Abstract
Some strains of mycoplasmas and ureaplasmas (family Mycoplasmataceae) are associated with nongonococcal urethritis (NGU) or other genitourinary infections. We have developed a rapid and reliable method of identifying the presence and prevalence of mycoplasmas and ureaplasmas in men with NGU. This method is based on the amplification of a part of the 16S rRNA gene by PCR and phylogenetic analysis. A portion of the 16S rRNA gene from 15 prototype strains was amplified with a set of common primers, and their nucleotides were sequenced. The nucleotide sequence of the V4 and V5 regions was analyzed by the neighbor-joining method. The 15 prototype strains were grouped into three distinct clusters, allowing us to clearly segregate the strains into distinct lineages. To determine the prevalence of these pathogens among patients with NGU, this protocol was tested with 148 urine samples. Amplifications were observed for 42 samples, and their nucleotide sequences were analyzed along with those of the 15 prototype strains. The phylogenetic tree thus constructed indicated that 15 of the 42 formed a cluster with Mycoplasma genitalium. Among the remaining specimens, 2 formed a cluster with Mycoplasma hominis, 19 with Ureaplasma urealyticum, and 5 with Ureaplasma parvum; the remaining sample contained both M. genitalium and U. urealyticum. This phylogeny-based identification of mycoplasmas and ureaplasmas provides not only a powerful tool for rapid diagnosis but also the basis for etiological studies of these pathogens.
Mycoplasmas and ureaplasmas, members of the family Mycoplasmataceae of the class Mollicutes, are widely distributed in humans, mammals, birds, reptiles, fish, and other vertebrates as well as plants (23). Up to now, 13 species of mycoplasmas, M. buccale, M. faucium, M. fermentans, M. genitalium, M. hominis, M. lipophilum, M. orale, M. penetrans, M. pirum, M. pneumoniae, M. primatum, M. salivarium and M. spermatophilum, and 2 species of ureaplasmas, U. parvum and U. urealyticum, have been isolated in humans (30, 31). Three of these species, M. genitalium, U. parvum, and U. urealyticum, are thought to be associated with genitourinary infections (26, 29, 30). In experimentally infected chimpanzees, M. genitalium has been shown to induce symptomatic genital infections with inflammatory and antibody responses, suggesting that M. genitalium may be a pathogen of nongonococcal urethritis (NGU) (32). Because it has been extremely difficult to isolate, M. genitalium has not been clearly designated as an etiologic agent in NGU. With the development of PCR-based techniques (14, 20), M. genitalium has been detected to a significantly greater extent in symptomatic males than in asymptomatic males, which suggests that M. genitalium is likely to be an etiologic agent in some cases of NGU (8, 11, 13, 18). For ureaplasmas, several lines of evidence have suggested that U. urealyticum also plays an important role in the etiology of male NGU. However, the colonization rates in asymptomatic males make it difficult to reach an unequivocal conclusion with respect to the etiologic role of U. urealyticum (15, 26, 30). Previously, U. urealyticum has been differentiated into biovars 1 and 2. Biovar 1 is composed of serovars 1, 3, 6, and 14, and biovar 2 is composed of serovars 2, 4, 5, and 7 to 13 (4, 22, 24). In 1998, U. urealyticum biovars 1 and 2 were classified into U. parvum and U. urealyticum, respectively (17). M. hominis has been associated with bacterial vaginosis, pelvic inflammatory disease, postpartum fever, and postabortal fever, as well as a number of gynecological infections (9, 27, 30).
