Abstract
The cause(s) of sarcoidosis is unknown. Mycobacterium spp. are suspected in Europe and Propionibacterium spp. are suspected in Japan. The present international collaboration evaluated the possible etiological links between sarcoidosis and the suspected bacterial species. Formalin-fixed and paraffin-embedded sections of biopsy samples of lymph nodes, one from each of 108 patients with sarcoidosis and 65 patients with tuberculosis, together with 86 control samples, were collected from two institutes in Japan and three institutes in Italy, Germany, and England. Genomes of Propionibacterium acnes, Propionibacterium granulosum, Mycobacterium tuberculosis, Mycobacterium avium subsp. paratuberculosis, and Escherichia coli (as the control) were counted by quantitative real-time PCR. Either P. acnes or P. granulosum was found in all but two of the sarcoid samples. M. avium subsp. paratuberculosis was found in no sarcoid sample. M. tuberculosis was found in 0 to 9% of the sarcoid samples but in 65 to 100% of the tuberculosis samples. In sarcoid lymph nodes, the total numbers of genomes of P. acnes or P. granulosum were far more than those of M. tuberculosis. P. acnes or P. granulosum was found in 0 to 60% of the tuberculosis and control samples, but the total numbers of genomes of P. acnes or P. granulosum in such samples were less than those in sarcoid samples. Propionibacterium spp. are more likely than Mycobacteria spp. to be involved in the etiology of sarcoidosis, not only in Japanese but also in European patients with sarcoidosis.
More than 100 years have passed since the first description of sarcoid lesions by Jonathon Hutchinson, but the cause of this systemic granulomatous disease is still unknown. Sarcoidosis seems to result from exposure of a genetically susceptible subject to a specific environmental agent(s) (13). Although an infectious agent is suspected, none has been identified. Similarities of clinical, histologic, and immunologic features between sarcoidosis and tuberculosis have led some to think that sarcoidosis may be an atypical kind of tuberculosis; however, Mycobacterium tuberculosis has not been isolated in culture from sarcoid lesions. Using PCR, some investigators in Europe detected mycobacterial DNA in samples of affected tissue from patients with sarcoidosis (9, 23, 26), but others did not (2, 25, 29). Other evidence of mycobacterial involvement in sarcoidosis was found by Graham et al., who obtained spheroplasts (bacteria with defective cell walls) that reverted to acid-fast bacteria by culture from skin biopsy samples from patients with sarcoidosis (10); five of the six cultured isolates were found to be Mycobacterium avium subsp. paratuberculosis by PCR (8).
In Japan, Propionibacterium acnes has been isolated in culture from biopsy samples of 31 (78%) of 40 lymph nodes from 40 patients with sarcoidosis (1). By quantitative PCR, many genomes of P. acnes or Propionibacterium granulosum were detected in all 15 biopsied lymph nodes from 15 Japanese patients with sarcoidosis (15). These results point to an etiological link between propionibacteria and sarcoidosis. Indeed, P. acnes, a strong adjuvant, causes granulomas when injected into sensitized rats (31) and rabbits (14). However, P. acnes is indigenous to the skin and intestines of healthy humans. It has been isolated in culture from 21% of 180 tissue samples from patients with diseases other than sarcoidosis (1) and detected in small amounts in 2 (13%) of 15 tuberculous lymph nodes and in 3 (20%) of 15 control lymph nodes by quantitative PCR (15).
In this study, biopsy samples of lymph nodes from patients with sarcoidosis and tuberculosis, together with control samples, were collected from two Japanese institutes in Tokyo and Kumamoto and three European institutes in Italy, Germany, and England. Genomes of P. acnes, P. granulosum, M. tuberculosis, M. avium subsp. paratuberculosis, and Escherichia coli (as the control) were counted. This collaboration compares the frequencies of detection and amounts of mycobacterial and propionibacterial DNA in samples from the different institutes and evaluates the possible etiological links between sarcoidosis and these bacteria.
MATERIALS AND METHODS
Tissue samples.
In Japan, samples were collected from Tokyo and Kumamoto, which are separated by some 910 km. In Europe, samples were collected at the Niguarda Hospital in Milan, Italy; the Ruhrlandklinik in Essen, Germany; and the Royal Brompton Hospital in London, England. Formalin-fixed and paraffin-embedded sections of biopsy samples from lymph nodes (one from each subject) were placed into tubes as described below. The tubes, without information attached about the diagnosis, were analyzed at Tokyo Medical and Dental University. The diagnosis in each case was established by one of the authors at the institute where the sample was obtained and was not communicated until all assays had been finished. The diagnosis of sarcoidosis was established by the clinical picture, no evidence of current infection by M. tuberculosis or other organisms known to produce granulomatous disease, and the presence of noncaseating granulomas in biopsy specimens of involved tissues. In one set of patients, tuberculosis was diagnosed by clinical findings, chest radiographs, and positive results of culture of sputum, bronchoalveolar lavage samples, or tissue samples affected by caseous necrosis. In subjects with suspected extrathoracic tuberculosis, however, enlarged lymph nodes were biopsied, and the histologic diagnosis was of tuberculosis if granulomas with central caseous necrosis were found. Information about the results of direct bacterial culture, if done, was not collected for either set of patients because it was not part of the criteria.
