Abstract
Glutathione monolayer-protected gold clusters were reacted by place exchange with 19- or 20-residue thiolated oligonucleotides. The resulting DNA/nanoparticle conjugates could be separated on the basis of the number of bound oligonucleotides by gel electrophoresis and assembled with one another by DNA-DNA hybridization. This approach overcomes previous limitations of DNA/nanoparticle synthesis and yields conjugates that are precisely defined with respect to both gold and nucleic acid content.
Keywords: DNA-DNA hybridization, monolayer-protected gold clusters, nanostructure
DNA-DNA hybridization has been exploited in the assembly of nanostructures (1-3), including nanomechanical (4, 5) and nanoelectronic (6) devices, molecular computation devices (7), biosensors, and DNA scaffolds (8). Many of these applications involve the use of DNA oligonucleotides tethered to gold nanoparticles. Additional oligonucleotides may be hybridized to these DNA/nanoparticle conjugates, or nanoparticles may be hybridized with one another.
Two types of DNA/nanoparticle conjugates have been developed for these purposes. Both types entail the coupling of oligonucleotides through terminal thiol groups to colloidal gold particles. In one case, the oligonucleotides formed the entire monolayer coating the particles (3), whereas in the other case, the oligonucleotides were incorporated in a phosphine monolayer, and particles containing discrete numbers of oligonucleotides were separated by gel electrophoresis (2, 9). A minimal length of ≈50 residues was required, both for separation by electrophoresis and hybridization with complementary DNA sequences. These limitations of shorter oligonucleotides were attributed to interaction between the DNA and the gold surface (10, 11), with the DNA “intrinsically bent” (10) and wrapped around a colloidal particle (11), and with greatest affinity for C and G residues and least for A and T residues (12).
Materials and Methods
Oligonucleotides, synthesized by MWG Biotech, High Point, NC (www.mwgbiotech.com), were as follows: 20 residues, 5′SHACAACTTTCAACAGTCTAAC-3′; 19 residues, 5′-AGGCCGCACCTAGGACGGT-3′SH; and 39 residues, complementary to 19 and 20 residues, TGTTGAAAGTTGTCAGATTGTCCGGCGTGGATCCTGCCA. Glutathione monolayer-protected clusters (MPCs) were synthesized as described (13). Band 3, with a core of average mass 9 kDa, corresponding to 46 gold atoms, and diameter of 1.2 nm, was used. Oligonucleotides (10 μl of 500 μM) were reduced by treatment with 1 μl of 50 mM Tris(2-caroxyethyl)phosphine (Sigma 646547) for 30 min at room temperature. Glutathione MPC (10 μl of 1 mM) was added, followed by incubation for 1 h at 50°C.
All gels were 15T/5C acrylamide in TBE, run at a constant 100 V.
A model of double-helical DNA with the sequence used here was generated with the use of nucleic acid builder (14). The C6 5′-thiol and C3 3′-thiol linker regions were added to the models with pymol (15). Gold clusters were approximated as spheres. The DNA models were allowed to rotate about the axis of the thiol bond and attached to the gold clusters by using in-house software.
Before staining for gold, gels were soaked in 10 mM MgCl2, 60 mM NaCl in 95% ethanol. This step shrank the gels, often revealing bands without the need for further treatment. For gold staining, gels were washed six times for 15 min in 55% ethanol, followed by incubation in a mixture of 6 ml of Silver Enhancer A, 6 ml of Silver Enhancer B (Sigma S5020 and S5145), and 14 ml of ethanol. If bands developed within 30 min, the stain was removed and the gel was returned to 55% ethanol. If no bands appeared, the gel was washed briefly with 55% ethanol and fresh stain mixture was applied (reapplication every 30 min was necessary because the stain began to precipitate nonspecifically ≈30 min after solutions A and B were mixed).
