Abstract
The properties of 23 cell-detaching Escherichia coli strains that were isolated from stool specimens in Nigeria are described. Common properties of the strains included the presence of genes encoding α-hemolysin (100%), pyelonephritis-associated pili (100%), and cytotoxic necrotizing factor 1 (70%) as well as lactose negativity (70%) and multiple antibiotic resistance (74%). Antibiotic resistance was shown in most cases to be transferable and associated with the presence of class 1 integrons. Phenotypic properties and pulsed-field gel electrophoresis analysis demonstrated that the majority of the strains, particularly multiply resistant, lactose-negative O4:H40 strains, were closely related. Multiply-resistant cell-detaching E. coli strains may represent an important reservoir for antibiotic resistance genes.
Five categories of Escherichia coli have been consistently associated with diarrhea in epidemiologic studies or produced the signs and symptoms of diarrhea when administered to healthy volunteers (14). These classes, enteropathogenic, enterotoxigenic, enteroinvasive, enterohemorrhagic, and enteroaggregative E. coli, possess specific virulence factors that are responsible for their diarrheagenicity (14). Two other categories, diffusely adherent E. coli and cytolethal distending toxin E. coli, are considered possible diarrheagenic agents because they possess putative virulence genes, but neither has been consistently associated with diarrhea in epidemiologic surveys or volunteer challenge studies (14). These categories are identified by the presence of putative virulence genes and by specific phenotypes, particularly epithelial cell adherence patterns and toxin production (14). Cell-detaching E. coli strains (CDEC) are another putative class of diarrheagenic E. coli, originally proposed by Gunzburg et al., who found this class to be significantly associated with diarrhea in a cohort of Aborigine children (7).
CDEC were originally defined by their capacity to detach tissue culture cells from solid supports in adherence assays or in a cell-detaching assay (7). In a study of the pathogenesis of CDEC, Elliott et al. reported that such strains possessed pyelonephritis-associated pili (P-pili) and produced α-hemolysin as well as cytotoxic necrotizing factor 1 (CNF1) (4). Furthermore, animal studies demonstrated that CDEC were capable of eliciting diarrhea in the reversible intestinal tie adult rabbit diarrhea model (4). These authors suggested that the term diarrhea-associated hemolytic E. coli (DHEC) be used to describe this class of E. coli. Not many other epidemiological surveys have sought this relatively recently defined category of potentially diarrheagenic E. coli. Of those that have, most have found them not to be associated with diarrhea, and their significance remains unknown (9, 12, 21).
In a recent Nigerian case-control study, although they were not significantly associated with diarrhea, CDEC were the most frequently recovered pathotype after enteroaggregative E. coli (21). In that study, only heat-stable toxin-producing E. coli and enteroaggregative E. coli were significantly associated with diarrhea (21). Considerable variation was noted among strains belonging to other categories of diarrheagenic E. coli with respect to virulence factors, biochemical properties, and serotypes (21). In contrast, the CDEC were less diverse in their characteristics: They almost universally hybridized to α-hly, pap, and cnf1 probes (for hemolysin, P-pilus. and CNF1 genes, respectively) (21). In addition, the majority of the CDEC isolates (65.2% versus 5.2% of E. coli belonging to other groups) were lactose negative. We speculated that they might represent a clone or group of clones which may or may not be pathogenic but are efficiently disseminated within the study environment.
In this paper, we characterize these strains in detail and report O4 and other antibiotic-resistant serogroups. The terminology CDEC (versus DHEC) is retained because these strains were not significantly associated with diarrhea.
The CDEC isolates described in this paper were defined by their ability to detach cells in the cell-detaching assay described by Gunzburg and colleagues (7, 21). DNA probes for CDEC-associated virulence factors (α-hly, pap, and cnf1) as well as probes for virulence factors associated with other diarrheagenic E. coli strains were employed in hybridization experiments as described previously (21). Serotyping was performed as described by Ørskov and Ørskov (22) with antisera against E. coli O antigens 1 to 173 and H antigens 1 to 56. Phage typing was done according to the protocol of Milch (13), expanded by specific phages for K1 and K5 and employing a total of 46 phages.
