sp. nov. and S. psygmophilum sp. nov. (Dinophyceae), two dinoflagellates symbiotic with Cnidaria. J Phycol. 48(6):1380–1391. doi: 10.1111/j.1529-8817.2012.01217.x. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch AR, Leitch IJ. 2008. Genomic plasticity and the diversity of polyploid plants. Science. 320(5875):481–483. doi: 10.1126/science.1153585. [DOI] [PubMed] [Google Scholar]
- Lian H, Zang R, Wei J, Ye W, Hu M-M, Chen Y-D, Zhang X-N, Guo Y, Lei C-Q, Yang Q, et al. 2018. The zinc-finger protein ZCCHC3 binds RNA and facilitates viral RNA sensing and activation of the RIG-I-like receptors. Immunity. 49(3):438–448.e5. doi: 10.1016/j.immuni.2018.08.014. [DOI] [PubMed] [Google Scholar]
- Lin S, Cheng S, Song B, Zhong X, Lin X, Li W, Li L, Zhang Y, Zhang H, Ji Z, et al. 2015. The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science. 350(6261):691–694. doi: 10.1126/science.aad0408. [DOI] [PubMed] [Google Scholar]
- Liu D, Hunt M, Tsai IJ. 2018a. Inferring synteny between genome assemblies: a systematic evaluation. BMC Bioinformatics. 19(1):26. doi: 10.1186/s12859-018-2026-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Stephens TG, González-Pech RA, Beltran VH, Lapeyre B, Bongaerts P, Cooke I, Aranda M, Bourne DG, Forêt S, et al. 2018b. Symbiodinium genomes reveal adaptive evolution of functions related to coral-dinoflagellate symbiosis. Commun Biol. 1(1):95. doi: 10.1038/s42003-018-0098-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locatelli NS, Kitchen SA, Stankiewicz KH, Osborne CC, Dellaert Z, Elder H, Kamel B, Koch HR, Fogarty ND, Baums IB. 2024. Chromosome-level genome assemblies and genetic maps reveal heterochiasmy and macrosynteny in endangered Atlantic Acropora. BMC Genomics. 25(1):1119. doi: 10.1186/s12864-024-11025-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyola A, Almouzni G. 2007. Marking histone H3 variants: how, when and why? Trends Biochem Sci. 32(9):425–433. doi: 10.1016/j.tibs.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Lukashin AV, Borodovsky M. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26(4):1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehara K, Harada A, Sato Y, Matsumoto M, Nakayama KI, Kimura H, Ohkawa Y. 2015. Tissue-specific expression of histone H3 variants diversified after species separation. Epigenetics Chromatin. 8(1):35. doi: 10.1186/s13072-015-0027-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Satoh N. 2019. A likely ancient genome duplication in the speciose reef-building coral genus, Acropora. iScience. 13:20–32. doi: 10.1016/j.isci.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapleson D, Garcia Accinelli G, Kettleborough G, Wright J, Clavijo BJ. 2017. KAT: a K-mer analysis toolkit to quality control NGS datasets and genome assemblies. Bioinformatics (Oxford, England). 33(4):574–576. doi: 10.1093/bioinformatics/btw663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17(1):3. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- Mat Razali N, Cheah BH, Nadarajah K. 2019. Transposable elements adaptive role in genome plasticity, pathogenicity and evolution in fungal phytopathogens. Int J Mol Sci. 20(14):3597. doi: 10.3390/ijms20143597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthey-Doret C, Baudry L, Bignaud A, Cournac A, Remi M, Guiglielmoni N. 2020. hicstuff: simple library/pipeline to generate and handle Hi-C data. Version v2.3.1. Zenodo. doi: 10.5281/zenodo.4066363. [DOI] [Google Scholar]
- Matzke MA, Mette MF, Matzke AJM. 2000. Transgene silencing by the host genome defense: implications for the evolution of epigenetic control mechanisms in plants and vertebrates. Plant Mol Biol. 43(2/3):401–415. doi: 10.1023/A:1006484806925. [DOI] [PubMed] [Google Scholar]
- McFadden CS, Quattrini AM, Brugler MR, Cowman PF, Dueñas LF, Kitahara MV, Paz-García DA, Reimer JD, Rodríguez E. 2021. Phylogenomics, origin, and diversification of Anthozoans (Phylum Cnidaria). Syst Biol. 70(4):635–647. doi: 10.1093/sysbio/syaa103. [DOI] [PubMed] [Google Scholar]
- McGhee JD, Felsenfeld G. 1980. Nucleosome structure. Annu Rev Biochem. 49(1):1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- Muir PR, Wallace CC, Pichon M, Bongaerts P. 2018. High species richness and lineage diversity of reef corals in the mesophotic zone. Proc R Soc Lond B Biol Sci. 285(1893):20181987. doi: 10.1098/rspb.2018.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff EP. 2020. The quest for an animal model of coral health and disease. Lab Anim (NY). 49(2):37–41. doi: 10.1038/s41684-019-0467-7. [DOI] [PubMed] [Google Scholar]