Many laboratories have developed species-specific PCR techniques to detect mycoplasmas and ureaplasmas (14, 17, 20, 33) and to evaluate their ability to cause disease in various genital and urinary tract infections. However, using several species-specific PCRs to determine all human mycoplasmas and ureaplasmas associated with urethritis would be a complicated undertaking. To date, amplification of the genome by PCR, followed by determination of the nucleotide sequences and phylogenetic analysis has become a generalized technique for both the classification and identification of etiological agents (2, 3, 12, 21, 28). When the nucleotide sequences of 16S rRNA genes were compared by van Kuppeveld et al. (33), the 5′ flanking region of the variable V3 region and 3′ flanking region of the variable V6 region were well conserved among the 15 prototype strains of mycoplasmas and ureaplasmas. Therefore, we selected these conserved regions for the primers to amplify all human mycoplasmas and ureaplasmas by PCR. Consequently, a region of approximately 250 bases of V4 and V5 was employed for the phylogeny.
In the present communication, we determined the nucleotide sequences, including the V4 and V5 regions of 15 prototype strains, using PCR products. The partial 16S rRNA gene database thus constructed was used for a phylogeny-based identification of PCR products from patients with urethritis.
MATERIALS AND METHODS
Reference strains.
The prototype strains of 15 species of human mycoplasmas and ureaplasmas—M. buccale, M. faucium, M. fermentans, M. genitalium, M. hominis, M. lipophilum, M. orale, M. penetrans, M. pirum, M. pneumoniae, M. primatum, M. salivarium, M. spermatophilum, U. parvum, and U. urealyticum—were obtained from the National Institute of Infectious Diseases, Tokyo, Japan (Tsuguo Sasaki), or from the American Type Culture Collection. These strains were directly used for DNA extraction without further propagation.
Clinical specimens.
Between July 1999 and July 2000, urine samples were obtained from 148 male patients with urethritis and 42 asymptomatic males in the Department of Urology, Toyota Memorial Hospital, Toyota City, Japan. A diagnosis of urethritis was based on the observation of five or more polymorphonuclear leukocytes per oil-immersion field (magnification, × 1,000) in a Gram stain of an endourethral swab specimen. All urine specimens obtained from the symptomatic and the asymptomatic men were subjected to an AMPLICOR STD-1 assay for Chlamydia trachomatis (target sequence, 207-bp fragment in the cryptic plasmid) and for Neisseria gonorrhea (target sequence, 201-bp fragment in the putative cytosine methyltransferase gene) (Roche Diagnostics Systems, Indianapolis, Ind.) as described in the manufacturer’s instructions (5). Among the 148 urethritis patients, 55 had been diagnosed with gonococcal urethritis (GU) based on a culture positive for N. gonorrhoeae, and 47 of the remaining 93 patients were positive for Chlamydia trachomatis. The last 46 had been diagnosed with Chlamydia trachomatis-negative NGU.
DNA extraction.
The precipitates from 1 ml of urine were harvested by centrifugation at 5,000 × g for 30 min and washed with 0.9 ml of phosphate-buffered saline (pH 7.4). The precipitates were treated with proteinase K (700 μg/ml) at 55°C for 2 h in 500 μl of digestion buffer (10 mM Tris-HCl [pH 8.0], 50 mM KCl, 1.5 mM MgCl2, 0.01% gelatin, 0.45% NP-40, 0.45% Tween 20, 0.5% sodium dodecyl sulfate), and the DNA was extracted using phenol-chloroform methods (19). After ethanol precipitation, DNA was collected by centrifugation and then dissolved in 50 μl of TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA).
PCR.