All the CDEC examined hybridized with the probes for pap and α-hemolysin. As shown in Table 1, most of the strains hybridized with the cnf1 probe, but this factor was not found significantly more commonly in strains from children with diarrhea than in those from healthy controls (P > 0.05). The presence of aggregative adherence plasmids in two strains and stx genes in two strains could account for the diarrheagenicity of three of these four strains irrespective of any cell-detaching properties that they may also possess.
TABLE 1.
Probes hybridizing to colony blots of CDEC isolates
| Target gene | Properties of target gene | Description of probe (reference) | Positive controls | No. (%) of CDEC strains carrying target gene
|
||
|---|---|---|---|---|---|---|
| From children with diarrhea | From healthy controls | Total | ||||
| hlyA | Hemolysin | 2.2-kb HindIII fragment from pANN215 (6) | Uropathogenic E. coli strain 536, CDEC strain A70.1 | 15 (100) | 8 (100) | 23 (100) |
| papA | Adhesin | 3-kb HindIII fragment from pRHU845 | 536, A70.1 | 15 (100) | 8 (100) | 23 (100) |
| cnfI | Cytotoxic necrotizing toxin 1 | 950-bp BamHI fragment from pSE266 (4) | 536, A70.1 | 11 (73.3) | 5 (50) | 16 (69.6) |
| stx1 and stx2 | Shiga toxins 1 and 2 | pNN110-18 (SmaI/PstI), pJN37-19 (BamHI) (17) | Enterohemorrhagic E. coli strain EDL933 | 1 (6.5) | 1 (12.5) | 2 (8.7) |
| CVD432, astA, pet | Enteroaggregative E. coli plasmid (pAA) probe; enteroaggregative heat-stable enterotoxin; and enteroaggregative plasmid-encoded enterotoxin | 0.9-kb EcoRI-PstI fragment (1); 250-bp NruI-ClaI fragment from pJPN61 (15); 2.5-kb MluI-PstI fragment from pJPN205 (5) | Enteroaggregative E. coli strain 042 | 2 (13.0) | 0 (0) | 2 (8.7) |
| intI1 | Integrase associated with class 1 integrons | 1,175-bp RsaI-BamHI fragment from Tn21 (24) | K-12 strain C600 carrying plasmid R100 | 10 (66.7) | 4 (50.0) | 14 (60.9) |
| intI2 | Integrase associated with class 2 integrons | 1,560 bp SphI-HpaI fragment from Tn7 were used as intI1 and intI2 probes (26). | K-12 strain C600 carrying plasmid R483 | 1 (6.7) | 0 (0) | 1 (4.3) |
Antibiotic susceptibility testing was done according to the standard disk diffusion method approved by the National Committee for Clinical Laboratory Standards (16). The agents tested were ampicillin (10 μg), chloramphenicol (30 μg), spectinomycin (25 μg), gentamicin (30 μg), nalidixic acid (30 μg), ofloxacin (5 μg), trimethoprim (5 μg) (Oxoid), ciprofloxacin (10 μg), sulfisomidine (250 μg), trimethoprim/sulfamethoxazole (23.8/1.2 μg), tetracycline (30 μg), mezlocillin (30 μg), piperacillin (30 μg), cefotaxime (30 μg), and amikacin (30 μg) (AB Biodisk).
Resistant strains were mated with E. coli K-12 strain C600 (supE44 hsdR thi-1 thr-1 leuB6 lacY1 tonA21 Nalr Fur) to determine if resistance markers were conjugatable (27). Transconjugants were selected on plates containing nalidixic acid (40 mg/liter) (Sigma Chemical Company, St. Louis, Mo.) with either trimethoprim (10 mg/liter) as the lactate (Wellcome), sulfathiazole (40 mg/liter) as the sodium salt (Sigma), tetracycline (40 mg/liter) as the hydrochloride (Pfizer Pharmaceuticals, Lagos, Nigeria), or chloramphenicol (40 mg/liter) (Sigma).
