Abstract
The need to support bioinformatics training has been widely recognized by scientists, industry, and government institutions. However, the discussion of instructional methods for teaching bioinformatics is only beginning. Here we report on a systematic attempt to design two bioinformatics workshops for graduate biology students on the basis of Gagne's Conditions of Learning instructional design theory. This theory, although first published in the early 1970s, is still fundamental in instructional design and instructional technology. First, top-level as well as prerequisite learning objectives for a microarray analysis workshop and a primer design workshop were defined. Then a hierarchy of objectives for each workshop was created. Hands-on tutorials were designed to meet these objectives. Finally, events of learning proposed by Gagne's theory were incorporated into the hands-on tutorials. The resultant manuals were tested on a small number of trainees, revised, and applied in 1-day bioinformatics workshops. Based on this experience and on observations made during the workshops, we conclude that Gagne's Conditions of Learning instructional design theory provides a useful framework for developing bioinformatics training, but may not be optimal as a method for teaching it.
Keywords: instructional design theory, Gagne, conditions of learning, bioinformatics, graduate
INTRODUCTION
In the past few years, interest in bioinformatics has grown dramatically, with many universities, for-profit organizations, and nonprofit organizations now offering programs and courses in bioinformatics (for review, see Hemminger, 2004; Luo, 2002; Samish, 2003). Behind this growing interest is a real need for newcomers to develop the emerging technology and for existing scientists to utilize it effectively in their job (Ben-Dor et al., 2003). Education and training are essential in order to fill in these needs.
The need to support bioinformatics education has been widely recognized by scientists and industry, as well as by government institutes (e.g., Altman, 1998; Brass, 2000; Gavaghan, 2000; MacLean and Miles, 1999). In a 1998 report submitted to the White House Office of Science and Technology Policy, it was declared that “There is a national need for training and education in bioinformatics” (Bioinformatics in the 21st century, 1998). In 2001, the National Institutes of Health (NIH) and the National Science Foundation (NSF) conducted a workshop in an attempt to assess needs in bioinformatics research, training, education, and career development and to develop a list of recommendations to address identified gaps (Swaja et al., 2001, p. 1). In Israel, where the program described in this essay took place, the Ministry of Science and Technology supports a national Center of Knowledge for Bioinformatics Infrastructure (COBI), which provides training, consultation, and support services and maintains infrastructure for bioinformatics research (Center of Knowledge for Bioinformatics Infrastructure, 2004). Editorials (e.g., Brass, 2000; Gavaghan, 2000; Pearson, 2001) and scientific conferences (e.g., Workshop on Education in Bioinformatics) also discuss bioinformatics education.
The literature on bioinformatics education often covers topics at a macro level, such as integrating bioinformatics into undergraduate and graduate programs, the desired contents of bioinformatics curricula (Altman, 1998; Feig and Jabri, 2002; Honts, 2003; Salter, 1998), what audiences should be trained, and what resources should be devoted to bioinformatics education (Swaja et al., 2001). Others provide examples of courses and ongoing bioinformatics programs (e.g., Altman and Koza, 1996; Campbell, 2003; Feig and Jabri, 2002; Jenkins, 2000; Kim, 2000; Magee et al., 2001). Delivery methods, especially distance learning, are also a major concern (e.g., Brass, 2002; Cheng, 2002).
Developing new and better instructional methods is one of the challenges facing bioinformatics educators and support services (Ben-Dor et al., 2003). Yet this issue has been largely overlooked in the scholarly discourse on bioinformatics education. A few exceptions include Abbot (2002), Cheng (2002), Choo et al. (2004), Courtois and Handel (1998), and Kim (2000). These exceptions stress the dearth in the literature rather than satisfy the need to develop better instructional methods for bioinformatics.
