Genetic identity testing involves identifying the patterns of genetic material that are unique to almost every individual. Although over 99% of the DNA sequences in the human genome are identical between individuals, a small number of sequence differences are used to distinguish all humans (1). Those different sequences are usually targeted for identity testing. The techniques that are applied in identity testing are DNA fingerprinting, DNA profiling, and DNA typing. Although there are some technical differences between these tests, the terms have been used interchangeably.
DISCOVERY OF THE DNA FINGERPRINT
Historically, identity testing in the forensic field started with the analysis of the ABO blood group system. Later, new markers for identity and paternity identification were based on variations of serum proteins and red blood cell enzymes; eventually the human leukocyte antigen system was used (2). It was not until 20 years ago that Sir Alec Jeffreys, professor and geneticist at the University of Leicester in the United Kingdom (UK), pioneered DNA-based identity testing (3).
Professor Jeffreys was interested in studying the genetic variation between individuals and had done some of the early work to detect genetic differences in humans. However, the answer did not come to him on the initial project he was interested in but rather on an unrelated project: analysis of the myoglobin gene in seal meat at the headquarters of the British Antarctic Survey. When he and his colleagues compared the myoglobin gene in seals with the human counterpart, they found that some short repeating sequences were homologous between seals and humans. When they compared those sequences with the published sequences of tandem repeats called minisatellites, they found that they were the same (4). Professor Jeffreys recognized that the repeating sequences “could be highly variable, informative genetic markers” (3). His group developed a radioactive probe, made up of short sequences, that could latch onto those repeating sequences and ultimately reveal patterns that were unique to each individual: a DNA “fingerprint” (5).
The steps involved in DNA fingerprinting are as follows. First, the DNA is extracted from the specimen (i.e., blood, semen, skin, hair). After DNA extraction, restriction enzymes are added, which work like scissors to cut the DNA into the smaller segments that are different between individuals. The DNA seg-ments are sorted by agarose gel electrophoresis and visualized by staining with ethidium bromide. A Southern blot is performed to transfer the DNA onto a membrane. A radioactive probe is applied to the membrane, and the pattern of DNA is detected by exposing the membrane to x-ray film. The result is a pattern of DNA bands that looks like a supermarket bar code. Each individual has a signature fingerprint (5).
Professor Jeffreys looked at a DNA fingerprint of a human family; he also looked at the fingerprint of a cow, a baboon, a mouse, and a tobacco plant. The pattern of DNA segments, composed of perhaps 15 to 20 bands, was different for each one. However, closer inspection of the patterns of the human family revealed that the mother and the father each had their own pattern and that the child had a composite of both, having inherited an allele from the father and the mother.
In the spring of 1985, Professor Jeffreys and his colleagues published their first article on DNA fingerprinting and saw the utility of it in the forensic sciences and in paternity identification (6). Newspapers publicized the findings, and shortly thereafter a lawyer became interested in the test and saw its applicability in one of the cases she was representing.
DNA AS A PROBLEM-SOLVING TOOL
The first case: an immigration dispute
A family from Ghana immigrated to the UK and became citizens. However, one of the sons went back to Ghana and was stopped from returning to the UK because he had a forged passport. The family's lawyer contacted Professor Jeffreys and asked whether he could confirm that the boy was in fact the mother's son and not her nephew (she had several sisters in Ghana). The situation was complicated by the absence of the father.
Samples of DNA were taken from the mother, from the son whose identity was disputed, and from the mother's three undisputed children. The patterns confirmed the relationship between the mother and the son in question. Moreover, the testing confirmed that all four children had the same father (7). This immigration case opened the door for using DNA fingerprinting in forensic cases and for identity determination.
Refining the assay
After the success of that case, Professor Jeffreys was bombarded with many inquiries. In 1986, he received a phone call from law enforcement officials in Leicestershire, UK, requesting his help in solving a double-murder case. Professor Jeffreys believed that his initial DNA fingerprinting methods would not work in a criminal case because of the large amount of DNA material required for the test to be successful. If the amount of DNA evidence is small and only 15 to 20 DNA bands are examined, it would be impossible to know where the DNA bands are coming from and which minisatellite regions are involved. The process is very cumbersome.
In the beginning Professor Jeffreys used a probe that had sequences that latched to different minisatellite loci. To simplify his DNA fingerprinting assay, he developed a probe that latched to a single minisatellite locus. A single-locus probe recognizes at most two DNA segments in an individual, corresponding to two alleles: one inherited from the mother and the other from the father. Professor Jeffreys used this new technique to solve the double-murder case in 1986 (8).