The susceptibility of transconjugants to ampicillin, chloramphenicol, spectinomycin, gentamicin, trimethoprim, sulfisomidine, trimethoprim/sulfamethoxazole, and tetracycline was determined by the NCCLS disk diffusion method. In these in vitro transconjugation experiments, 12 of the 13 isolates showing resistance to more than six antibiotics were capable of transferring most of these resistances horizontally (Table 2). The presence of a class 1 integron was demonstrated in 13 of 15 strains that were resistant to three or more agents by hybridization with a probe specific for the intI1 gene encoding class 1 integrons (Table 2). A class 2 integron was detected in one strain with the intI2 probe.
TABLE 2.
Properties of CDEC isolates
| Strain | Source | Serotype | Phage typea | Bio-type | Lactose fermen-tation | Colicin produc-tion | Virulence genesb | Antibiotic resistance profilec | Conjugative transfer of antibiotic resistance | intI class | Class 1 integron insertion amplified by PCR (kb) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| C06b | Diarrhea, urban | O1:K1:H7 | 3, 4 | 4a/716 | − | + | cnf1 | Cm, Te, Tp, TS, Su, St | 1 | 1.6 | |
| D03b | Diarrhea, urban | O1:K1:H7 | 4a, 4b, 15, 33 | 4a/717 | + | + | No product | ||||
| G167b | Control, rural | O2:K5:H4 | 34 | 23a/752 | − | + | cnf1, stx1, stx2 | Te | No product | ||
| G131a | Control, rural | O4:H1 | 16 | 4b/717 | − | − | cnf1 | AC, Ap, Cm, Te, Tp, TS, Sh, St, Su | Ap, Te, Tp, TS, Sh, Su, St | 1 | No product |
| E15 | Diarrhea, urban | O4:H5 | 20 | 4b/717 | + | − | cnf1 | AC, Ap, Mz, Pi, Te, Tp, TS, Sh, St, Su | Ap, Te, Tp, TS, Sh, Su, St | 1 | 1.6 |
| E01b | Diarrhea, urban | O4:H40 | 16 | 4b/717 | − | − | cnf1 | Ap, Mz, Pi, Te, Tp, TS, Sh, St, Su | Ap, Te, Tp, TS, Sh, Su, St | 1 | 1.6 |
| E26 | Diarrhea, urban | O4:H40 | 16 | 4b/717 | − | − | cnf1 | AC, Ap, Mz, Pi, Te, Tp, TS, St, Su | Ap, Tp, TS, Su, St | 1 | 1.6 |
| G30a | Diarrhea, rural | O4:H40 | 4b/717 | − | − | cnf1 | AC, Ap, Mz, Pi, Te, Tp, TS, Sh, St, Su | Ap, Te, Tp, TS, Sh, Su, St | 1 | 1.6 | |
| C62a | Control, urban | O4:H40 | 4b/717 | − | − | AC, Ap, Mz, Pi, Te, Tp, TS, Sh, St, Su | Ap, Tp, TS, Su | 1 | 1.6 | ||
| C81 | Control, urban | O4:H40 | 4b/717 | − | − | cnf1 | AC, Ap, Mz, Pi, Te, Tp, TS, Sh, St, Su | Ap, Tp, TS, Su | 1 | 1.6 | |
| E58 | Control, urban | O4:H40 | 4b/717 | − | − | cnf1 | AC, Ap, Mz, Pi, Te, Tp, TS, Sh, St, Su | Ap, Te, Tp, TS, Sh, Su, St | 1 | 1.6 | |