Results and Discussion
We report here on the preparation of DNA/nanoparticle conjugates from thiol MPCs (16). Inasmuch as thiols have a higher affinity for gold colloids than citrate or phosphines (17), we presumed that interactions between DNA and a gold surface would be better suppressed by thiols. From a set of 16 thiol MPCs produced by a modified Brust synthesis (18), we have so far attempted the derivatization of glutathione-protected (Fig. 1), thioglucose-protected (see Figs. 5 and 6 which are published as supporting information on the PNAS web site), and 4-mercaptobenzoic acid-protected (data not shown) MPCs with thiolated DNA. Derivatization was effected by Murray Place Exchange (19) and was successful in all cases. The resulting nanoparticle conjugates proved free from the limitations of short oligonucleotides described above. Derivatization of other thiol MPCs has not yet been attempted, but we anticipate the reaction with thiolated DNA will be generally applicable to water-soluble MPCs.
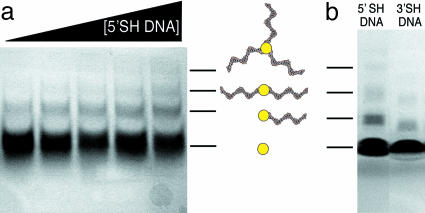
Fig. 1.
Purification of DNA/nanoparticle conjugates. (a) Reaction mixtures containing gold nanoparticles (500 μM) and increasing concentrations of 5′-thiolated 20-residue oligonucleotide (from left to right, 50, 100, 150, 200, and 250 μM) were subjected to electrophoresis in a polyacrylamide gel (15T/5C) in TBE buffer (89 mM Tris, 89 mM Borate, 2 mM EDTA, pH 8.3). (b) Reaction mixtures containing gold nanoparticles (500 μM) and either 5′-thiolated 20-residue oligonucleotide (250 μM, Left) or 3′thiolated 19-residue oligonucleotide (250 μM, Right) were subjected to electrophoresis as in a. Both gels are shown unstained. In both cases, gold staining reveals quadruple and quintuple conjugate bands (data not shown).
We selected a size-fractionated, glutathione-protected gold cluster with an ≈1.2-nm gold core for detailed analysis. The MPC was reacted with a 20-residue oligodeoxyribonucleotide, modified with an SH group on a six-carbon linker at the 5′ end. Purification and complete separation of all conjugates were achieved by preparative PAGE (Fig. 1a). Similar results were obtained with a 19-residue oligonucleotide, modified with an SH group on a three-carbon linker at the 3′ end (Fig. 1b). The reactivity of the 19-mer was slightly less, both because of the shorter linker and the shorter length of DNA. In both cases, the majority of the gold clusters did not react (Fig. 1), even in the presence of a molar excess of thiolated oligonucleotide (data not shown). Increasing the concentration of oligonucleotide increased the yield of product, yet most of the oligonucleotide and gold cluster remained unreacted (Fig. 1a). The separation by gel electrophoresis was noteworthy because of the limitation in previous work to oligonucleotides >50 residues. The glutathione monolayer evidently suppressed the DNA-gold interactions believed to interfere with separation in previous work.
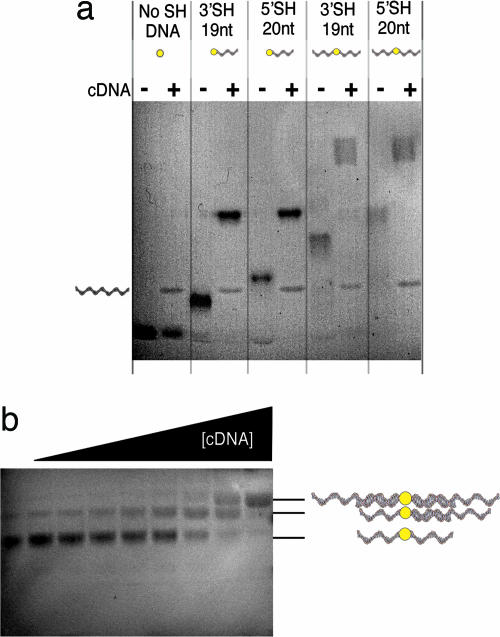
DNA/nanoparticle conjugates were hybridized with complementary DNA. For this purpose, a 39-residue oligonucleotide containing complete sequences complementary to both the 19- and 20-residue oligonucleotides was added before gel electrophoresis. Hybridization was revealed by a shift of the MPC band toward lower electrophoretic mobility (Fig. 2a). In the case of a DNA/nanoparticle conjugate believed to bear two 20-residue oligonucleotides, two shifted bands were formed (Fig. 2b). The relative intensities of the bands varied with increasing amounts of complementary oligonucleotide as expected for two oligonucleotides in the conjugate. Again, the glutathione monolayer apparently suppressed DNA-gold interactions, enabling the reactivity of oligonucleotides shorter than the limit of 50 residues reported previously.