Identifying a killer
Two 15-year-old girls in Leicestershire had been raped and murdered. Although the attacks had occurred 3 years apart, similarities led the police to believe that one person was responsible for both. A suspect in custody confessed to the most recent murder but not the earlier one, making the investigation even more complicated. Professor Jeffreys was asked to do DNA profiling on a blood specimen that was collected from the suspect and on tissue specimens and semen collected from the two victims.
The DNA profiling revealed that the semen from both victims was identical, proving that one person had committed both murders. In a surprising twist, the results also proved that the suspect in police custody was not the murderer, so he was released and became the first suspect to be cleared of a crime by DNA evidence.
A large-scale manhunt was then launched to find the person whose DNA profile matched that of the killer's semen. All adult men who lived in the area were asked to give blood or saliva specimens for testing. More than 5000 specimens were collected and DNA profiling carried out on the 10% of men who had the same blood type as the killer, but no match was found. The police and the public were disappointed that this new and sophisticated test was unable to identify the killer.
Six months after the initial investigation, a woman reported overhearing a man who claimed to have given blood on behalf of a colleague, Colin Pitchfork. Pitchfork was apprehended and his blood tested; the long-sought DNA match was made, and Pitchfork was convicted of both murders. This successful outcome established DNA profiling as a valuable tool in solving crimes.
Newer methods and the “Angel of Death”
By the end of 1986, DNA profiling was being used all over the world. DNA typing was refined with the introduction of the polymerase chain reaction (PCR) together with the discovery of different repeating sequences called microsatellites. DNA amplification by PCR provides increased sensitivity, thus allowing small amounts of DNA to be analyzed, even from archival and partially degraded samples.
Minisatellites, also known as variable numbers of tandem repeats (VNTR), are made up of repeated sequences that can vary in unit length from 6 to 100 bases. These units can be repeated two to several hundred times at each minisatellite. Thousands of different minisatellites are scattered throughout the genome, but they are often clustered near the telomere, or end of the chromosome (9–11).
Microsatellites, also known as short tandem repeats (STR), are made up of a unit that can vary in length from 1 to 7 bases. This unit is repeated 5 to 100 times at each microsatellite locus. Thousands of different microsatellites are randomly scattered throughout the genome, but not in a specific area (12, 13).
PCR-based DNA typing was used to end the 40-year hunt for Nazi prison camp doctor Josef Mengele, who escaped from the Allies at the end of World War II. Nicknamed the “Angel of Death” at Auschwitz, Mengele was thought to have fled to South America. Police were eventually given a tip that Mengele had drowned at sea in 1979 and was buried in Brazil. In 1985, the badly decomposed remains were exhumed so that DNA samples could be taken, but the specimens were so poor that Professor Jeffreys resorted to what amounted to reverse paternity testing: he used blood specimens from Mengele's wife and son to reconstitute Mengele's DNA pattern. In 1992, the remains were confirmed to be those of Mengele (14).
Development of a DNA database
In the USA, the DNA Identification Act of 1994 authorized the Federal Bureau of Investigation to expand a pilot project into a national DNA database, the Combined DNA Index System (CODIS), as a tool for solving violent crimes. CODIS combines DNA analysis with computer technology to enable crime laboratories at the local, state, and national levels to exchange and compare DNA profiles electronically (15). The database comprises two indexes: the Forensic Index, which contains DNA profiles from crime scene evidence, and the Offender Index, which contains profiles from those convicted of felony sex offenses and other violent crimes. The system is based on the amplification of 13 core STR loci, as well as the amelogenin gene, which is located on the X and Y chromosomes and is useful in determining the sex of an individual (16).
CLINICAL APPLICATIONS OF IDENTITY TESTING
Today, DNA identity testing is widely used in the field of forensics and paternity identification. Other clinical applications are based upon the methods developed for forensic testing. They include assessment of donor hematopoietic engraftment after bone marrow transplantation and chimerism analysis after solid organ transplantation. Other uses include confirming a diagnosis of hydatidiform mole and resolving issues of specimen identity in cases of specimen mislabeling or misidentification. Finally, DNA identity testing can be used to evaluate tumor transmission after transplantation and thus determine whether a malignancy is of donor or recipient origin.
VNTR and STR are useful for identity testing because they are polymorphic and are inherited in a mendelian fashion. Each individual inherits one paternal and one maternal allele of a specific STR locus, leading to further diversity in STR pattern (Figure 1).