A forward primer, GPO-1 (5′-ACTCCTACGGGAGGCAGCAGTA-3′), and a reverse primer, MGSO (5′-TGCACCATCTGTCACTCTGTTAACCTC-3′), were used to amplify an approximately 700-bp length (703 to 713 bp) of the 16S rRNA gene (33). Ten microliters of DNA was added to 40 μl of 1× Taq buffer (10 mM Tris-HCl [pH 8.3], 1.5 mM MgCl2, 50 mM KCl, and 0.001% gelatin) containing 0.5 μM concentrations of each of the GPO-1 and MGSO primers, 0.2 mM concentrations of deoxynucleoside triphosphates, and 1.25 U of AmpliTaq DNA polymerase (Roche Diagnostics Systems). The PCR was performed with the GeneAmp PCR System 9600 (Applied Biosystems, Foster City, Calif.) under the following conditions: 94°C for 30 s (denaturation), 64°C for 30 s (annealing), and 72°C for 60 s (elongation) for 40 cycles. To improve the efficiency of PCR, a newly designed forward primer, My-ins (5′-GTAATACATAGGTCGCAAGCGTTATC-3′), and a reverse primer, MGSO, were used for seminested PCR; 2 μl of the first PCR product was added to 48 μl of 1× Taq buffer containing 0.5 μM concentrations of each primer, 0.2 mM concentrations of deoxynucleoside triphosphates, and 1.25 U of Taq DNA polymerase. The PCR was performed under the following conditions: 94°C for 30 s (denaturation), 60°C for 30 s (annealing), and 72°C for 60 s (elongation) for 35 cycles. The PCR products were separated in 3% agarose gel and purified with a QIAquick PCR Purification kit (Qiagen, Inc., Hilden, Germany). The nucleotide sequence was determined by using a 373A DNA auto sequencer (Applied Biosystems) with an ABI Prism Dye Terminator Cycle Sequencing FS Ready Reaction kit (Applied Biosystems) and a reverse primer, My-seq2 (5′-TGCGGGTCCCCGTCAATTCC-3′). Nucleotide sequences of approximately 250 bp were determined in this study with M. buccale, M. faucium, M. fermentans, M. genitalium, M. hominis, M. lipophilum, M. orale, M. penetrans, M. pirum, M. pneumoniae, M. primatum, M. salivarium, and M. spermatophilum, U. parvum, and U. urealyticum. The 771-bp DNA fragment, including the first PCR target region of M. genitalium, was amplified and introduced into pT7Blue T-vector (Novagen, Inc., Madison, Wis.) to construct a control plasmid, pMyg16S. After purification of the plasmid using a QIAprep Spin Miniprep Kit (Qiagen), the amount of DNA was quantified by measuring the optical density at 260 nm, and the copy number was calculated. This plasmid was used to estimate the sensitivity of the PCR.
Phylogenetic analysis.
The 250-bp nucleotide sequences including the V4 and V5 regions were phylogenetically analyzed by using SINCA (Fujitsu Limited, Tokyo, Japan), a software program for genetic analysis. The evolutionary distances were estimated by using Kimura’s two-parameter method (16), and unrooted phylogenetic trees were constructed using the neighbor-joining method (25). Bootstrap analyses were performed by 1,000 resamplings of the data sets. Bootstrap values of 70% or higher were considered statistically significant for the grouping (7).
Nucleotide sequence accession numbers.
The GenBank accession numbers of the nucleotide sequences presented in this paper are AB069810 to AB069824.
RESULTS
Amplification of prototype strains.
Comparison of the nucleotide sequences of the 16S rRNA genome among the 15 prototype strains of human mycoplasmas and ureaplasmas demonstrated that the flanking regions of 5′ V3 are highly conserved among prokaryotes and that the flanking regions of 3′ V3 are conserved among mycoplasmas and ureaplasmas. Twenty-two bases in the flanking region of 5′ V3 were selected for the forward primer GPO-1, and 26 bases in the flanking region of 5′ V4 were selected for the seminested forward primer My-ins. The MGSO primer was used as a reverse primer designed specific to mycoplasmas and ureaplasmas in the flanking regions of 3′ V6 of the 16S rRNA gene as previously reported (33). The seminested PCR with the set of common primers allowed us to amplify approximately 511 to 520 bp, including the V4 and V5 regions, from the 15 prototype strains of all human mycoplasmas and ureaplasmas. The seminested PCR were specific to mycoplasmas and ureaplasmas, so there was no cross-amplification with a variety of genitourinary tract contagions, including Chlamydia trachomatis, Neisseria gonorrhoeae, Candida albicans, herpes simplex virus types 1 and 2, and adenovirus types 40 and 41 (data not shown). The sensitivity of the seminested PCR, as determined by a 520-bp band after the staining, appeared to be 10 copies per PCR tube when 10-fold serial dilutions of the plasmid pMyg16S were used for the template (data not shown). These results indicate that the seminested PCR with a set of common primers is specific to mycoplasmas and ureaplasmas and is capable of amplifying all 15 prototype strains of human mycoplasmas and ureaplasmas with high sensitivity.