Biopsy samples from Tokyo, Kumamoto, and Italy were of superficial lymph nodes taken by incisional biopsy. In Tokyo, there were 24 patients with sarcoidosis, 13 patients with tuberculosis, and 25 patients with nonspecific lymphadenitis. In Kumamoto, there were 19 patients with sarcoidosis, 15 patients with tuberculosis, and 15 patients with nonspecific lymphadenitis. In Italy, there were 17 patients with sarcoidosis, 17 patients with tuberculosis, and 16 patients with nonspecific lymphadenitis. Samples from Germany and England were of mediastinal lymph nodes taken during mediastinoscopy. In Germany, there were 33 patients with sarcoidosis, 5 patients with tuberculosis, and 15 patients with lung cancer without metastasis. In Italy, there were 15 patients with sarcoidosis, 15 patients with tuberculosis, and 15 patients with lung cancer without metastasis.
Procedures.
Three 10-μm-thick sections cut from a paraffin block into autoclaved test tubes were treated with xylene and then with ethanol. The tissue pellets recovered were suspended in buffer ATL (Qiagen, Valencia, Calif.) and digested with proteinase K (2 mg/ml; Wako, Osaka, Japan) at 37°C overnight. DNA was extracted with a QIAamp tissue kit (Qiagen) as described in the manufacturer’s instructions. We measured the concentration of DNA by spectrophotometry. About 500 ng of DNA from each extract was used for PCR.
We used primers for real-time (TaqMan) PCR quantification to amplify 16S rRNA of P. acnes, P. granulosum, and E. coli; the insertion sequence 6110 of M. tuberculosis; the insertion sequence 900 of M. avium subsp. paratuberculosis; and the human β-globin gene. Primers PA-F (5′-GCGTGAGTGACGGTAATGGGTA-3′) and PA-R (5′-TTCCG ACGCGATCAACCA-3′) were designed to amplify a 131-bp portion of P. acnes 16S rRNA. Primers PG-F (5′-ACATGGATCCGGGAGCTTC-3′) and PG-R (5′-ACCCAAC ATCTCACGACACG-3′) were designed to amplify a 102-bp portion of P. granulosum 16S rRNA. Primers EC-F (5′-CATGCAAGTCGAACGGTAACAG-3′) and EC-R (5′-GCGACGTTATGCGGTATTAGC-3′) were designed to amplify a 135-bp portion of E. coli 16S rRNA. Primers MT-F (5′-TCCTATGACAATGCACTAGCCG-3′) and MT-R (5′-GCCAACTCGACATCCTCGAT-3′) were designed to amplify a 101-bp portion of M. tuberculosis insertion sequence 6110 (9). Primers MP-F (5′-GTTCGGGGCCGTCGC TTAGG-3′) and MP-R (5′-GCGGGCGGCCAATCTCCTT-3′) were designed to amplify a 99-bp portion of M. avium subsp. paratuberculosis insertion sequence 900 (8). Primers BG-F (5′-TGCCTATCAGAAAGTGGTGGCT-3′) and BG-R (5′-GCTCAAGGCCCTTCATAATATCC-3′) were designed to amplify a 150-bp portion of the human β-globin gene. TaqMan probes PA-TAQ (5′-AGCGTTGTCCGGATTTATTGGGCG-3′), PG-TAQ (5′-CGGTTCACAGGTGGTGCATTGGC-3′), EC-TAQ (5′-TGCTTTGCTGACGAGT GGCGGA-3′), MT-TAQ (5′-TGATCAAACCCGGCAAGCCCTGT-3′), MP-TAQ (5′-TGCCCAGGGACGTCGGGTATGG-3′), and BG-TAQ (5′-TGGCTAATGCCCTGGCCCACAA-3′) were designed to hybridize with the PCR product of P. acnes, P. granulosum, E. coli, M. tuberculosis, M. avium subsp. paratuberculosis, and human β-globin DNA, respectively. These probes were labeled with 6-carboxyfluorescein on the 5′-end and 6-carboxytetramethylrhodamine (TAMRA) on the 3′ end.
PCR was done in 50 μl of a mixture containing 5 μl of a DNA sample, 5 pmol of each primer needed, 2 pmol of a TaqMan probe, 10 nmol of each of the four deoxynucleotides, 175 nmol of MgCl2, 1.25 U of AmpliTaq Gold (PE Biosystems, Foster City, Calif.), and 1× TaqMan buffer A (PE Biosystems) to make 50 μl. Amplification and detection were done with a detection system (GeneAmp 5700; PE Biosystems) with the profile of 95°C for 5 min and 50 cycles of 95°C for 15 s and 60°C for 1 min. The TAMRA signal was used for standardization of the results. The number of bacterial genomes in tissue samples was estimated with an internal standard curve for each species, prepared with three replicates of four concentrations (10 ng, 1 ng, 100 pg, and 10 pg) of bacterial or human DNA. The concentration of DNA was expressed in terms of the number of bacterial genomes with 2.5 × 109 Da per genome, or of human genomes with 1.8 × 1012 Da per genome, used in the conversion. Finally, the result of quantitative PCR for each sample was expressed as the number of bacterial or human genomes in 500 ng of total DNA from the sample. Negative controls without bacterial DNA were included in every PCR. Assays were done three times for each sample, and the mean number of genomes was calculated.