In this paper we report a systematic attempt to design bioinformatics training on the basis of Robert Gagne's Conditions of Learning instructional design theory (Gagne, 1977; Gagne and Briggs, 1974). To better assess the feasibility of applying this theory to bioinformatics training, two workshops were designed: a microarray analysis workshop (Shachak et al., 2003) and a primer design workshop. We begin this paper by reviewing the characteristics of instructional design theories in general and their plausible importance for bioinformatics education. Then a brief description of Gagne's theory is provided. The instructional design process is illustrated using examples from the two workshops. Finally, some qualitative empirical findings are provided and the applicability of Gagne's theory to bioinformatics education is discussed.
BACKGROUND
Instructional Design Theories: Implications for Bioinformatics Education
Instructional design theory is defined as “a theory that offers explicit guidance on how to better help people learn and develop” (Reigeluth, 1999, p. 5). Unlike other theories, instructional design theories are design oriented in nature rather than descriptive or explanatory, but they often build upon learning theories, which are explanatory. Instructional design theories provide guidelines about what methods to use in what situations (Reigeluth, 1999).
As the definition above implies, applying instructional design theories might improve teaching and learning in general. This is especially important for bioinformatics, since bioinformatics poses special challenges to instructors and developers of educational and training programs. First, unlike many other topics, bioinformatics education requires hands-on exercises. Sitting in a classroom and listening to lectures is certainly not sufficient to qualify biologists in the use of bioinformatics applications (Ben-Dor et al., 2003). Second, bioinformatics combines exact sciences (mathematics, computer science) and an empirical science (biology). Therefore, it requires biologists not only to adopt the use of new laboratory and information tools, but also to integrate theories and models rooted in other disciplines. The use of instructional design theories might help bioinformatics instructors to face these challenges by structuring educational activities and adapting teaching methods to various situations and cognitive learning processes.
Gagne's Conditions of Learning Instructional Design Theory
Conditions of Learning is an instructional design theory developed by Robert M. Gagne as an attempt to make teaching a systematic, rule-guided process (Zemke, 1999). Although first published more than 35 years ago, it is still fundamental in the fields of instructional design and instructional technology (Merrill, 2000); parts of it were included in newer theories, such as Instructional Transaction Theory (Merrill, 1999).
Gagne (1977, p. 3) defines learning as “a change in human disposition or capability which persists over a period of time, and which is not simply ascribable to process of growth.” Taking a behaviorist perspective, he argues that the best indication that learning has occurred is a change in performance. Performances can be categorized into five domains of learned capabilities: motor skills, intellectual skills, cognitive strategies, verbal information, and attitudes. According to the Conditions of Learning theory, different internal and external conditions are necessary for each learned capability. For example, to learn a new motor skill the learner must acquire other motor skills of which it is composed (internal condition) and receive feedback (external condition). Lower-level intellectual skills (e.g., concepts, rules) and information must be acquired to learn high-level intellectual skills (e.g., problem solving). Regardless of the domain of performance, Gagne (1977) argues there are eight basic forms of learning that compose all learning performances. Those basic forms of learning are extracted from previous works of behaviorists such as Pavlov, Skinner, and Thorndike (for review, Zemke, 1999).
The most significant contribution of Gagne's theory, however, is nine instructional events outlined by it. Each corresponds to a different cognitive process (Zemke, 1999). These instructional events should satisfy or provide the necessary conditions for learning and serve as the basis for designing instruction and selecting appropriate media (Gagne, 1977). The nine events are: 1) gaining attention, 2) informing the learner of the objective, 3) stimulating recall of prior knowledge, 4) presenting the stimulus, 5) providing learning guidance, 6) eliciting the performance, 7) providing feedback about performance correctness, 8) assessing performance, and 9) enhancing retention and transfer (Gagne and Briggs, 1974).
These three components of Gagne's theory—learned capabilities, the basic forms of learning, and instructional events—provide the basis for defining learning objectives and creating a sequence of instruction (Gagne and Briggs, 1974). It will be further discussed and illustrated in the following sections.