Phylogenetic analysis of prototype strains.
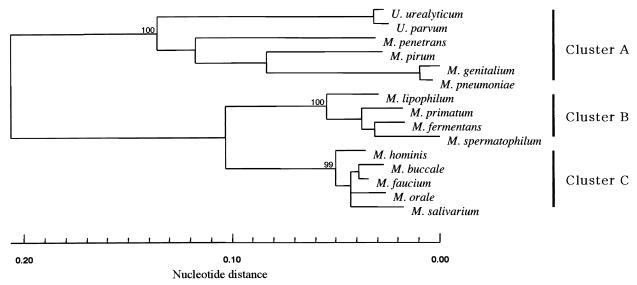
An approximately 250-bp length that included the V4 and V5 regions of each 16S rRNA gene of the 15 prototypes was sequenced, and the nucleotide sequences were aligned using the SINCA genetic software program. It appeared that there were several deletions or insertions among the prototype strains, ranging from 244 to 252 bp. To evaluate the reliability of the phylogeny-based identification of mycoplasmas and ureaplasmas, a phylogenetic tree based on partial 16S rRNA gene sequences of approximately 250 nucleotides each was constructed and was compared with those based on approximately 1,000 bp of 16S rRNA genes. As depicted in Fig. 1, human mycoplasmas and ureaplasmas were segregated into three major clusters based on partial sequences of the 16S rRNA genes. Cluster A was composed of six strains, M. pneumoniae, M. genitalium, M. pirum, M. penetrans, U. urealyticum, and U. parvum. Cluster B was composed of four strains, M. spermatophilum, M. fermentans, M. primatum, and M. lipophilum. Cluster C was composed of five strains, M. faucium, M. buccale, M. hominis, M. orale, and M. salivarium. The constellation of each strain appeared to be the same as that based on approximately 1,000 bp of the 16S rRNA gene nucleotide sequences (data not shown). The genetic relationships of the 15 prototype strains within a cluster and between clusters were further analyzed by comparing the nucleotide sequences of all these sequence pairs. The nucleotide identity within the 15 prototypes ranged from 62.7 to 98.8%, with an average nucleotide identity of 77.4%. These results indicate that the phylogenetic analysis with a partial 16S rRNA gene is capable of distinguishing among the 15 prototype strains of all human mycoplasmas and ureaplasmas.
FIG. 1.
Phylogenetic analysis of prototype strains of human mycoplasmas and ureaplasmas. The nucleotide sequences including the V4 to V5 regions (approximately 250 bp) from the 15 prototype strains were analyzed by the neighbor-joining method. The numbers at the nodes represent the percentage of 1,000 bootstrap pseudoreplicates that contained the cluster distal to the node.
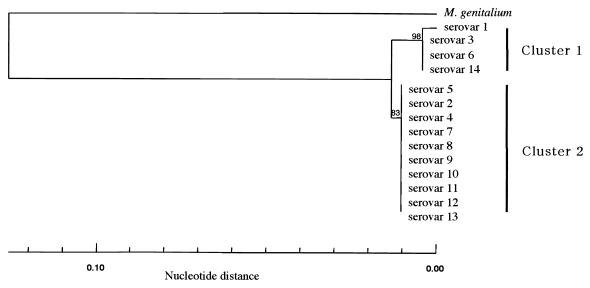
To assess genetic relationships among the 14 serovars of U. urealyticum and U. parvum, which were previously designated 14 serovars of U. urealyticum, a phylogenetic tree based on approximately 250-nucleotide sequences of the V4 and V5 regions was constructed. As depicted in Fig. 2, the 14 serovars of U. urealyticum and U. parvum were segregated into two major clusters. Cluster 1 included three serovars (serovars 3, 6, and 14) showing 100% nucleotide identity, as well as serovar 1, showing 99.6% nucleotide identity with the other serovars in this cluster. Cluster 2 included 10 serovars (serovars 2, 4, 5, and 7 to 13) with 100% nucleotide identity. Interestingly, each cluster corresponded well with the two newly designated species of ureaplasmas, U. parvum and U. urealyticum.