Fig. 2.
Hybridization of DNA/nanoparticle conjugates with complementary DNA. (a) Nanoparticle conjugates containing one or two of the thiolated oligonucleotides indicated were purified from a gel as in Fig. 1a, mixed with an equal volume (5 μl) of complementary DNA (cDNA, 100 μM) or not as indicated, and subjected to electrophoresis as in Fig. 1. The gel was gold-stained. (b) Nanoparticle conjugate containing two 20-residue oligonucleotides was purified from a gel as in Fig. 1a, mixed with an equal volume (5 μl) of complementary DNA (from left to right, 0, 4 μM, 8 μM, 16 μM, 31 μM, 62 μM, 125 mM, 250 mM, and 500 mM), and subjected to electrophoresis as in Fig. 1. The gel was not stained.
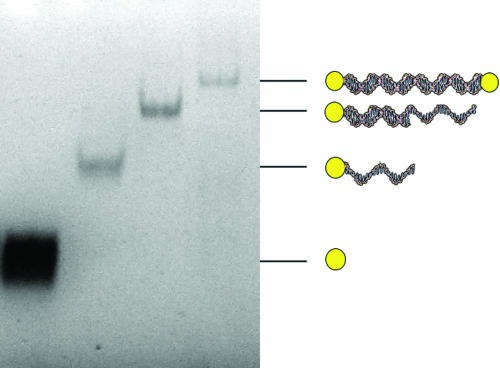
DNA/nanoparticle conjugates could be joined by hybridization to one another. For example, 19- and 20-residue oligonucleotide/nanoparticle conjugates could be joined by hybridization to the same 39-residue oligonucleotide to form a single, larger conjugate (Fig. 3). DNA/nanoparticle conjugates bearing multiple oligonucleotides can doubtless be connected in this manner to form chains and networks, although these possibilities have not yet been tested.
Fig. 3.
DNA-mediated assembly of nanoparticles. Gold nanoparticles (first lane at the left) were reacted with 20-residue oligonucleotide. The nanoparticle conjugate bearing a single DNA molecule was gel-purified (second lane), hybridized with 39-residue complementary oligonucleotide and gel-purified again (third lane), and finally combined with a slight excess of nanoparticles bearing a single 19-residue single conjugate (fourth lane). Gel electrophoresis was as in Fig. 1. The gel was not stained.
Others have suggested the use of DNA-modified nanoparticles as nanoscale construction units, with conjugates bearing four or more oligonucleotides serving as vertices, triple conjugates as corners, double conjugates as lines, and single conjugates as termini (9). Advantages of MPCs over gold colloids for this purpose include smaller size (0.8-2.5 nm in diameter) and translational mobility of monolayer components (19).
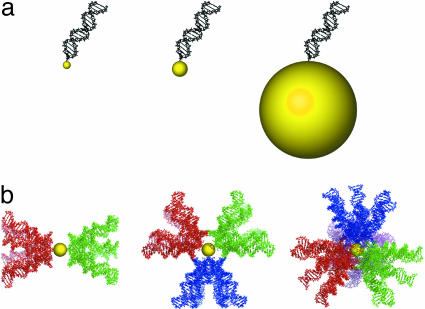
The smaller size of MPCs is advantageous because the diameters of the particles are comparable to that of duplex DNA (Fig. 4a). When multiple oligonucleotides are attached to an MPC, their locations are restricted by steric hindrance with one another. This constraint will be most pronounced for the smallest nanoparticles [0.8-nm glutathione-protected MPCs (20)]. In contrast, there is little, if any, steric limitation on the locations of oligonucleotides attached to a larger colloidal gold particle (Fig. 4a). We investigated the steric constraint in DNA/MPCs by molecular modeling. As the oligonucleotides of interest here are shorter than the persistence length of DNA, movement of an oligonucleotide relative to the gold cluster could be modeled by rotation in the linker between the DNA and gold cluster. Rotation about a single bond in the linker results in a large swept volume of the DNA (Fig. 4b). In the case of 1.2-nm nanoparticles, a colinear arrangement of two duplex DNAs is favored to minimize interaction. Three DNAs will tend to a trigonal planar arrangement and four DNAs to a tetrahedral arrangement.