The specificities of primers and probes for the bacterial and human DNAs tested were examined with the same procedures as for tissue samples. TaqMan analysis with each set of primers and probes for P. acnes, P. granulosum, M. tuberculosis, M. avium subsp. paratuberculosis, E. coli, or the human β-globin gene was done with 10 ng of bacterial DNA extracted from P. acnes (ATCC 6919), P. granulosum (ATCC 25564), M. tuberculosis (RIMD 1337002), M. avium subsp. paratuberculosis (ATCC 19698), E. coli (ATCC 11775), Propionibacterium avidum (ATCC 25577), Propionibacterium lymphophilum (ATCC 27520), M. avium (IID 585), Bacteroides vulgatus (ATCC 8482), Fusobacterium varium (JCM 3722), and Helicobacter pylori (ATCC 43504D) and 10 ng of human DNA extracted from normal spleen tissue.
The sensitivity and reproducibility of TaqMan PCR were evaluated with eight replicates of 10-fold serial dilutions of each bacterial DNA or human DNA.
Statistical analysis.
The chi-square test was used for analysis of the differences in frequency of detection between pairs of groups. The Mann-Whitney U test was used to compare the numbers of genomes in samples from different institutes and in samples from patients with different diseases. Differences with a P value of <0.05 were considered to be statistically significant.
RESULTS
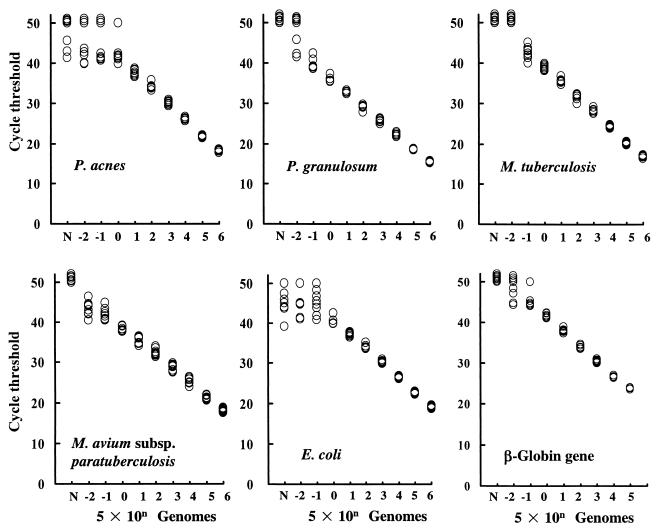
The sensitivity and reproducibility of TaqMan PCR are shown in Fig. 1. The results were reproducible for 5 or more genomes (per PCR) of P. granulosum, M. tuberculosis, M. avium subsp. paratuberculosis, and the β-globin gene and with 50 or more genomes (per PCR) of P. acnes and E. coli. No genomes of P. granulosum, M. tuberculosis, or M. avium subsp. paratuberculosis were detected in any negative controls, but fewer than 50 genomes of P. acnes or E. coli were detected in some negative controls. We therefore considered the lower limit of detection to be 5 genomes for P. granulosum, M. tuberculosis, and M. avium subsp. paratuberculosis; 5 genome equivalents for the β-globin gene; and 50 genomes for P. acnes and E. coli. TaqMan PCR specifically amplified the target bacterial DNA, giving about 2 × 106 genomes per PCR, while background values for DNA of other bacteria were below the limit of detection. TaqMan PCR of the β-globin gene specifically amplified human DNA to 3 × 103 genome equivalents per PCR, and bacterial genomes were not detected.
FIG. 1.
Standard curves of real-time (TaqMan) PCR for bacterial and human DNA. Eight samples of a given amount of bacterial DNA (from 5 × 106 to 5 × 10−2 genomes per PCR) or human DNA (from 5 × 105 to 5 × 10−2 genome equivalents per PCR) were amplified by 50 cycles of TaqMan PCR. The cycle threshold for each sample was calculated. Some datum points overlap. Abbreviations: N, negative control; n, exponent of 10.
Genome equivalents of the β-globin gene in samples ranged from 40 to 2,400 per 500 ng of total DNA, but there was no significant difference in the mean numbers detected in samples from different institutes, with means of 450 for Tokyo, 290 for Kumamoto, 860 for Italy, 180 for Germany, and 310 for England.
Results for sarcoidosis samples were as follows. Genomes of P. acnes were found in 35 (81%) Japanese, 16 (94%) Italian, 27 (82%) German, and 15 (100%) English patients, and genomes of P. granulosum were found in 23 (53%) Japanese, 14 (82%) Italian, 12 (36%) German, and 10 (67%) English patients (Table 1). There was no difference in the detection rate of P. acnes or P. granulosum between samples from the two areas of Japan and among samples from the four other countries, except for a higher frequency of P. granulosum detected in Italian patients with sarcoidosis. Genomes of M. tuberculosis were found in none to three patients with sarcoidosis in each of these institutes. Genomes of E. coli were found in lesions of two patients with sarcoidosis.
TABLE 1.