FIG. 2.
Phylogenetic analyses of 14 serovars of ureaplasmas. The nucleotide sequences including the V4 to V5 regions of the isolates were determined and then analyzed by the neighbor-joining method. The numbers at the nodes represent the percentage of 1,000 bootstrap pseudoreplicates that contained the cluster distal to the node. Fourteen serovars of ureaplasmas formed two distinct clusters, U. parvum (previously U. urealyticum biovar 1) and U. urealyticum (previously U. urealyticum biovar 2).
Amplification and identification of clinical isolates.
To apply the seminested PCR to amplify mycoplasmas and ureaplasmas from urine specimens, 148 urine specimens from urethritis patients and 42 urine specimens from asymptomatic men were used to amplify partial nucleotide sequences of the 16S rRNA gene. Gonococcus was found in 55 of the 148 urine specimens from the urethritis patients (GU), and C. trachomatis was detected in 47 of the residual 93 NGU specimens, by AMPLICOR STD-1 (Table 1). No gonococcus or C. trachomatis was detected in the 42 urine specimens from asymptomatic men. When approximately 520-bp lengths of mycoplasmas or ureaplasmas were amplified by the seminested PCR from the 148 urine specimens from symptomatic men, 42 specimens were positive: 9 were from gonococcus-positive urine specimens (16.4%), and 33 were from urine specimens of patients with NGU (35.5%). Among the 33 mycoplasma- or ureaplasma-positive urine specimens from NGU patients, 12 were from C. trachomatis-positive urine (25.5%) and the remaining 21 were from C. trachomatis-negative urine (45.7%). Fifteen of 42 urine samples (35.7%) collected from asymptomatic men were positive for mycoplasmas or ureaplasmas (Table 1). These results demonstrate that one seminested PCR with a set of common primers is able to amplify mycoplasmas and ureaplasmas from urine specimens as well as prototype strains.
TABLE 1.
Amplification and identification of clinical isolates
| Detected organism | No. (%) of specimens from indicated patients
|
Asymptomatic
|
||
|---|---|---|---|---|
| GU | NGU | |||
| C. trachomatis positive | C. trachomatis negative | |||
| M. genitalium | 2 (3.6) | 4 (8.5) | 9 (19.6) | 1 (2.4) |
| M. hominis | 0 (0.0) | 1 (2.1) | 1 (2.2) | 0 (0.0) |
| M. faucium | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (2.4) |
| U. urealyticum and M. genitalium | 0 (0.0) | 1 (2.1) | 0 (0.0) | 0 (0.0) |
| U. parvum | 1 (1.8) | 1 (2.1) | 3 (6.5) | 9 (21.4) |
| U. urealyticum | 6 (10.9) | 5 (10.6) | 8 (17.4) | 4 (9.5) |
| Mycoplasma or ureaplasma | 9 (16.4) | 12 (25.5) | 21 (45.7) | 15 (35.7) |
| Total | 55 | 47 | 46 | 42 |
To completely identify 57 PCR products, 42 from urethritis patients and 15 from asymptomatic men, the DNA fragments were subjected to sequence analysis. When the nucleotide sequences of internal regions of approximately 250 bp were compared with those of 15 prototype strains of human mycoplasmas and ureaplasmas, all but five specimens showed 100% identity with one of the 15 prototype strains: 16 for M. genitalium, 2 for M. hominis, 1 for M. faucium, 23 for U. urealyticum, 14 for U. parvum, and 1 for M. genitalium and U. urealyticum (Table 1). The other five showed their highest identity (99.6%) to M. genitalium and second-highest identity (98.0%) to M. pneumoniae. None of the DNA sequences showed higher identity than M. genitalium or M. pneumoniae to these five DNA sequences when the database was searched with Blast. The phylogenetic analyses based on partial 16S rRNA genes successfully separated 57 isolates into distinct monophyletic clusters with 1 of the 15 prototype strains with 80 to 99% bootstrap support, allowing us to