WORKSHOPS DESIGN PROCESS
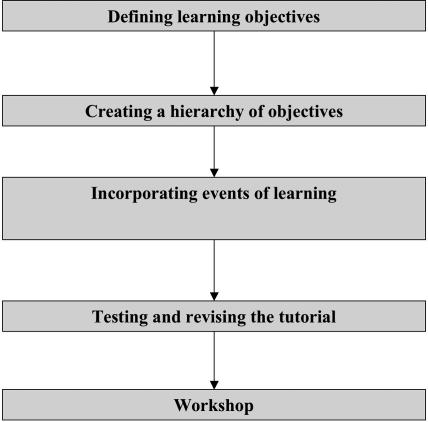
A five-step process was employed to design two bioinformatics workshops on the basis of Conditions of Learning. The process is shown in Figure 1, and each step is described in detail in the following paragraphs.
Figure 1. The process of designing a bioinformatics workshop on the basis of “Conditions of Learning.”.
Step 1: Defining Learning Objectives
The first step in developing training materials on the basis of Gagne's Conditions of Learning was to define top-level learning objectives for each workshop. Learning objectives describe what the learner should be able to do after completing the instruction. Thus, they define the type of learned capability or, in other words, the domain of performance.
According to Gagne, the only proof of learning is a change in performance. In order to clearly define the desired change in performance, learning objectives have to be observable and measurable. Gagne and Briggs (1974) suggest that all learning objectives include the following five elements: situation, learned capability, object, action, tools and other constraints. Though this exact scheme was not followed, the defined learning objectives contained most of these elements. Example 1 from the microarray analysis workshop demonstrates that.
Example 1: Learning Objective of the Microarray Analysis Workshop.
[situation] Given gene expression data, [object] the learners [learned capability: problem solving] will be able to correctly analyze the data [tools and constraints] using GeneSpring™ software.
In this example, the learned capability is described in general terms “correctly analyze.” To make this objective observable and measurable, evaluation criteria for correct analysis must be defined. Specifying learning objectives for prerequisite information and skills, as described below, can help in defining such criteria.
Step 2: Creating a Hierarchy of Objectives
The design of learning objectives is a top-to-bottom process. Example 1 above demonstrates a learned capability of higher-level intellectual skill (i.e., problem solving). Learning of intellectual skills requires that “presentation of learning situation for each new skill should be preceded by prior mastery of subordinate skills” (Gagne and Briggs, 1974, p. 105) and that “information relevant to the learning of each new skill should be previously learned or presented in instruction” (Gagne and Briggs, 1974, p. 105). Therefore, the next step was to define learning objectives for prerequisite information and skills. For each of these prerequisite objectives, the process repeated itself until reaching a level assumed to be known by all students.
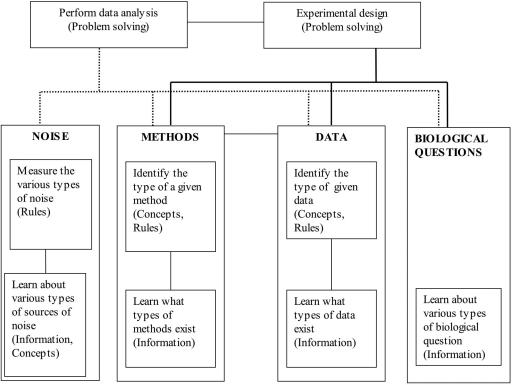
The following example from the microarray analysis workshop demonstrates this principle: in order to be able to perform microarray data analysis, one needs to measure and reduce the noise contained in microarray experimental data. This implies a learned capability of applying rules. To do that, one needs to know what creates the noise in microarray data and discriminate systemic noise from random noise. This defines learned capabilities of acquiring concepts (systemic/random noise) and recall of information. The same principle applies for methods, data, and biological questions. In addition, all four “branches” are interconnected. The result of this process is a complex hierarchy,1 as shown in Figure 2. This hierarchy of objectives provides a general framework for the microarray analysis workshop instructional sequence.
Figure 2. A hierarchy of learning objectives for the microarray analysis workshop. Topic areas are in bold uppercase, types of learned capabilities in parentheses.