| G76 | Diarrhea, rural | O5:H4 | 22, 24, 33 | 9b/615 | + | − | Ap, Te, Tp, TS, Sh, St, Su | Ap, Te, Tp | 1 | 1.6 | |
| G75 | Diarrhea, urban | O15:H18 | 15 | 10a/676 | + | − | cnf1 | Te, Tp, TS, Sh, St, Su | No product | ||
| C51 | Control, urban | O16:K1:H- | 22, 24, 33 | 1b/713 | + | + | Te, Su | No product | |||
| C03 | Diarrhea, urban | O16:K1:H6 | (4a), 4b, 24, 33 | 1b/713 | + | + | cnf1 | Te, Sh, Su, St | 2 | No product | |
| C11a | Diarrhea, urban | O21:H5 | 4a, 4b | 25a/776 | − | − | cnf1 | AC, Ap, Cm, Mz, Pi, Te, Tp, TS, Sh, St, Su | Ap, Tp | 1 | 0.75 |
| G40a | Diarrhea, urban | O21:H5 | 4a, 4b | 9a/714 | − | − | cnf1 | Ap, Cm, Mz, Pi, Te, Tp, TS, Sh, St, Su | 1 | 0.75 | |
| G104a | Control, rural | O51:H10 | 2a/210 | − | − | cnf1 | No product | ||||
| E05a | Diarrhea, urban | O95:H10 | 3,6,15, 29,30 | 2b/611 HCA | − | + | cnf1 | Cm, Te | No product | ||
| E08a | Diarrhea, urban | O158:K1:H14 | 4a, 4b, 33 | 9a/634 | + | + | cnf1, stx1, stx2 | Cm, Te, St, Su | No product | ||
| E29a | Diarrhea, urban | Ont:H4 | 4a, 4b, 6 | 16a/736 | − | − | pet, astA, aafII | AC, Ap, Cm, Mz, Pi, Te, Tp, TS, Sh, Su, St | Ap, Tp, TS, Su, St | 1 | No product |
| G02a | Diarrhea, rural | Ont:H4 | 4a, 4b, (6) | 16a/736 | − | − | pet, astA, aafII | AC, Ap, Cm, Mz, Pi, Te, Tp, TS, Su, St | Ap, Tp, TS, Su, St | 1 | No product |
| G106b | Control, rural | Ont:H10 | 24 | 9b/215 | − | + | No product |
Parentheses indicate a weakly lytic reaction (10 to 50 plaques).
Other than hlyA and pap.
AC, amoxicillin-clavulanic acid; Ap, ampicillin; Mz, mezlocillin; Pi, piperacillin; Te, tetracycline; Tp, trimethoprim; TS, trimethoprim-sulfamethoxazole; Sh, spectinomycin; St, streptomycin; Su, sulfonamide.
Insert sizes in class 1 integrons were determined by amplification with primers that annealed to the 5′ and 3′ conserved ends (5′-GGC ATC CAA GCA GCA AG-3′ and 5′-AAG CAG ACT TGA CCT GA-3′, respectively) as described by Levesque et al. (11), using E. coli C600 carrying plasmid R100 as a positive control. Nine of the isolates yielded a 1.6-kb product with one identical internal HincII site, and two gave a 750-bp product. Three strains that hybridized with the intI1 probe and all those that did not failed to yield any product. Colony hybridization of other E. coli isolates recovered during the same study demonstrated that CDEC had the highest prevalence of intI1 and that strains isolated from the same host as a multiply-resistant CDEC invariably carried class 1 integrons (Table 3).
TABLE 3.