Patients in whom bacterial DNA was detected in lymph nodes
| Location of institute | Sarcoidosis patients
|
Tuberculosis patients
|
Controls
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total no. | No. (%) having:
|
Total no. | No. (%) having:
|
Total no. | No. (%) having:
|
||||||||||
| P. acnes | P. granulosum | M. tuberculosis | E. coli | P. acnes | P. granulosum | M. tuberculosis | E. coli | P. acnes | P. granulosum | M. tuberculosis | E. coli | ||||
| Tokyo | 24 | 19 (79) | 11 (46) | 0 (0) | 0 (0) | 13 | 1 (8) | 0 (0) | 13 (100)a | 0 (0) | 25 | 7 (28) | 0 (0)b | 0 (0) | 0 (0) |
| Kumamoto | 19 | 16 (84) | 12 (63) | 0 (0) | 1 (5) | 15 | 1 (7) | 1 (7) | 10 (67) | 1 (7) | 15 | 1 (7) | 0 (0)b | 0 (0) | 0 (0) |
| Italy | 17 | 16 (94) | 14 (82)c | 1 (6) | 0 (0) | 17 | 5 (29) | 2 (12) | 11 (65) | 0 (0) | 16 | 3 (19) | 3 (19) | 2 (13) | 0 (0) |
| Germany | 33 | 27 (82) | 12 (36) | 3 (9) | 1 (3) | 5 | 1 (20) | 0 (0) | 4 (80) | 0 (0) | 15 | 5 (33) | 3 (20) | 0 (0) | 0 (0) |
| England | 15 | 15 (100) | 10 (67) | 1 (7) | 0 (0) | 15 | 5 (33) | 5 (33) | 11 (73) | 0 (0) | 15 | 9 (60)d | 4 (27) | 0 (0) | 0 (0) |
The frequency of tuberculosis samples with M. tuberculosis detected in Tokyo was higher than in Kumamoto (P = 0.022), Italy (P = 0.017), and England (P = 0.044).
The frequencies of control samples with P. granulosum detected in Tokyo and Kumamoto were lower than in Italy (P = 0.25 and 0.076), Germany (P = 0.020 and 0.068), and England (P = 0.0065 and 0.032), respectively.
The frequency of sarcoidosis samples with P. granulosum detected in Italy was higher than in Tokyo (P = 0.018) and Germany (P = 0.0020).
The frequency of control samples with P. acnes detected in England was higher than in Tokyo (P = 0.046), Kumamoto (P = 0.0019), and Italy (P = 0.019) (chi-square test of proportions, pairwise).
Either P. acnes or P. granulosum was found in all but two of the patients with sarcoidosis; P. acnes but no P. granulosum was detected in about 20 to 65% of the patients with sarcoidosis, and P. granulosum but no P. acnes was detected in smaller proportions or none in these patients (Table 2). For samples with DNAs from both propionibacteria detected, the frequency of more P. acnes genomes being dominant was higher in England than in Italy and Germany, and the frequency of more P. granulosum genomes being dominant was higher in Italy than elsewhere.
TABLE 2.
Patterns of detection of different propionibacterial DNA in lymph nodes from patients with sarcoidosis
| Location of institute | na | Species detected [no. (%)]
|
||||
|---|---|---|---|---|---|---|
| Bothb
|
P. acnes onlyc | P. granulosum onlyc | Neitherd | |||
| P. acnes dominant | P. granulosum dominant | |||||
| Tokyo | 24 | 5 (20) | 1 (4) | 13 (54) | 5 (21) | 0 (0) |
| Kumamoto | 19 | 5 (26) | 5 (26) | 6 (32) | 2 (11) | 1 (5) |
| Italy | 17 | 1 (6) | 12 (71)e | 3 (18) | 1 (6) | 0 (0) |
| Germany | 33 | 2 (6) | 4 (12) | 21 (64)f | 5 (15) | 1 (3) |
| England | 15 | 6 (40)g | 4 (27) | 5 (33) | 0 (0) | 0 (0) |
n, number of patients.
“P. acnes dominant” and “P. granulosum dominant” mean that both P. acnes and P. granulosum genomes were detected but that the number of genomes of one of these species was much greater than that of the other species.
Only genomes of the indicated species were detected.
Neither P. acnes nor P. granulosum genomes were detected.
The frequency of P. granulosum-dominant samples in Italy was higher than in Tokyo (P < 0.001), Kumamoto (P =0.0079), Germany (P < 0.001), and England (P = 0.013).
Total frequency of P. acnes only in Germany was higher than in Kumamoto (P = 0.026), Italy (P = 0.0020), and England (P = 0.0064).
The frequency of P. acnes-dominant samples in England was higher than in Italy (P = 0.019) and Germany (P = 0.0035) (chi-square test of proportions, pairwise).
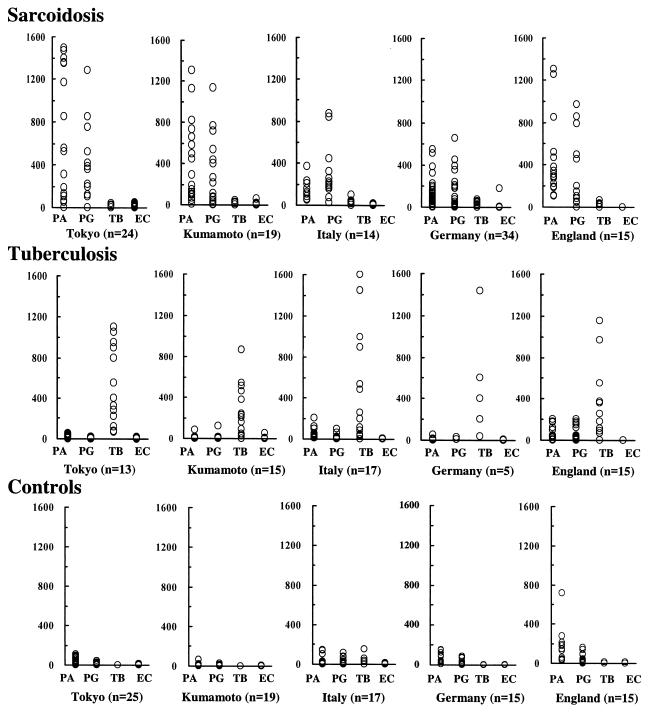
The total numbers of genomes of P. acnes or P. granulosum detected in sarcoid lymph nodes were much higher than those of M. tuberculosis detected in the same institutes (P < 0.001 in all institutes; Fig. 2).