Hierarchies of objectives were created for the two workshops. As described below, these hierarchies were used to design lectures and modify existing hands-on tutorials—a previously designed tutorial for the microarray analysis workshop (Ron Ophir, unpublished), and the commercial tutorial provided with the Oligo®6 demo software for the primer design workshop.
Step 3: Incorporating the Events of Learning into the Hands-on Tutorials
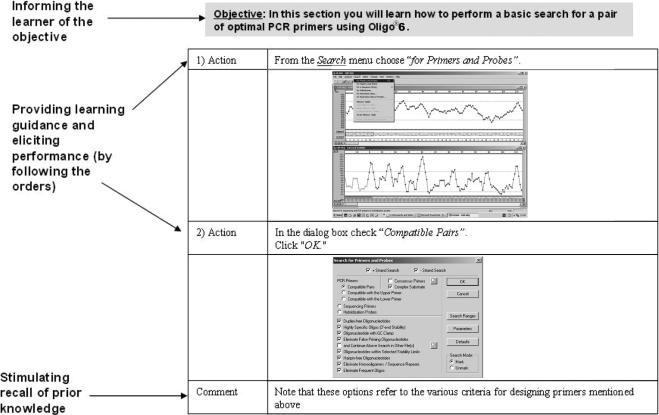
At this step, events of learning proposed by the Conditions of Learning theory were incorporated into the modified hands-on tutorials. Figure 3 demonstrates the incorporation of some learning events into the primer design hands-on tutorial.
Figure 3. Learning events as they appear in the primer design tutorial. Searching for primer pairs within a DNA sequence using Oligo®6 is illustrated. Learning objective, actions that provide learning guidance and comments to stimulate recall of prior knowledge, are labeled. To demonstrate the actions and their results, screen captures of Oligo®6 are presented.
Since the tutorials were designed for adult experienced learners, we assumed that some of the learning events are inherent in the tutorials or can be supplied by the learners themselves. For example, the instructions given in the tutorials present the stimulus material and provide learning guidance. By following these instructions the learner elicits the performance. Indeed, it has been suggested that “[a]n older, more sophisticated learner may supply most of these (learning) events by his own study effort” (Gagne and Briggs 1974, p. 135).
The following three events were deliberately incorporated into the tutorial manual: 1) informing the learner of the objective, 2) stimulating recall of prerequisite learning, and 3) providing feedback about performance correctness. The last two of Gagne's proposed events—assessing the performance and enhancing retention and transfer—were ignored because they did not fit with the voluntary nature of participating in the workshops or with the limited time frame. Incorporation of the three learning events mentioned above resulted in a first draft of the hands-on tutorial, which was tested and revised as described below.
Step 4: Testing and Revising the Tutorials
At this stage, the first draft of the tutorials was tested on a small number of trainees and revised. The microarray analysis tutorial was tested on a group of students participating in a microarray laboratory course. Two groups of two and three students were closely observed. For the primer design tutorial, it was impossible to perform the test in a real classroom situation. Therefore, it was tested by two volunteers. For each tutorial, the following information was recorded: 1) inconsistencies between the tutorial and the screen, 2) questions addressed to the instructor, 3) comments on the work process made by the students to each other and to the observer,2 4) errors made by the learners,3 and 5) suggestions for changes in the tutorial.
These observations allowed us to find errors and improve the tutorials. For example:
Some of the orders in the microarray analysis tutorial were phrased “write x,y,z in the file name line.” One observant commented that it should be reversed: “in the file name line write x,y,z.” This comment reflected that the natural cognitive process is first to identify the window elements, then to move the cursor to the right position, and finally to write the file name. Therefore, the tutorial was revised to support this cognitive process.
One of the volunteers who tested the primer design tutorial commented that working with the tutorial made him feel he did not complete the tasks. Thus, the sequence of tasks was modified to reflect a complete and systematic process.