Fig. 4.
Modeling of DNA/nanoparticle conjugates. (a) A 19-residue duplex DNA molecule and 0.8-, 1.2-, and 10-nm-diameter gold particles drawn to scale. (b) Scale drawing as in a, illustrating partial swept volumes for two, three, and four DNA molecules in colinear, trigonal planar, and tetrahedral arrangements. See Tables 1-3, which are published as supporting information on the PNAS web site, for the Protein Data Bank coordinates for these models.
The small size and uniformity of MPCs enables the separation of DNA/nanoparticle conjugates on the basis of DNA length at single nucleotide resolution. For example, 19- and 20-residue DNA/nanoparticle conjugates differed in electrophoretic mobility in our gels (Figs. 1b and 2a). This finding raises the possibility of separating mixed DNA/nanoparticle conjugates on the basis of sequence, so long as the DNAs of different sequence also differ in length.
Translational mobility of monolayer components is advantageous when DNA/nanoparticle conjugates are assembled into larger multiparticle structures. Mobility permits monolayer components, such as DNA, to adopt both favorable and unfavorable configurations. In the absence of external constraints, the DNAs in a triple conjugate will tend to a trigonal planar arrangement, as already mentioned. As part of a larger structure, however, the triple conjugate could form a corner.
As this work was nearing completion, a report appeared on thiol treatment for the control of DNA conformation on colloidal gold particles (21). A 15-residue oligonucleotide bearing a 5′ SH group on a six-carbon linker was adsorbed to a 9.4-nm-diameter gold particle. Passivation with 6-mercapto-1-hexanol caused a slight decrease in electrophoretic mobility, indicative of diminished direct DNA-gold interaction, and enabled hybridization with complementary oligonucleotide. Our work with glutathione-protected gold MPCs confirms the capacity of thiols to suppress DNA-gold interaction and confers the advantages of MPCs over colloidal gold mentioned above.
The chemistry we report for assembling DNA/gold nanoparticles opens the way to many applications proposed or under-taken for nanostructures in the past. The nanostructures may use gold nanoparticles as single-electron transistors (22) and have many of the topological features of integrated electronics (7, 23, 24). They may be metallized (6, 24) to create negative refractive index materials (25, 26). They may be useful as building blocks for NanoPutians (27).
Supplementary Material
Acknowledgments
This research was supported by National Institutes of Health Grants GM63025 and AI21144 (to R.D.K.), National Institutes of Health Grant GM41455 and a Howard Hughes Predoctoral Fellowship (to M.T.S.), and National Institutes of Health Training Grant T32 GM08294 (to C.J.A.).
Abbreviation: MPC, monolayer-protected cluster.