Figure 1.
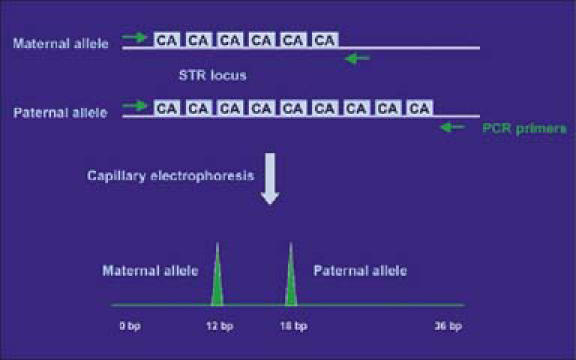
Schematic of short tandem repeat (STR) markers. In this heterozygous example, differences in the number of CA repeats (6 vs 9) between the two alleles result in two distinct polymerase chain reaction peaks by capillary electrophoresis. These peaks can be used as genetic markers of individual identity.
STR analysis in transplantation
Allogeneic bone marrow transplantation is being used to treat a variety of hematological malignancies. The aim is to reconstitute the depleted recipient's bone marrow with the donor's stem cells, thus establishing a complete engraftment. Relapse of the patient's original disease or rejection of the transplanted stem cells can occur. Thus, monitoring the recipient's peripheral blood and bone marrow for the presence of donor or recipients cells is of clinical use (17).
In the molecular pathology laboratory at Baylor University Medical Center, STR analysis is performed using a commercially available kit for simultaneous amplification of 9 STR loci, as well as the amelogenin gene on the X and Y chromosomes. Several loci are examined so that chromosomal rearrangement or deletion, which is frequently observed in leukemia patients, is less likely to affect the results. To assess the posttransplant engraftment status, the STR profiles of the donor and the recipient are first established before transplantation (Figure 2). PCR analysis of STR loci highlights differences among pretransplant donor and recipient leukocytes. Loci that are different between donor and recipient are considered informative and are subsequently monitored after transplantation to determine engraftment status. The STR profile of the posttransplantation sample is compared with that of the donor sample and the original recipient sample. The presence of only donor alleles indicates a complete engraftment, whereas a mixed chimerism shows the presence of recipient and donor markers at the different loci.
Figure 2.
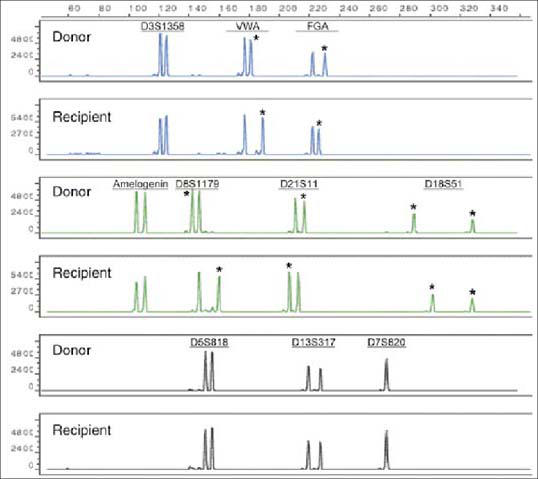
STR profiles of the recipient and donor before transplantation. The asterisks indicate the informative alleles at each locus.
The STR assay is also useful in chimerism analysis after liver transplantation. Four percent of donor lymphocytes (transient lymphocytes) are present for up to 3 weeks after transplantation. If the donor lymphocytes persist, they can cause graft-versus-host disease (GVHD), which is an underdiagnosed and often fatal complication that occurs in approximately 1% of cases, usually 2 to 6 weeks after transplantation. The following cases illustrate the utility of the STR assay in chimerism analysis.
A liver transplant recipient presented with a skin rash 2 weeks after transplant, and a punch biopsy of the skin was obtained. The pathologist noted the presence of a lymphocyte infiltrate in the dermis. Possible causes included a drug reaction, a viral infection, or early GVHD. DNA was extracted from the area of lymphocyte infiltration, and STR analysis was performed. The results showed that some of the lymphocytes were of donor origin, confirming the diagnosis of early GVHD (Figure 3).
Figure 3.