Class 1 and 2 integrons in other categories of E. coli
| E. coli isolates | No. of isolates screened | No. (%) of isolates hybridizing to probes for:
|
||
|---|---|---|---|---|
| intI1 only | intI2 only | intI1 and intI2 | ||
| CDEC | 21 | 11 (52.4) | 1 (4.8) | 0 (0) |
| Enteroaggregative-cell-detaching E. colia | 2 | 2 (100) | 0 (0) | 0 (0) |
| Enteroaggregative E. coli | 129 | 36 (27.9) | 4 (3.1) | 5 (3.9) |
| Other diarrheagenic E. coli | 49 | 11 (22.4) | 0 (0) | 0 (0) |
| Other E. coli (normal flora) | 130 | 22 (16.9) | 5 (3.8) | 3 (2.3) |
| Total | 331 | 82 (24.8) | 10 (3.0) | 8 (2.5) |
CDEC Ont:H4 isolates E29a and G02a, which bear aggregative adherence plasmids.
Serotyping revealed that most of the CDEC belonged to serotypes associated with extraintestinal disease. Three strains were O-untypeable. The remaining 20 isolates belonged to 10 different O-groups (12 serotypes), and four O-groups, O1, O4, O16, and O21, accounted for 16 (69.6%) of the isolates. Eight (34.8%) of the CDEC strains belonged to the O4 serogroup, with the serotype O4:H40 (six strains) being the most predominant. Fourteen strains produced a capsule or exopolysaccharide visible by light microscopy. K1 antigens were detected in five and K5 antigens in one of these strains.
We noted that seven of the 15 CDEC isolates from cases and none from controls were lysogenized by phages 4a and 4b (P = 0.02, Fisher’s exact test), but we did not find any serotype or other subtype to be significantly associated with diarrhea. The significance of the phage type association is not clear, and we can therefore provide no more evidence that they may be enteric pathogens, although the small numbers of isolates in each group weaken the statistical analysis.
Interestingly, serogroups of the strains and the virulence factors they possess—α-hemolysin, P-pili, and CNF1—are associated with E. coli isolates from extraintestinal infections. Fecal E. coli strains such as these, which display characteristics typical of uropathogenic strains, have been described in several studies. In some studies, they are considered a pool of potential uropathogens (29) and therefore undesirable, even if they do not cause gastrointestinal disease.
We did not find that CDEC were significantly associated with diarrhea in our studies, but this class of E. coli was isolated more frequently from cases of diarrhea and showed a stronger correlation with gastrointestinal disease than did other acknowledged diarrheal pathogens such as enteropathogenic and enteroinvasive E. coli strains (21). Four of the CDEC isolates belonged to other pathotypes. They include two Ont:H4 enteroaggregative strains from children with diarrhea that possessed genes encoding enteroaggregative heat-stable toxin (astA), plasmid-encoded toxin (pet), and aggregative adherence fimbriae II (aaf) (20). Two other strains, one of which was isolated from a child with diarrhea, carried the stx1 and stx2 genes and could therefore be classified as Shiga toxin-producing Escherichia coli.
Marques et al. (12) reported that in addition to finding no association of this class of organisms with diarrhea in Brazil, CDEC were usually isolated from hosts with diarrhea along with another pathogen. This suggests that the organisms may not be diarrheagenic but may possess the ability to persist in a host with diarrhea. This hypothesis is consistent with the observations of Wold et al. (29, 30) that P-piliated strains adhere to colonic epithelial cells and are persistent gastrointestinal colonizers of persons with asymptomatic bacteriuria. They may therefore represent a significant proportion of the resident flora in children who suffer repeated diarrhea episodes. This could be one explanation for the association of CDEC with diarrhea observed by Nicoletti et al. (18) and Gunzburg et al. (7).
As we have shown here that they often carry multiple antibiotic resistance genes on mobile elements, it is possible that they serve as efficient reservoirs for resistance that could be transferred to other commensals or pathogens. If these strains do have the ability to persist in the gut, the chances that such transfers will occur is high. We probed other E. coli isolates recovered during the same study with the IntI1 and IntI2 probes. As shown in Table 3, class 1 integrons were more prevalent among pathogenic E. coli than normal flora, and the highest prevalence was seen among CDEC. We also found that all five E. coli isolates recovered from one subject with a multiply-resistant IntI1-positive strain hybridized with the IntI1 probe.