Step 5: Conducting the Workshops
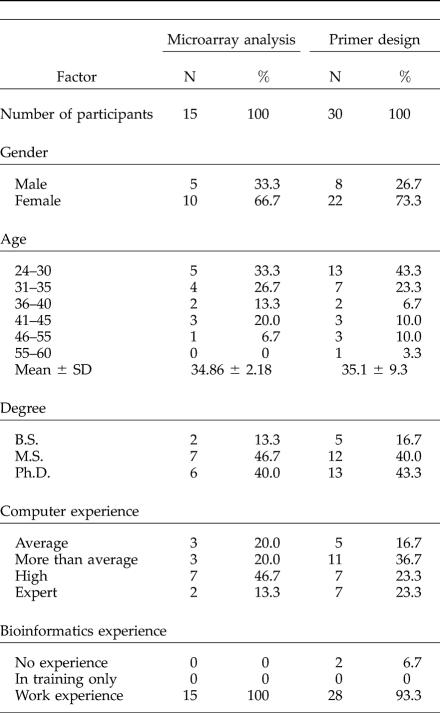
The final tutorials were employed in three 1-day workshops: one microarray analysis workshop conducted at the Weizmann Institute of Science, Israel, and two primer design workshops conducted at the Weizmann Institute and Tel-Aviv University, Israel. Each workshop consisted of introductory lectures and a hands-on session conducted in a computer laboratory. About 80 people participated in the introductory lectures for the microarray analysis workshop. Of them, 18 participated in the hands-on session. A total of 36 people attended the primer design workshops: 15 at the Weizmann Institute and 21 at Tel-Aviv University. All of them also took the hands-on session. During the workshops participants were asked to fill in questionnaires that included data on demographic variables (gender, age, degree, and role) and self-reported level of experience in using computers and bioinformatics applications. Fifteen participants in the microarray analysis workshop and 30 in the primer design workshops completed the questionnaires (response rates of 83.3 percent and 66.7 percent, respectively). Descriptive statistics for the two groups are presented in Table 1.
Table 1.
Descriptive statistics of participants in the microarray analysis and primer design workshops
During the hands-on session of the microarray analysis workshop, 5-min observations of several groups were conducted. The main findings of these observations were:
Students often skipped learning objectives, information provided to recall prior knowledge, or comments provided for feedback on performance.
Students tried to perform actions as soon as possible without taking the time to read long texts.
When a task repeated itself with modifications, students did not use the tutorial to follow the orders step-by-step, but tried to recall their previous actions from memory.
This exploratory way of learning often resulted in making errors, which required the instructor's assistance to solve.
In the primer design workshops it was impossible to conduct systematic observations. However, in one of these workshops the instructor's impression was similar to the above findings. In the other workshop, the impression was that most students did follow the tutorial step-by-step, without skipping any part of it. In that workshop, students rarely made errors and seldom needed the instructor's assistance. The implications of these findings are discussed below.
DISCUSSION
In this paper we report a systematic attempt to design bioinformatics training on the basis of Gagne's Conditions of Learning instructional design theory. We are not aware of any other attempt to apply instructional design theories to the teaching of bioinformatics. The appropriateness of Gagne's Conditions of Learning to bioinformatics training is discussed here from two different points of view: a designer's perspective and an instructor's perspective.
As mentioned earlier, newer instructional design theories build on Gagne, or have adopted some components of his theory. However, Conditions of Learning and related theories also have been criticized for being “very inefficient to use because an instructional designer must build every presentation from fundamental components” (Merrill et al., 1991). Our personal experience with designing two different bioinformatics tutorials does not support this criticism. From a designer's perspective we conclude that Conditions of Learning provides a practical framework for developing bioinformatics training in an academic setting. Specifically, defining learning objectives, top-to-bottom design, and creating a hierarchy of objectives were useful in defining priorities, structuring training activities, dividing the tutorial into units, and determining the sequence of instruction.