- Noel B, Denoeud F, Rouan A, Buitrago-López C, Capasso L, Poulain J, Boissin E, Pousse M, Da Silva C, Couloux A, et al. 2023. Pervasive tandem duplications and convergent evolution shape coral genomes. Genome Biol. 24(1):123. doi: 10.1186/s13059-023-02960-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by Gene Duplication. New York (NY): Springer-Verlag. [Google Scholar]
- Otero S, Desvoyes B, Gutierrez C. 2014. Histone H3 dynamics in plant cell cycle and development. Cytogenet Genome Res. 143(1–3):114–124. doi: 10.1159/000365264. [DOI] [PubMed] [Google Scholar]
- Park S, Sonn JY, Oh Y, Lim C, Choe J. 2014. SIFamide and SIFamide receptor defines a novel neuropeptide signaling to promote sleep in Drosophila. Mol Cells. 37(4):295–301. doi: 10.14348/molcells.2014.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JE, Baumgarten S, Michell CT, Baums IB, LaJeunesse TC, Voolstra CR. 2016. Gene expression variation resolves species and individual strains among coral-associated dinoflagellates within the genus Symbiodinium. Genome Biol Evol. 8(3):665–680. doi: 10.1093/gbe/evw019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters EC, Cairns SD, Pilson MEQ, Wells JW, Jaap WC, Lang JC, Vasleski CE, St Pierre Gollahon L. 1988. Nomenclature and biology of Astrangia poculata (=A. danae, =A. astreiformis) (Cnidaria: Anthozoa). Proc Biol Soc Wash. 2(101):234–250. [Google Scholar]
- Price KL, Peters EC. 2018. Histological Techniques for Corals. Annapolis (MD)/Annandale (VA): Glen Muir Technologies and Pathobiology Consulting Services. [Google Scholar]
- Reich HG, Kitchen SA, Stankiewicz KH, Devlin-Durante M, Fogarty ND, Baums IB. 2021. Genomic variation of an endosymbiotic dinoflagellate (Symbiodinium ‘fitti’) among closely related coral hosts. Mol Ecol. 30(14):3500–3514. doi: 10.1111/mec.15952. [DOI] [PubMed] [Google Scholar]
- Roach MJ, Schmidt SA, Borneman AR. 2018. Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinformatics. 19(1):460. doi: 10.1186/s12859-018-2485-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano SL, Palumbi SR. 1996. Evolution of scleractinian corals inferred from molecular systematics. Science. 271(5249):640–642. doi: 10.1126/science.271.5249.640. [DOI] [Google Scholar]
- Ruan J, Li H. 2020. Fast and accurate long-read assembly with wtdbg2. Nat Methods. 17(2):155–158. doi: 10.1038/s41592-019-0669-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K, Reinberg D. 2005. Histone variants meet their match. Nat Rev Mol Cell Biol. 6(2):139–149. doi: 10.1038/nrm1567. [DOI] [PubMed] [Google Scholar]
- Schwager EE, Sharma PP, Clarke T, Leite DJ, Wierschin T, Pechmann M, Akiyama-Oda Y, Esposito L, Bechsgaard J, Bilde T, et al. 2017. The house spider genome reveals an ancient whole-genome duplication during arachnid evolution. BMC Biol. 15(1):62. doi: 10.1186/s12915-017-0399-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp KH, Pratte ZA, Kerwin AH, Rotjan RD, Stewart FJ. 2017. Season, but not symbiont state, drives microbiome structure in the temperate coral Astrangia poculata. Microbiome. 5(1):120. doi: 10.1186/s40168-017-0329-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato C, Shoguchi E, Kawashima T, Hamada M, Hisata K, Tanaka M, Fujie M, Fujiwara M, Koyanagi R, Ikuta T, et al. 2011. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature. 476(7360):320. doi: 10.1038/nature10249. [DOI] [PubMed] [Google Scholar]
- Shoguchi E, Shinzato C, Kawashima T, Gyoja F, Mungpakdee S, Koyanagi R, Takeuchi T, Hisata K, Tanaka M, Fujiwara M, et al. 2013. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr Biol. 23(15):1399–1408. doi: 10.1016/j.cub.2013.05.062. [DOI] [PubMed] [Google Scholar]
- Shumaker A, Putnam HM, Qiu H, Price DC, Zelzion E, Harel A, Wagner NE, Gates RD, Yoon HS, Bhattacharya D. 2019. Genome analysis of the rice coral Montipora capitata. Sci Rep. 9(1):2571. doi: 10.1038/s41598-019-39274-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Slater GSC, Birney E. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 6(1):31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A, Hubley R. (2008). RepeatModeler Open-1.0. Available from http://www.repeatmasker.org. Accessed April 2020.