Strains of serotypes O4:H40, O21:H5, and Ont:H4 were in all cases lactose negative, colicin negative, and resistant to 10 or more antimicrobials. The resistance was associated with the presence of class 1 integrons and transferable in mating experiments. All class 1 integrons identified in all O4:H40 isolates and one O1:K1:H7 isolate had 1.6-kb insertions indistinguishable by HincII restriction analysis. O21:H5 class 1 integrons had 750-bp insertions, while the insertions in the O4:H1 and Ont:H4 strains could not be amplified under the conditions used.
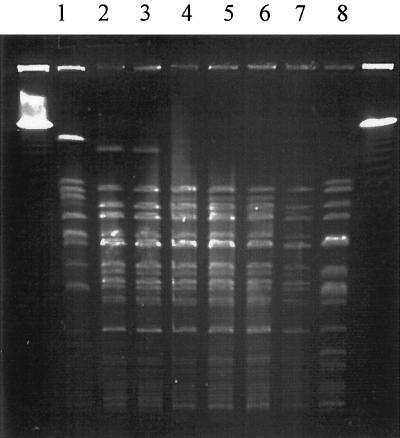
Class 1 integrons, the prototype being that borne on Tn21, have been demonstrated to be important in the evolution and dissemination of antibiotic resistance genes in E. coli from many parts of the world (8). Pulsed-field gel electrophoresis (PFGE) of SfiI-digested agarose-embedded DNA was carried out as described by Rios et al. (23), and four of the O4:H40 strains (G30a, C62a, C81, and E58) showed profiles that were indistinguishable (Fig. 1), suggesting that they are clonal (28). The other two O4 strains, E01b and E26, showed a pattern that differed only in the loss of one restriction site and could therefore be considered closely related (28).
FIG. 1.
PFGE of SfiI-digested genomic DNA from CDEC isolates. Lanes: 1, O4:H5 strain E15; 2 to 7, O4:H40 strains E01b, E26, G30a, C62a, C81, and E58, respectively; 8, O4:H1 strain G131a. Size markers are the λ pulsed-field ladder (Bio-Rad).
O4 isolates with properties similar to those of these strains have been reported from diarrhea cases elsewhere. Nicoletti et al. (18) recovered 23 lactose-negative, α-hemolytic, multiply-resistant O4 E. coli from children with diarrhea in Somalia. As in this study, they were unable to present any evidence that the strains were themselves pathogens, but they appeared to be closely related or clonal (3). An O4:H10 isolate for which no conventional virulence factors were found was also reported by Sullivan et al. (25) to be recovered from a case of persistent diarrhea in the Gambia, and a multiply-resistant, hemolytic O4 atypical enteroaggregative isolate capable of mannose-resistant hemagglutination caused an outbreak in a Serbian neonatal ward (2).
We also found indistinguishable electrophoretic patterns for both O21:H5 isolates and both O1:K1:H7 isolates (data not shown). All the isolates that showed a PFGE pattern that was seen more than once were resistant to multiple antibiotics. These data suggest that they may represent successful, multiply-resistant clones distributed within the study area. Interestingly, 11 of these 12 isolates (compared to 5 of the other 11; P < 0.02) were isolated from children residing in urban areas, where the population density is considerably greater than in rural locations. Although these strains were not associated with diarrhea, the dissemination of strains such as these is one possible factor that could account for the high prevalence of antibiotic-resistant enteric bacteria within the study area and other urban locations (10, 19). The strains also appear to strongly resemble uropathogenic E. coli, and the possibility that they have the potential to cause disease in other niches is worth examining.
Acknowledgments
This work was funded by grants from the International Program in the Chemical Sciences (NIG01) to A.L. and I.N.O. and the National Institutes of Health (AI21657 and AI41325) to J.B.K. I.N.O. thanks the Fulbright Commission and USIA for a fellowship.