As a way of teaching, however, there are mixed findings regarding the effectiveness of the theory. While students in one of the primer design workshops followed the tutorial step-by-step, did not skip any parts of it, and hardly made errors, students in the two other workshops often skipped long reading paragraphs, tried to learn by exploration, and often needed the instructor's assistance to recover from errors. The instructors, especially in the microarray analysis workshop, felt they had spent more time answering questions, helping people recover from errors, and explaining issues already described in the tutorials than they initially expected.
A possible explanation for the observations in the microarray analysis workshop and one of the primer design workshops is that participants were highly experienced learners. As seen in Table 1, all participants had a B.S. degree or higher, they had more than average computer experience, and the vast majority of them had working experience with at least one bioinformatics tool, most commonly BLAST—87 percent of the microarray analysis and 83 percent of the primer design trainees used it. It is possible that such experienced learners prefer learning by exploration over structured step-by-step, although this way of learning is not supported by the tutorials described here. A similar proposition was made by researchers in human–computer interaction, who studied the way office workers learned word processing (Carroll et al., 1988). Nevertheless, the difference between the various workshops remains unexplained.
Instructional design theorists proposed that instruction based on Conditions of Learning and on related theories is often passive rather than interactive (Merrill et al., 1991). As discussed above, bioinformatics education requires interactivity in using computer applications. Together with the observations and instructors' perceptions described above, this criticism suggests that Conditions of Learning is not an optimal way to teach bioinformatics. Other learning and instructional design theories, which better support exploration and interactivity, may be sought. Some promising directions are Problem Based Learning (for review, Allen and Tanner, 2003), which has been recently applied to bioinformatics education (Choo et al., 2004), and Minimalism—a theory derived from the field of human–computer interaction, which was specifically designed to support the way people learn to use computer applications (Carroll et al., 1987–8). The feasibility and effectiveness of applying these approaches to bioinformatics teaching has yet to be verified.
LIMITATIONS AND DIRECTIONS FOR FUTURE RESEARCH
We reported here that Conditions of Learning–based instruction may not be optimal for teaching bioinformatics. This suggestion is based on observations and instructors' impressions. However, because of time limitation and the voluntary nature of participation in the workshop, summative evaluation of learning outcomes such as the assimilation of learning materials, performance with the tools, skills improved, or the transfer of knowledge to similar tasks could not be performed. Our personal experience indicates that some step-by-step tutorials of bioinformatics applications are highly effective, especially for self-study. Learning outcomes as well as the conditions under which step-by-step tutorials are useful (e.g., application or student characteristics) have yet to be studied.
Another possible direction for future research is that proposed at the end of the Discussion section: to study the effectiveness of exploration-based approaches for teaching bioinformatics such as Problem Based Learning (Allen and Tanner, 2003) or Minimalism (Carroll et al, 1987–8).
ACCESSING MATERIALS
The final microarray analysis and primer design tutorials described in this essay can be accessed from http://bioportal.weizmann.ac.il/ws/02–03/270303_ro/abstract_ro.html and http://cgr.harvard.edu/compbio/erubin/Oligo_gagne_final.pdf, respectively.
Acknowledgments
The preparation of the workshops described in this manuscript is part of a doctoral dissertation conducted at Bar-Ilan University, Israel. The workshops were hosted by the bioinformatics units of Tel-Aviv University and the Weizmann Institute of Science, Israel, and supported by COBI. The authors would like to thank Dr. Shifra Ben-Dor for her assistance in preparing the primer design workshops.
Footnotes
1The term hierarchy used by Gagne and Briggs (1974) does not necessarily imply a linear sequence of instruction. Rather, it often refers to the organization of learning objectives in a complex, networked structure as is the case here.
2The students had been informed of the observation's purpose and knew they were being watched. Though they were not requested to, the students often made comments and suggestions to the observer about the tutorial.