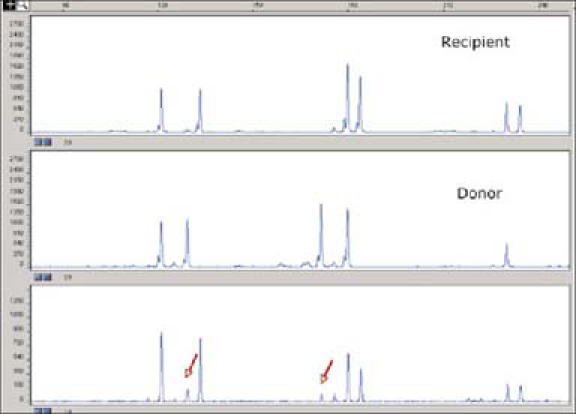
Evaluation of chimerism status by STR analysis after liver transplantation. Rows 1 and 2 show the STR profiles of the recipient and donor, respectively. Arrows indicate the presence of donor alleles in the recipient blood sample in row 3. This result supports the diagnosis of graft-versus-host disease.
In another scenario, a liver transplant recipient was diagnosed with a more severe case of GVHD a few weeks after transplantation. A blood sample from the recipient was obtained; CD3+, CD8+, and CD15+ cells were sorted; and STR analysis was performed on the DNA extracted from the fractionated cells. The recipient's T-cell profile showed 36% donor DNA, while the profile for the CD8 subset showed 84% donor DNA. The myeloid cells, however, were not of donor origin and showed no donor DNA (Figure 4). These findings confirmed the diagnosis of GVHD.
Figure 4.
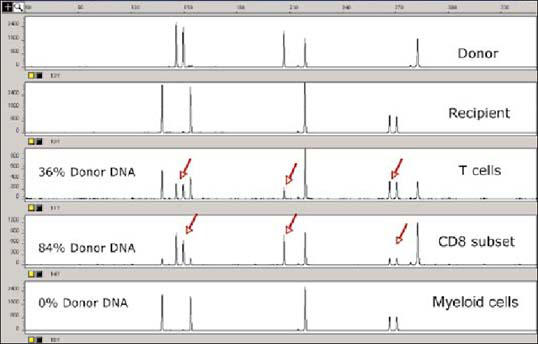
Evaluation of chimerism status by STR analysis after liver transplantation of a patient with severe graft-versus-host disease. Row 1 shows the STR profile of the donor and row 2, the recipient. After transplantation, the recipient's T-cell profile showed 36% donor DNA (row 3), while the CDS subset profile showed 84% donor DNA (row 4). The myeloid cells, however, were not of donor origin and showed no donor DNA (row 5).
Resolving specimen identity
In anatomic pathology, DNA identity testing can be used to determine the origin of mislabeled or mishandled specimens. Even though there are strict policies for proper specimen labeling and handling, unlabeled or mislabeled samples are sometimes received. In the pathology laboratory, “floaters,” or contaminating fragments of tissue, can be transferred accidentally to the paraffinembedded tissue of another patient. The College of American Pathologists reviewed over 1 million cases from 417 institutions to determine the frequency of mislabeling and misidentification of specimens. Errors were identified in 5000 cases (18).
Two cases illustrate the use of STR analysis to resolve questions about specimen identification. In the first case a patient underwent cecum endoscopic biopsy. Histological examination of the embedded tissue showed a fragment of adenomatous tissue with high-grade dysplastic glands. Also present in the biopsy were fragments of unremarkable colonic mucosa. The pathofogist was unsure whether all the tissues had come from the same individual.
In the second case a patient had polyps removed endoscopically. Histological examination of the sigmoid polyps showed benign mucosal polypoid lesions composed of serrated glands and one fragment of invasive adenocarcinoma. The pathologist was not convinced that the fragment had come from the same individual as the rest of the specimen.
In both cases, the specimens were sent for DNA typing by PCR amplification of several STR loci to determine whether the fragment in question and the other fragments had come from the same individual. In case 1, analysis confirmed that all of the tissues were from the same patient. In case 2, the tissue in question had a different DNA pattern than the other tissues, indicating that it was a “floater” that was transferred to the paraffin section.
To confirm the findings, a blood specimen from the patient was analyzed and the STR markers were compared with those of the tissue in question; the tissue was clearly a contaminant.
CONCLUSION
Twenty years after the development of DNA fingerprinting, DNA analysis remains the key to linking suspects to biological evidence and to identifying individuals in crimes and disasters. Another important use is the establishment of paternity in custody and child support litigation. DNA profiling is used to diagnose inherited disorders and human diseases.
The list of additional uses for DNA fingerprinting continues to grow. For example, DNA markers have proven to be powerful in the study of population genetics. Molecular markers are used to detect sudden changes in populations, effects of population fragmentation, and interaction of different populations.