Tuberculosis samples gave the following results. Genomes of P. acnes were found in two (7%) Japanese, five (29%) Italian, one (20%) German, and five (33%) English patients; genomes of P. granulosum were found in one (4%) Japanese, two (12%) Italian, no German, and five (33%) English patients. In control samples, genomes of P. acnes were found in eight (20%) Japanese, three (19%) Italian, five (33%) German, and nine (60%) English patients; genomes of P. granulosum were found in no Japanese, three (19%) Italian, three (20%) German, and four (27%) English patients. The frequency of detection of P. acnes in control samples was higher in England than in Tokyo, Kumamoto, or Italy.
The total numbers of genomes of P. acnes or P. granulosum detected in samples from patients with tuberculosis and control samples were lower than those detected in samples from patients with sarcoidosis, except for P. granulosum in German samples. Many genomes of P. acnes were detected in one English control sample.
Genomes of M. tuberculosis were found in none of the Japanese sarcoid samples and in 6 to 9% of European sarcoid samples, with no significant differences between institutes. Genomes of M. tuberculosis were found in all of the Tokyo patients with tuberculosis and in 65 to 80% of the tuberculosis samples from other institutes.
Genomes of M. avium subsp. paratuberculosis were not found.
DISCUSSION
There are many infectious agents that cause granulomas resembling those of sarcoidosis, including mycobacteria, herpesviruses, Histoplasma capsulatum, Treponema pallidum, Sporothrix schenckii, Coccidioides immitis, Schistosoma japonicum, Listeria monocytogenes, Rhodococcus spp., and the agent of Whipple’s disease. Sarcoidosis has a worldwide distribution, so the putative agent(s) that causes sarcoidosis also must be widely distributed.
Many causative agents of sarcoidosis have been proposed: chemical constituents of pine pollen (4), acid-fast bacilli (28), mycobacteriophage (19), some unidentified transmissible agent (20), Borrelia burgdorferi (11), M. tuberculosis (9, 23, 26), and human herpesvirus 8 (5). However, none of these proposed substances or organisms has been isolated from sarcoid lesions so far. B. burgdorferi was proposed as a possible agent of sarcoidosis by a Chinese group (11) on the basis of the elevated titers of antibodies in serum in sarcoid patients, but the same group later concluded that the elevated titers are a nonspecific response (30). Human herpesvirus 8 was proposed by a English group (5) to be a cause of sarcoidosis on the basis of the results of nested PCR, but later research on these lines by other groups (6, 16, 18) gave negative results. For these reasons, we did not search for B. burgdorferi or human herpesvirus 8 in our study.
Graham and his group isolated cell wall-defective bacteria (spheroplasts) that reverted to an acid-fast bacillus, M. avium subsp. paratuberculosis, from skin biopsy samples from six of nine patients with sarcoidosis (8, 10). In contrast, we detected genomes of M. avium subsp. paratuberculosis in none of the samples, regardless of their country of origin. It is unlikely that we failed to detect the DNA of this bacterial species because of low sensitivity of the detection method. The analytical sensitivity of TaqMan PCR is close to that of the nested PCR; both methods can detect as few as 10 copies of a standard plasmid (24). The sensitivity of the TaqMan PCR may account for the detection of a few genomes of P. acnes and E. coli in some negative controls, because traces of DNA of these indigenous microbes can be present in the reaction mixture, as suggested by Moore (22) in the case of the nested PCR of human herpesvirus 8. It is unlikely that the positive results in negative controls were caused by contamination, because amplified samples are not handled further in TaqMan PCR: the risk of contamination is lower than in conventional PCR.
P. acnes has been isolated in culture from biopsy samples of lymph nodes from Japanese patients with sarcoidosis (1). In this study, P. acnes or P. granulosum was detected in samples from Europe as well as Japan, with only two exceptions among the 108 samples from patients with sarcoidosis. One possible explanation of the exceptions is that they originally contained such genomes but the DNA denatured during preparation; consistent with this possible explanation, PCR of the β-globin gene in these two samples gave the two lowest numbers of genome equivalents of all 108 samples examined. The propionibacteria in question were found also in all 12 lymph node samples from five American patients with sarcoidosis provided by one of the authors. There was no relationship between the frequencies of detection, or numbers of propionibacterial genomes, in groups of samples from Japanese patients and the age or sex of the patient.
P. acnes and P. granulosum are indigenous bacteria found on healthy skin, and P. acnes was first isolated from a lesion of acne vulgaris. Although it is not the direct cause of acne, there are many P. acnes cells in sebaceous follicles. The number of P. acnes cells on a square centimeter of skin is generally from 102 to 106. P. granulosum has the same distribution as P. acnes but is present in smaller numbers. The possibility of contamination by propionibacteria from the skin during biopsy was excluded because DNA was extracted from histologic sections and because propionibacterial genomes were abundant in the sarcoid lesions only. The presence of organisms in tissues is considered incidental if only a few genomes are detected. The possibility that propionibacteria can be present in tissue without pathological effects was suggested by results of bacterial culture (1). Genomes of E. coli were examined as a control to eliminate the possibility that the propionibacteria detected were not specific to the disease but were simply residing in the host. Only 2 of the 108 samples from patients with sarcoidosis contained genomes of E. coli, and those patients were few. Whether or not there is an etiological connection, many propionibacteria being found only in sarcoid lesions suggests that such bacteria may be present abnormally or may proliferate ectopically in such lesions. It is unlikely that granuloma formation may bring about secondary proliferation of the bacteria in the lesions, because few propionibacteria were detected in lesions with tuberculosis, which contained granulomas similar to those of sarcoidosis.