We thank Michelle Trucksis, Peter Chapman, and Ola Sköld for access to facilities and helpful suggestions.
REFERENCES
- 1.Baudry, B., S. J. Savarino, P. Vial, J. B. Kaper, and M. M. Levine. 1990. A sensitive and specific DNA probe to identify enteroaggregative Escherichia coli, a recently discovered diarrheal pathogen. J. Infect. Dis 161:1249–1251. [DOI] [PubMed] [Google Scholar]
- 2.Èobeljiã, M., B. Miljkoviã-Selimoviã, D. Paunovi¿-Todosijeviã, Z. Velièkoviã, Z. Lepđanoviã, N. Zec, D. Saviã, R. Iliã, S. Konstantinoviã, B. Jovanoviã, and V. Kostiã. 1996. Enteroaggregative Escherichia coli associated with an outbreak of diarrhoea in a neonatal nursery ward. Epidemiol. Infect. 117:11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colonna, B., L. Ranucci, P. A. Fradiani, M. Casalino, A. Calconi, and M. Nicoletti. 1992. Organization of aerobactin, hemolysin, and antibacterial resistance genes in lactose-negative Escherichia coli strains of serotype O4 isolated from children with diarrhea. Infect. Immun. 60:5224–5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elliott, S. J., S. Srinivas, M. J. Albert, K. Alam, R. M. Robins-Browne, S. T. Gunzburg, B. J. Mee, and B. J. Chang. 1998. Characterization of the roles of hemolysin and other toxins in enteropathy caused by alpha-hemolytic Escherichia coli linked to human diarrhea. Infect. Immun. 66:2040–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eslava, C., F. Navarro-Garcia, J. R. Czeczulin, I. R. Henderson, A. Cravioto, and J. P. Nataro. 1998. Pet, an autotransporter enterotoxin from enteroaggregative Escherichia coli. Infect. Immun. 66:3155–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goebel, W., and J. Hedgpeth. 1982. Cloning and functional characterization of the plasmid-encoded hemolysin determinant of Escherichia coli. J. Bacteriol. 151:1290–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gunzburg, S. T., B. J. Chang, S. J. Elliott, V. Burke, and M. Gracey. 1993. Diffuse and enteroaggregative patterns of adherence of enteric Escherichia coli isolated from aboriginal children from the Kimberley region of Western Australia. J. Infect. Dis 167:755–758. [DOI] [PubMed] [Google Scholar]
- 8.Huovinen, P., L. Sundström, G. Swedberg, and O. Sköld. 1995. Trimethoprim and sulfonamide resistance. Antimicrob. Agents Chemother. 39:279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knutton, S., R. Shaw, A. D. Phillips, H. R. Smith, G. A. Willshaw, P. Watson, and E. Price. 2001. Phenotypic and genetic analysis of diarrhea-associated Escherichia coli isolated from children in the United Kingdom. J. Pediatr. Gastroenterol. Nutr. 33:32–40. [DOI] [PubMed] [Google Scholar]
- 10.Lamikanra, A., and I. N. Okeke. 1997. A study of the effect of the urban/rural divide on the incidence of antibiotic resistance in Escherichia coli. Biomed. Lett. 55:91–97. [Google Scholar]
- 11.Lévesque, C., L. Piché, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marques, L. R., C. M. Abe, P. M. Griffin, and T. A. Gomes. 1995. Association between alpha-hemolysin production and HeLa cell-detaching activity in fecal isolates of Escherichia coli. J. Clin. Microbiol. 33:2707–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milch, H. 1978. Phage typing of Escherichia coli, p.87–155. In T. Bergan and J. R. Norris (ed.), Methods in microbiology, vol. 11. Academic Press, New York, N.Y.