References
- 1.Zanchet, D., Micheel, C. M., Parak, W. J., Gerion, D., Williams, S. C. & Alivisatos, A. P. (2002) J. Phys. Chem. B 106, 11758-11763. [DOI] [PubMed] [Google Scholar]
- 2.Alivisatos, A. P., Johnsson, K. P., Peng, X., Wilson, T. E., Loweth, C. J., Bruchez, M. P., Jr. & Schultz, P. G. (1996) Nature 382, 609-611. [DOI] [PubMed] [Google Scholar]
- 3.Mirkin, C. A., Letsinger, R. L., Mucic, R. C. & Storhoff, J. J. (1996) Nature 382, 607-609. [DOI] [PubMed] [Google Scholar]
- 4.Hamad-Schifferli, K., Schwartz, J., Santos, A. T., Zhang, S. & Jacobson, J. M. (2002) Nature 415, 152-155. [DOI] [PubMed] [Google Scholar]
- 5.Yurke, B., Turberfield, A. J., Mills, A. P., Jr., Simmel, F. C. & Neumann, J. L. (2000) Nature 406, 605-608. [DOI] [PubMed] [Google Scholar]
- 6.Keren, K., Krueger, M., Gilad, R., Ben-Yoseph, G., Sivan, U. & Braun, E. (2002) Science 297, 72-75. [DOI] [PubMed] [Google Scholar]
- 7.Mao, C., LaBean, T. H., Reif, J. H. & Seeman, N. C. (2000) Nature 407, 493-496. [DOI] [PubMed] [Google Scholar]
- 8.Yan, H., Park, S. H., Finkelstein, G., Reif, J. H. & LaBean, T. H. (2003) Science 301, 1882-1884. [DOI] [PubMed] [Google Scholar]
- 9.Zanchet, D., Micheel, C. M., Parak, W. J., Gerion, D. & Alivisatos, A. P. (2001) Nano Lett. 1, 32-35. [Google Scholar]
- 10.Gearheart, L. A., Ploehn, H. J. & Murphy, C. J. (2001) J. Phys. Chem. B 105, 12609-12615. [Google Scholar]
- 11.Parak, W. J., Pellegrino, T., Micheel, C. M., Gerion, D., Williams, S. C. & Alivisatos, A. P. (2003) Nano Lett. 3, 33-36. [Google Scholar]
- 12.Storhoff, J. J., Elghanian, R., Mirkin, C. A. & Letsinger, R. L. (2002) Langmuir 18, 6666-6670. [Google Scholar]
- 13.Schaaff, T. G. & Whetten, R. L. (2000) J. Phys. Chem. B 104, 2630-2641. [Google Scholar]
- 14.Macke, T. & Case, D. A. (1998) in Molecular Modeling of Nucleic Acids, eds. Leontes, N. B. & SantaLucia, J. (Am. Chem. Soc., Washington, DC), pp. 379-393.
- 15.DeLano, W. L. (2002) ThepymolMolecular Graphics System (Delano Scientific, San Carlos, CA).
- 16.Templeton, A. C., Wuelfing, W. P. & Murray, R. W. (2000) Acc. Chem. Res. 33, 27-36. [DOI] [PubMed] [Google Scholar]
- 17.Warner, M. G., Reed, S. M. & Hutchison, J. E. (2000) Chem. Mater. 12, 3316-3320. [Google Scholar]
- 18.Ackerson, C. J., Jadzinsky, P. D. & Kornberg, R. D. (2005) J. Am. Chem. Soc. 127, 6550-6551. [DOI] [PubMed] [Google Scholar]
- 19.Hostetler, M. J., Templeton, A. C. & Murray, R. W. (1999) Langmuir 15, 3782-3789. [Google Scholar]
- 20.Negishi, Y., Takasugi, Y., Sato, S., Yao, H., Kimura, K. & Tsukuda, T. (2004) J. Am. Chem. Soc. 126, 6518-6519. [DOI] [PubMed] [Google Scholar]
- 21.Park, S., Brown, K. A. & Hamad-Schifferli, K. (2004) Nano Lett. 4, 1925-1929. [Google Scholar]
- 22.Sato, T., Ahmed, H., Brown, D. & Johnson, B. (1997) J. Appl. Phys. 82, 696-701. [Google Scholar]
- 23.Keren, K. & Braun, E. (2004) Chem. Eng. Technol. 27, 447-452. [Google Scholar]
- 24.Li, H., Park, S. H., Reif, J. H., LaBean, T. H. & Yan, H. (2004) J. Am. Chem. Soc. 126, 418-419. [DOI] [PubMed] [Google Scholar]
- 25.Smith, D. R., Pendry, J. B. & Wiltshire, M. C. K. (2004) Science 305, 788-792. [DOI] [PubMed] [Google Scholar]
- 26.Pendry, J. B., Martin-Moreno, L. & Garcia-Vidal, F. J. (2004) Science 305, 847-849. [DOI] [PubMed] [Google Scholar]
- 27.Chanteau, S. H. & Tour, J. M. (2003) J. Organ. Chem. 68, 8750-8766. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.