3Errors may be categorized as semantic errors, which arise from the use of inappropriate method in a particular situation; syntactic errors, which occur when a correct action is used improperly; and slips, such as typing mistakes (Lazonder and van der Meij, 1995). For example, in GeneSpring™, folders are displayed on the main screen navigation panel, but also in dialog windows that open. A common semantic error was selecting a folder from the main navigation panel instead of from the dialog window. In Oligo®6, a common syntactic error was that after defining the ranges in which primers are searched, people clicked “OK” instead of defining search parameters by clicking “Parameters.” As a result, the display on the screen was different than that described in the tutorial.
REFERENCES
- Abbot C. Teaching bioinformatics at the undergraduate level: is it a form of Problem Based Learning (PBL)? (2002).. Presentation at the Workshop on Education in Bioinformatics, Edmonton, Canada.
- Allen D, Tanner K.(2003).Approaches to cell biology teaching: learning content in context—problem-based learning Cell Biol. Educ 2 (2) 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman R.B.(1998).A curriculum for bioinformatics: the time is ripe Bioinformatics 14 (7) 549–550. [DOI] [PubMed] [Google Scholar]
- Altman R.B, Koza J. A programming course in bioinformatics for computer and information science students. Pac. Symp. Biocomput. (1996).. pp. 73–84. In Proc. [PubMed]
- Ben-Dor S, Butcher S, Iyer K.L, Kelso J, Littlejohn T, Shachak A, Rubin E. Training and support for bioinformatics: theoretical and practical aspects. (2003).. Paper presented at the Intelligent Systems for Molecular Biology (ISMB) conference, Brisbane, Australia.
- Bioinformatics in the 21st century. A report to the Research Resources and Infrastructure Working Group, Subcommittee on Biotechnology, National Science and Technology Council, White House Office of Science and Technology Policy. (1998).. http://clinton4.nara.gov/WH/EOP/OSTP/NSTC/html/bioinformaticsreport.html#Training (accessed 26 October 2004).
- Brass A.(2000).Bioinformatics education—a UK perspective Bioinformatics 16 (2) 77–78. [DOI] [PubMed] [Google Scholar]
- Brass A. Experiences from developing and delivering the current programme in bioinformatics by distance learning. (2002).. Presentation at the Workshop on Education in Bioinformatics (WEB), Edmonton, Canada.
- Campbell M.A.(2003).Public access for teaching genomics, proteomics, and bioinformatics Cell Biol. Educ 2 (2) 98–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll J.M, Mack R.L, Lewis C.H, Grischkowsky N.L, Robertson S.R. Exploring a wordprocessor. Effective Documentation: What We Have Learned from Research. (1988).. In: S. Doheny-Farina, ed. Cambridge, MA: The MIT Press, 103–126.
- Carroll J.M, Smith-Kerker P.L, Ford J.R, Mazur-Rimetz S.A. The minimal manual. Hum. Comput. Interact. (1987–8).;3:123–153. [Google Scholar]
- Center of Knowledge for Bioinformatics Infrastructure. (2004).. http://cobi.org.il/ (accessed 26 October 2004).
- Cheng B. The third degree: distance education and biomedical informatics. (2002).. Presentation at the Workshop on Education in Bioinformatics (WEB), Edmonton, Canada.
- Choo K.H, Tong J.C, Tan T.W, Ranganathan S. Application of Problem Based Learning pedagogy in global online bioinformatics education Georgia Tech. (2004).. Presentation at the Workshop on Education in Bioinformatics (WEB), Glasgow, Scotland.
- Courtois M.P, Handel M.A.(1998).A collaborative approach to teaching genetics information sources Res. Strat 16 (3) 211–220. [Google Scholar]
- Feig A.L, Jabri E.(2002).Incorporation of bioinformatics exercises into the undergraduate biochemistry curriculum Biochem. Mol. Biol. Educ 30 (4) 224–231. [Google Scholar]
- Gagne R.M. Conditions of Learning, 3rd ed. New York: Holt, Rinehart and Winston; (1977).. [Google Scholar]
- Gagne R.M, Briggs L.J. Principles of Instructional Design. New York: Holt, Rinehart and Winston; (1974).. [Google Scholar]
- Gavaghan H.(2000).Europe seeks solution to bioinformatics shortfall Nature 404 (6778) 687–688. [DOI] [PubMed] [Google Scholar]
- Hemminger B. Bioinformatics program summary. (2004).. http://ils.unc.edu/bmh/bioinfo/ASIST02_bioinformatics_programs_summary.html (accessed 17 November 2004).