Differences among institutes in frequencies of samples with P. acnes or P. granulosum detected in sarcoid samples did not suggest the predominance of either species of propionibacterium in sarcoid samples from patients with different genetic or environmental backgrounds. In the sarcoid samples in which we detected genomes of P. acnes or P. granulosum, histologic features were the same regardless of the species.
The greater frequencies and larger numbers of propionibacterial genomes in tuberculous and control samples from Germany and England than in those from Japan and Italy might be related to the fact that only the German and English samples were taken from mediastinal lymph nodes. We examined another 42 samples of axillary lymph nodes (with and without cancer metastasis) taken from 21 German patients with breast cancer and detected a few genomes of P. acnes in two (5%) of the samples; no P. granulosum genomes were detected. This comparison of biopsy samples of mediastinal and axillary lymph nodes from the same institute suggested that propionibacteria were detected at a higher frequency in mediastinal lymph nodes draining from the lungs, with numbers of propionibacterial genomes, when detected, being greater than in superficial lymph nodes, whether sarcoid granulomas were found or not. The finding suggests that propionibacteria reside normally not only on the skin but also in lungs in individuals without sarcoidosis, which may be why lungs and mediastinal (hilar) lymph nodes are so frequently involved in sarcoidosis.
Genomes of M. tuberculosis were not found in any of the 43 samples from Japanese patients with sarcoidosis, but a few were found in 5 (8%) of the 65 samples from European patients with sarcoidosis. In sarcoid samples containing M. tuberculosis genomes, mycobacteria might have a role in the disease, but propionibacteria are more likely to cause inflammation, because the total number of propionibacterial genomes was much larger than that of mycobacterial genomes. Small amounts of DNA unrelated to the disease also can be found by PCR. The few mycobacterial genomes found in the patients without tuberculosis might arise from bacillus Calmette-Guérin vaccination or a latent infection. The amount of DNA on the threshold of PCR detection could be affected by incidental differences between samples. Therefore, the previous results (9, 17, 23, 26) of successful detection of mycobacterial DNA by PCR may need to be reevaluated by quantitative PCR.
Many genomes of M. tuberculosis were found in 65 to 100% of samples from patients with tuberculosis, and the numbers of M. tuberculosis genomes in those samples were not very different from those of P. acnes or P. granulosum genomes in samples from patients with sarcoidosis. Failure to detect genomes of M. tuberculosis in some samples from patients with tuberculosis might be ascribed to differences in the total volumes of caseating granulomas in samples from each institute, because samples from examined patients with tuberculosis had different proportions of tuberculous lesions with central necrosis. In all samples of lymph nodes affected by tuberculosis taken in Tokyo, caseating granulomas occupied at least half of the volume and genomes of M. tuberculosis were detected without exception.
The basic concepts of virulence and pathogenicity of microbes have recently been redefined by Casadevall and Pirofski (3). They emphasize that the usual definition of a pathogen as a microorganism that causes disease in a host is inadequate; some microbes do not cause clinically evident disease in all hosts. They suggested a classification system for such pathogens based on their ability to cause damage as a function of the host’s immune response. To find the cause of sarcoidosis, we need to look at not only pathogenic bacteria such as mycobacteria but also indigenous bacteria of low virulence such as propionibacteria. Koch’s postulates for exogenous infection cannot be applied to a disease caused by endogenous bacteria. Immunologic reactions of Coombs’ type IV hypersensitivity to an antigen of indigenous bacteria may cause granuloma formation.
The inflammatory response in sarcoidosis involves many activated T cells and macrophages (12), with a pattern of cytokine production in the lungs consistent with a helper T-cell type 1 (Th1) immune response triggered by a still-undefined antigen(s) (22). If a propionibacterium caused sarcoidosis, it is likely that an antigen arising from the bacterium gave rise to a Th1 immune response in the subject. Ebe and colleagues (7) recently reported that a recombinant protein from a P. acnes DNA expression library causes a cellular immune response in some patients with sarcoidosis but not in subjects without sarcoidosis. Host factors may be more critical than agent factors in the etiology of sarcoidosis, as already suggested from the phenomenon of the Kveim test (27), in which a suspension of sarcoid tissues injected subcutaneously causes sarcoid granulomas in patients with sarcoidosis but not in healthy people or patients with other diseases. Sarcoidosis may arise from a Th1 immune response to one or more antigens of propionibacteria in an individual with a hereditary or acquired abnormality of the immune system.
FIG. 2.