References
- 1.Cooper DN, Smith BA, Cooke HJ, Niemann S, Schmidtke J. An estimate of unique DNA sequence heterozygosity in the human genome. Hum Genet. 1985;69:201–205. doi: 10.1007/BF00293024. [DOI] [PubMed] [Google Scholar]
- 2.Weedn VW. Forensic DNA tests. Clin Lab Med. 1996;16:187–196. [PubMed] [Google Scholar]
- 3.Jeffreys AJ, Wilson V, Thein SL. Hypervariable ‘minisatellite’ regions in human DNA. Nature. 1985;314:67–73. doi: 10.1038/314067a0. [DOI] [PubMed] [Google Scholar]
- 4.Wyman AR, White R. A highly polymorphic locus in human DNA. Proc Nat Acad Sci USA. 1980;77:6754–6758. doi: 10.1073/pnas.77.11.6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeffreys AJ, Wilson V, Thein SL. Individual-specific ‘fingerprints’ of human DNA. Nature. 1985;316:76–79. doi: 10.1038/316076a0. [DOI] [PubMed] [Google Scholar]
- 6.Gill P, Jeffreys AJ, Werrett DJ. Forensic application of DNA ‘fingerprints.’. Nature. 1985;318:577–579. doi: 10.1038/318577a0. [DOI] [PubMed] [Google Scholar]
- 7.Jeffreys AJ, Brookfield JF, Semeonoff R. Positive identification of an immigration test-case using human DNA fingerprints. Nature. 1985;317:818–819. doi: 10.1038/317818a0. [DOI] [PubMed] [Google Scholar]
- 8.Wambaugh J. The Blooding. New York: Bantam Books; 1989. [Google Scholar]
- 9.Tautz D. Notes on the definition and nomenclature of tandemly repetitive DNA sequences. EXS. 1993;67:21–28. doi: 10.1007/978-3-0348-8583-6_2. [DOI] [PubMed] [Google Scholar]
- 10.Katti MV, Ranjekar PK, Gupta VP. Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol Biol Evol. 2001;18:1161–1167. doi: 10.1093/oxfordjournals.molbev.a003903. [DOI] [PubMed] [Google Scholar]
- 11.Bennett P. Demystified … microsatellites. Mol Pathol. 2000;53:177–183. doi: 10.1136/mp.53.4.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber JL. Human DNA polymorphisms and methods of analysis. Curr Opin Biotechnol. 1990;1:166–171. doi: 10.1016/0958-1669(90)90026-h. [DOI] [PubMed] [Google Scholar]
- 13.Koreth J, O'Leary JJ, O'DMcGee J. Microsatellites and PCR genomic analysis. J Pathol. 1996;178:239–248. doi: 10.1002/(SICI)1096-9896(199603)178:3<239::AID-PATH506>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 14.Jeffreys AJ, Allen MJ, Hagelberg E, Sonnberg A. Identification of the skeletal remains of Josef Mengele by DNA analysis. Forensic Sci Int. 1992;56:65–76. doi: 10.1016/0379-0738(92)90148-p. [DOI] [PubMed] [Google Scholar]
- 15.McEwen JE. Forensic DNA data banking by state crime laboratories. Am J Hum Genet. 1995;56:1487–1492. [PMC free article] [PubMed] [Google Scholar]
- 16.Moretti TR, Baumstark AL, Defenbaugh DA, Keys KM, Smerick JB, Budowle B. Validation of short tandem repeats (STRs) for forensic usage: performance testing of fluorescent multiplex STR systems and analysis of authentic and simulated forensic samples. J Forensic Sci. 2001;46:647–660. [PubMed] [Google Scholar]
- 17.Antin JH, Childs R, Filipovich AH, Giralt S, Mackinnon S, Spitzer T, Weisdorf D. Establishment of complete and mixed donor chimerism after allogeneic lymphohematopoietic transplantation: recommendations from a workshop at the 2001 Tandem Meetings of the International Bone Mar row Transplant Registry and the American Society of Blood and Marrow Transplantation. Biol Blood Marrow Transplant. 2001;7:473–485. doi: 10.1053/bbmt.2001.v7.pm11669214. [DOI] [PubMed] [Google Scholar]
- 18.Nakhleh RE, Zarbo RJ. Surgical pathology specimen identification and ac cessioning: a College of American Pathologists Q-Probes Study of 1,004,115 cases from 417 institutions. Arch Pathol Lab Med. 1996;120:227–233. [PubMed] [Google Scholar]