- Honts J.E.(2003).Evolving strategies for the incorporation of bioinformatics within the undergraduate cell biology curriculum Cell Biol. Educ 2 (4) 233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins R.O. Editorial: meeting the demand for bioinformaticians. Biochem. Mol. Biol. Educ. (2000).;28:272–273. [Google Scholar]
- Kim T.D. PCR primer design: an inquiry-based introduction to bioinformatics on the World Wide Web. Biochem. Mol. Biol. Educ. (2000).;28:274–276. [Google Scholar]
- Lazonder A.W, van der Meij H.(1995).Error-information in tutorial documentation: supporting users' errors to facilitate initial skill learning. Int. J. Hum-Comput St 42 (2) 185–206. [Google Scholar]
- Luo J.(2002).Bioinformatics service, education and research: the EMBnet and CBI In Silico Biol 2 (0016) http://www.bioinfo.de/isb/2002/02/0016/main.html (accessed 26 October 2004). [PubMed] [Google Scholar]
- MacLean M, Miles C.(1999).Swift action needed to close the skills gap in bioinformatics Nature 401 (6748) 10. [DOI] [PubMed] [Google Scholar]
- Magee J, Gordon J.I, Whelan A.(2001).Bringing the human genome and the revolution in bioinformatics to the medical school classroom: a case report from Washington University School of Medicine Acad. Med 76 (8) 852–855. [DOI] [PubMed] [Google Scholar]
- Merrill M.D. Instructional Transaction Theory (ITT): instructional design based on knowledge objects. Instructional-Design Theories and Models: A New Paradigm of Instructional Theory. (1999).. In: Vol. 2, ed. C.M. Reigeluth. Mahwah, NJ: Lawrence Erlbaum Associates, 397–424.
- Merrill M.D. Suggested self-study program for instructional systems development (ISD) (2000).. http://www.id2.usu.edu/MDavidMerrill/IDREAD.PDF (accessed 17 November 2004).
- Merrill M.D, Li Z, Jones M.K.(1991).Limitations of first generation instructional design (ID1) Educ. Technol 30 (1) 7–11. [Google Scholar]
- Pearson W.R.(2001).Editorial: training for bioinformatics and computational biology Bioinformatics 17 (9) 761–762. [DOI] [PubMed] [Google Scholar]
- Reigeluth C.M. What is instructional-design theory and how is it changing? Instructional-Design Theories and Models: A New Paradigm of Instructional Theory. (1999).. In: Vol. 2, ed. C.M. Reigeluth. Mahwah, NJ: Lawrence Erlbaum Associates, 5–29.
- Salter H.(1998).Teaching bioinformatics Biochem. Educ 26 (1) 3–10. [Google Scholar]
- Samish I. Bioinformatics education in Israel: academic, private and unique retraining programs. (2003).. Presentation at the Workshop on Education in Bioinformatics (WEB03), Brisbane, Australia.
- Shachak A, Ophir R, Rubin E, Fine S.F. Instructional-design theories in bioinformatics education: an application of Gagne's “Conditions of Learning” to microarray analysis training. (2003).. Presentation at the Workshop on Education in Bioinformatics (WEB), Brisbane, Australia.
- Swaja R.E, Rastegar S, Griffith L.G, Sachs M.B, Subramaniam S. Assessing bioengineering and bioinformatics research training, education and career development: opportunities for NIH and NSF collaboration. (2001).. http://www.bisti.nih.gov/NSFNIHFinalReport824.pdf (accessed 17 November 2004).
- Zemke R.(1999).Toward a science of training Training 36 (7) 32–36. [Google Scholar]