TaqMan measurement of bacterial DNAs from lymph nodes of patients with sarcoidosis or tuberculosis or from control lymph nodes. The y axis shows the numbers of bacterial genomes in 500 ng of total tissue DNA extracted from samples collected at different institutes. Some datum points overlap. Groups were compared as follows (by the Mann-Whitney U test): P. acnes and M. tuberculosis, P < 0.001 in all institutes; P. granulosum and M. tuberculosis, P < 0.001 in all institutes; P. acnes, sarcoidosis, and tuberculosis, P = 0.0040 in Tokyo, P = 0.0029 in Germany, and P < 0.001 in Kumamoto, Italy, and England; P. acnes, sarcoidosis, and control, P = 0.0033 in Tokyo, P = 0.0090 in Germany, P = 0.0013 in England, and P < 0.001 in Kumamoto and Italy; P. granulosum, sarcoidosis, and tuberculosis, P = 0.23 in Germany, P = 0.085 in England, and P < 0.001 in Tokyo, Kumamoto, and Italy; P. granulosum, sarcoidosis, and control, P = 0.17 in Germany, P = 0.017 in England, and P < 0.001 in Tokyo, Kumamoto, and Italy. Abbreviations: PA, P. acnes; PG, P. granulosum; TB, M. tuberculosis; EC, E. coli.
Acknowledgments
We thank T. Suzuki of Daktari Hospital for helpful advice and discussion and J. Minami, Y. Cho, N. Soejima, N. Ando, M. Sekine, and Y. Suzuki of the Department of Human Pathology, Tokyo Medical and Dental University, for technical assistance.
This work was supported in part by grants from the Ministry of Health and Welfare, Tokyo, Japan.
REFERENCES
- 1.Abe, C., K. Iwai, R. Mikami, and Y. Hosoda. 1984. Frequent isolation of Propionibacterium acnes from sarcoidosis lymph nodes. Zentbl. Bakteriol. Mikrobiol. Hyg. A 256:541–547. [DOI] [PubMed] [Google Scholar]
- 2.Bocart, D., D. Lecossier, A. De Lassence, D. Valeyre, J. P. Battesti, and A. J. Hance. 1992. A search for mycobacterial DNA in granulomatous tissues from patients with sarcoidosis using the polymerase chain reaction. Am. Rev. Respir. Dis. 145:1142–1148. [DOI] [PubMed] [Google Scholar]
- 3.Casadevall, A., and L. A. Pirofski. 1999. Host-pathogen interactions: redefining the basic concepts of virulence and pathogenicity. Infect. Immun. 67:3703–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cummings, M. M., and P. C. Hudgins. 1958. Chemical constituents of pine pollen and their possible relationship to sarcoidosis. Am. J. Med. Sci. 236:311–317. [PubMed] [Google Scholar]
- 5.Di Alberti, L., A. Piattelli, L. Artese, G. Favia, S. Patel, N. Saunders, S. R. Porter, C. M. Scully, S. L. Ngui, and C. G. Teo. 1997. Human herpesvirus 8 variants in sarcoid tissues. Lancet 350:1655–1661. [DOI] [PubMed] [Google Scholar]
- 6.di Gennaro, G., V. Canzonieri, O. Schioppa, G. Nasti, A. Carbone, and U. Tirelli. 2000. Discordant HHV 8 detection in a young HIV-negative patient with Kaposi’s sarcoma and sarcoidosis. Clin. Infect. Dis. 32:1100–1102. [DOI] [PubMed] [Google Scholar]
- 7.Ebe, Y., S. Ikushima, T. Yamaguchi, K. Kohno, A. Azuma, K. Sato, I. Ishige, Y. Usui, T. Takemura, and Y. Eishi. 2000. Proliferative response of peripheral blood mononuclear cells and levels of antibody to recombinant protein from Propionibacterium acnes DNA expression library in Japanese patients with sarcoidosis. Sarcoidosis Vasc. Diffuse Lung Dis. 17:256–265. [PubMed] [Google Scholar]
- 8.el-Zaatari, F. A. K., S. A. Naser, D. C. Markesich, D. C. Kalter, L. Engstand, and D. Y. Graham. 1996. Identification of Mycobacterium avium complex in sarcoidosis. J. Clin. Microbiol. 34:2240–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fidler, H. M., G. A. Rook, N. M. Johnson, and J. McFadden. 1993. Mycobacterium tuberculosis DNA in tissue affected by sarcoidosis. Br. Med. J. 306:546–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham, D. Y., D. C. Markesich, D. C. Kalter, and H. H. Yoshimura. 1988. Isolation of cell wall-defective acid-fast bacteria from skin lesions of patients with sarcoidosis, p.161–164. In C. Grassi, G. Rizzato, and E. Pozzi (ed.), Sarcoidosis and other granulomatous disorders. Elsevier, Amsterdam, The Netherlands.