- Smit A, Hubley R, Green P. 2015. RepeatMasker Open-4.0. Available from http://www.repeatmasker.org. Accessed April 2020.
- Stephens TG, Lee J, Jeong Y, Yoon HS, Putnam HM, Majerová E, Bhattacharya D. 2022. High-quality genome assembles from key Hawaiian coral species. GigaScience. 11:giac098. doi: 10.1093/gigascience/giac098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolarski J, Kitahara MV, Miller DJ, Cairns SD, Mazur M, Meibom A. 2011. The ancient evolutionary origins of Scleractinia revealed by azooxanthellate corals. BMC Evol Biol. 11(1):316. doi: 10.1186/1471-2148-11-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szmant A, Yevich P, Pilson MEQ. 1980. Gametogenesis and early development of the temperate coral Astrangia danae (Anthozoa: Scleractinia). Biol Bull. (158):257–269. doi: 10.2307/1540935. [DOI] [Google Scholar]
- Takeuchi R, Jimbo M, Tanimoto F, Iijima M, Yamashita H, Suzuki G, Harii S, Nakano Y, Yasumoto K, Watabe S. 2021. N-Acetyl-d-glucosamine-binding lectin in Acropora tenuis attracts specific Symbiodiniaceae cell culture strains. Mar Drugs. 19(3):146. doi: 10.3390/md19030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teshima KM, Innan H. 2008. Neofunctionalization of duplicated genes under the pressure of gene conversion. Genetics. 178(3):1385–1398. doi: 10.1534/genetics.107.082933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oppen MJH, Medina M. 2020. Coral evolutionary responses to microbial symbioses. Philos Trans R Soc Lond B Biol Sci. 375(1808):20190591. doi: 10.1098/rstb.2019.0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaser R, Šikić M. 2019. Yet another de novo genome assembler. 2019 11th International Symposium on Image and Signal Processing and Analysis (ISPA). p. 147–151.
- Voolstra CR, Li Y, Liew YJ, Baumgarten S, Zoccola D, Flot J-F, Tambutté S, Allemand D, Aranda M. 2017. Comparative analysis of the genomes of Stylophora pistillata and Acropora digitifera provides evidence for extensive differences between species of corals. Sci Rep. 7(1):17583. doi: 10.1038/s41598-017-17484-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee T-h, Jin H, Marler B, Guo H, et al. 2012. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40(7):e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuitchik DM, Almanzar A, Benson BE, Brennan S, Chavez JD, Liesegang MB, Reavis JL, Reyes CL, Schniedewind MK, Trumble IF, et al. 2021. Characterizing environmental stress responses of aposymbiotic Astrangia poculata to divergent thermal challenges. Mol Ecol. 30(20):5064–5079. doi: 10.1111/mec.16108. [DOI] [PubMed] [Google Scholar]
- Xu S, Li L, Luo X, Chen M, Tang W, Zhan L, Dai Z, Lam TT, Guan Y, Yu G. 2022. Ggtree: a serialized data object for visualization of a phylogenetic tree and annotation data. Imeta. 1(4):e56. doi: 10.1002/imt2.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Cooke I, Sprungala S, Wang W, Hayward DC, Tang Y, Huttley G, Ball EE, Forêt S, Miller DJ. 2018. Comparative genomics reveals the distinct evolutionary trajectories of the robust and complex coral lineages. Genome Biol. 19(1):175. doi: 10.1186/s13059-018-1552-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang L-G, Han Y, He Q-Y. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Zhang X, Wang M, Sun Y, Liu C, Li S, Yu Y, Gao Y, Liu F, Zhang X, et al. 2021. Simple sequence repeats drive genome plasticity and promote adaptive evolution in penaeid shrimp. Commun Biol. 4(1):186. doi: 10.1038/s42003-021-01716-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Liu X, Luo M, Yang S, Wu K. 2013. Involvement of histone modifications in plant abiotic stress responses. J Integr Plant Biol. 55(10):892–901. doi: 10.1111/jipb.12060. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Zhao S, Ni J, Su Y, Wang L, Xu Y. 2018. Effects of environmental factors on C-type lectin recognition to zooxanthellae in the stony coral Pocillopora damicornis. Fish Shellfish Immunol. 79:228–233. doi: 10.1016/j.fsi.2018.05.026. [DOI] [PubMed] [Google Scholar]
- Zwaenepoel A, Van de Peer Y. 2018. wgd—simple command line tools for the analysis of ancient whole-genome duplications. Bioinformatics. 35(12):2153–2155. doi: 10.1093/bioinformatics/bty91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequence data and the genome assembly have been submitted to NCBI under BioProject PRJNA1123198. The genome assembly and annotation files are also available at https://zenodo.org/records/14110456. Supplemental files are available at https://zenodo.org/records/14226509. The scripts associated with the data analysis presented here are available at https://github.com/kate-stankiewicz/apoculata_genome_assembly.
Supplemental material available at G3 online.