- 14.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nataro, J. P., D. Yikang, J. A. Giron, S. J. Savarino, M. H. Kothary, and R. Hall. 1993. Aggregative adherence fimbria I expression in enteroaggregative Escherichia coli requires two unlinked plasmid regions. Infect. Immun. 61:1126–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.National Committee for Clinical Laboratory Standards. 1990. Performance standards for antimicrobial disk susceptibility tests, 4th edition: Approved Standard NCCLS Document M7–A4. National Committee for Clinical Laboratory Standards, Villanova, Pa.
- 17.Newland, J. W., and R. J. Neill. 1988. DNA probes for Shiga-like toxins I and II and for toxin-converting bacteriophages. J. Clin. Microbiol. 26:1292–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicoletti, M., F. Superti, C. Conti, A. Calconi, and C. Zagaglia. 1988. Virulence factors of lactose-negative Escherichia coli strains isolated from children with diarrhea in Somalia. J. Clin. Microbiol. 26:524–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okeke, I. N., S. T. Fayinka, and A. Lamikanra. 2000. Antibiotic resistance trends in Escherichia coli from apparently healthy Nigerian students (1986–1998). Emerg. Infect. Dis. 6:393–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okeke, I. N., A. Lamikanra, J. Czeczulin, F. Dubovsky, J. B. Kaper, and J. P. Nataro. 2000. Heterogeneous virulence of enteroaggregative Escherichia coli strains isolated from children in Southwest Nigeria. J. Infect. Dis. 181:252–260. [DOI] [PubMed] [Google Scholar]
- 21.Okeke, I. N., A. Lamikanra, H. Steinrück, and J. B. Kaper. 2000. Characterization of Escherichia coli strains from cases of childhood diarrhea in provincial southwestern Nigeria. J. Clin. Microbiol. 38:7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ørskov, I., and F. Ørskov. 1984. Serotyping of Escherichia coli, p.43–112. In T. Bergan and J. R. Norris (ed.), Methods in microbiology, vol. 14. Academic Press, Inc., New York, N.Y.
- 23.Rios, M., V. Prado, M. Trucksis, C. Arellano, C. Borie, M. Alexandre, A. Fica, and M. M. Levine. 1999. Clonal diversity of Chilean isolates of enterohemorrhagic Escherichia coli from patients with hemolytic-uremic syndrome, asymptomatic subjects, animal reservoirs, and food products. J. Clin. Microbiol. 37:778–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simonsen, C., E. Chen, and A. Levinson. 1983. Identification of the type I trimethoprim-resistant dihydrofolate reductase specified by the Escherichia coli R-plasmid R483: comparision with prokaryotic and eukaryotic dihydrofolate reductases. J. Bacteriol. 155:1001–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sullivan, P. B., M. A. Coles, G. Aberra, and A. Ljungh. 1994. Enteropathogenic and enteroadherent-aggregative Escherichia coli in children with persistent diarrhoea and malnutrition. Ann. Trop. Paediatr. 14:105–110. [DOI] [PubMed] [Google Scholar]
- 26.Sundström, L., P. Rådström, G. Swedberg, and O. Sköld. 1988. Site-specific recombination promotes linkage between trimethoprim- and sulfonamide resistance genes: sequence characterization of dhfrV and sulI and a recombination active locus of Tn21. Mol. Gen. Genet. 213:191–201. [DOI] [PubMed] [Google Scholar]
- 27.Sundström, L., T. Vinayagamoorthy, and O. Sköld. 1987. Novel type of plasmid-borne resistance to trimethoprim. Antimicrob. Agents Chemother. 31:60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wold, A., D. Caugant, G. Lidin-Janson, P. de Man, and C. Svanborg. 1992. Resident colonic Escherichia coli strains frequently display uropathogenic characteristics. J. Infect. Dis. 165:46–52. [DOI] [PubMed] [Google Scholar]
- 30.Wold, A., M. Thorssen, S. Hull, and C. Eden. 1988. Attachment of Escherichia coli via mannose- or Galα1→4Galβ-containing receptors to human colonic epithelial cells. Infect. Immun. 56:2531–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]