- 11.Hua, B., Q. D. Li, F. M. Wang, C. X. Ai, and W. C. Luo. 1992. Borrelia burgdorferi infection may be the cause of sarcoidosis. Chin. Med. J. 105:560–563. [PubMed] [Google Scholar]
- 12.Hunninghake, G. W., and R. G. Crystal. 1981. Pulmonary sarcoidosis: a disorder mediated by excess helper T-lymphocyte activity at sites of disease activity. N. Engl. J. Med. 305:429–434. [DOI] [PubMed] [Google Scholar]
- 13.Hunninghake, G. W., U. Costabel, M. Ando, R. Baughman, J. F. Cordier, R. du Bois, A. Eklund, M. Kitaichi, J. Lynch, G. Rizzato, C. Rose, O. Selroos, G. Semenzato, and O. P. Sharma. 1999. ATS/ERS/WASOG statement on sarcoidosis. Sarcoidosis Vasc. Diffuse Lung Dis. 16:149–173. [PubMed] [Google Scholar]
- 14.Ichiyasu, H., M. Suga, A. Matsukawa, K. Iyonaga, T. Mizobe, T. Takahashi, and M. Ando. 1999. Functional roles of MCP-1 in Propionibacterium acnes-induced, T cell-mediated pulmonary granulomatosis in rabbits. J. Leukoc. Biol. 65:482–491. [DOI] [PubMed] [Google Scholar]
- 15.Ishige, I., Y. Usui, T. Takemura, and Y. Eishi. 1999. Quantitative PCR of mycobacterial and propionibacterial DNA in lymph nodes of Japanese patients with sarcoidosis. Lancet 354:120–123. [DOI] [PubMed] [Google Scholar]
- 16.Lebbe, C., F. Agbalika, B. Flageul, C. Pellet, M. Rybojad, F. Cordoliani, D. Farge, M. D. Vignon-Pennamen, J. Sheldon, P. Morel, F. Calvo, and T. F. Schulz. 1999. No evidence for a role of human herpesvirus type 8 in sarcoidosis: molecular and serological analysis. Br. J. Dermatol. 141:492–496. [DOI] [PubMed] [Google Scholar]
- 17.Li, N., A. Bajoghli, A. Kubba, and J. Bhawan. 1999. Identification of mycobacterial DNA in cutaneous lesions of sarcoidosis. J. Cutan. Pathol. 26:271–278. [DOI] [PubMed] [Google Scholar]
- 18.Maeda, H., T. Niimi, S. Sato, H. Kawaguchi, Y. Sugiura, S. Mori, and R. Ueda. 2000. Human herpesvirus 8 is not associated with sarcoidosis in Japanese patients. Chest 118:923–927. [DOI] [PubMed] [Google Scholar]
- 19.Mankiewicz, E., and J. Béland. 1963. The role of mycobacteriophages and of cortisone in experimental tuberculosis and sarcoidosis. Am. Rev. Respir. Dis. 89:707–720. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell, D. N., and R. J. W. Rees. 1969. A transmissible agent from sarcoid tissue. Lancet ii:81–84. [DOI] [PubMed] [Google Scholar]
- 21.Moller, D. R., J. D. Forman, M. C. Liu, P. W. Noble, B. M. Greenlee, P. Vyas, D. A. Holden, J. M. Forrester, A. Lazarus, M. Wysocka, G. Trinchieri, and C. Karp. 1996. Enhanced expression of IL-12 associated with Th1 cytokine profiles in active pulmonary sarcoidosis. J. Immunol. 156:4952–4960. [PubMed] [Google Scholar]
- 22.Moore, P. S. 1998. Human herpesvirus 8 variants. Lancet 351:679–680. [DOI] [PubMed] [Google Scholar]
- 23.Popper, H. H., E. Winter, and G. Hofler. 1994. DNA of Mycobacterium tuberculosis in formalin-fixed, paraffin-embedded tissue in tuberculosis and sarcoidosis detected by polymerase chain reaction. Am. J. Clin. Pathol. 101:738–741. [DOI] [PubMed] [Google Scholar]
- 24.Pusterla, N., J. B. Huder, C. M. Leutenegger, U. Braun, J. E. Madigan, and H. Lutz. 1999. Quantitative real-time PCR for detection of members of the Ehrlichia phagocytophila genogroup in host animals and Ixodes ricinus ticks. J. Clin. Microbiol. 37:1329–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richter, E., U. Greinert, D. Kirsten, S. Rusch-Gerdes, C. Schluter, M. Duchrow, J. Galle, H. Magnussen, M. Schlaak, H. D. Flad, and J. Gerdes. 1996. Assessment of mycobacterial DNA in cells and tissues of mycobacterial and sarcoid lesions. Am. J. Respir. Crit. Care Med. 153:375–380. [DOI] [PubMed] [Google Scholar]
- 26.Saboor, S. A., N. M. Johnson, and J. McFadden. 1992. Detection of mycobacterial DNA in sarcoidosis and tuberculosis with polymerase chain reaction. Lancet 339:1012–1015. [DOI] [PubMed] [Google Scholar]
- 27.Siltzbach, L. E. 1961. The Kveim test in sarcoidosis: a study of 750 patients. JAMA 178:476–482. [DOI] [PubMed] [Google Scholar]
- 28.Vanek, J., and J. Schwarz. 1970. Demonstration of acid-fast rods in sarcoidosis. Am. Rev. Respir. Dis. 101:395–400. [DOI] [PubMed] [Google Scholar]
- 29.Vokurka, M., D. Lecossier, R. M. du Bois, B. Wallaert, M. Kambouchner, A. Tazi, and A. J. Hance. 1997. Absence of DNA from mycobacteria of the M. tuberculosis complex in sarcoidosis. Am. J. Respir. Crit. Care Med. 156:1000–1003. [DOI] [PubMed] [Google Scholar]
- 30.Xu, Z., D. Ma, and W. Luo. 1996. Detection of Borrelia burgdorferi DNA in granulomatous tissues from patients with sarcoidosis using polymerase chain reaction in-situ technique. Zhonghua Jie He He Hu Xi Za Zhi 19:279–281.(In Chinese with English abstract.) [PubMed] [Google Scholar]
- 31.Yi, E. S., H. Lee, Y. K. Suh, W. Tang, M. Qi, S. Yin, D. G. Remick, and T. R. Ulich. 1996. Experimental extrinsic allergic alveolitis and pulmonary angiitis induced by intratracheal or intravenous challenge with Corynebacterium parvum in sensitized rats. Am. J. Pathol. 149:1303–1312. [PMC free article] [PubMed] [Google Scholar]