identify these nucleotide sequences in each species (Fig. 3). The PCR products from one urethritis urine specimen showed the presence of mixed sequences in the sequence analysis. To determine each nucleotide sequence, the amplicon was subcloned into pT7Blue, and the nucleotide sequences were resequenced. The results revealed the presence of a mixed infection by M. genitalium and U. urealyticum in this sample. Together, these results demonstrated that PCR targeting all human mycoplasmas and ureaplasmas was significantly useful to amplify mycoplasmas and ureaplasmas from urethritis patients and that the phylogenetic clustering, based on sequences of approximately 250 nucleotides including the V4 and V5 regions of partial 16S rRNA genes, is a powerful tool for the typing of mycoplasmas and ureaplasmas.
FIG. 3.
Phylogenetic analyses of mycoplasmas and ureaplasmas from urine specimens. Eight representative nucleotide sequences of 57 DNA fragments from urine specimens were analyzed with the 15 prototype strains as for Fig. 1. The numbers at the nodes represent the percentages of 1,000 bootstrap pseudoreplicates that contained the cluster distal to the node. The numbers in the parentheses indicate the numbers of PCR fragments that showed nucleotide sequences identical to those from urine samples.
DISCUSSION
With the development of molecular-biological techniques, many laboratories have applied PCR to rapidly identify mycoplasmas and ureaplasmas. In most of these studies, PCR has been employed to target species-specific nucleotide sequences (14, 17, 20, 33). Species-specific PCR has proven successful in achieving rapid detection, but it requires several sets of primers, since several species of mycoplasmas or ureaplasmas have been implicated as the etiologic agents of urethritis. In contrast, Hoffman et al. (10) reported the use of a primer set, GPO-1 and MGSO, for detecting not only human mycoplasmas but also ureaplasmas. However, their PCR was not sensitive enough to detect low amounts of mycoplasmas or ureaplasmas in clinical samples. Therefore, we newly designed the My-ins primer and modified the PCR to create a seminested PCR with the new primer as a way to achieve sensitivity high enough to amplify all human mycoplasmas and ureaplasmas. Using the primer set of GPO-1, MGSO, and My-ins, the seminested PCR is over 100 times more sensitive than the original PCR. For example, the seminested PCR is capable of amplifying 10 copies per PCR tube of M. genitalium with at least the same sensitivity as the M. genitalium-specific PCR previously reported (6, 14, 20). When we applied this highly sensitive seminested PCR to amplify mycoplasmas and ureaplasmas using urine specimens from urethritis patients, the seminested PCR was able to amplify not only M. genitalium but also M. hominis, M. faucium, U. urealyticum, and U. parvum. Therefore, we expect that this seminested PCR with a set of common primers can amplify all human mycoplasmas and ureaplasmas, because the My-ins primer was designed on the basis of a highly conserved region among 15 human mycoplasmas and ureaplasmas.
Until now, the phylogenetic analysis of mycoplasmas and ureaplasmas has been based on approximately 1,000 bp of 16S rRNA gene (2, 3, 21). The phylogenetic analysis based on these sequences enables the prototype strains to be classified into species; however, this size, approximately 1,000 bp of a nucleotide sequence, is too long for the phylogeny-based rapid identification of PCR products to be applied with clinical materials. Therefore, we focused on a relatively short and variable region comprising 250-nucleotide sequences including variable V4 and V5 regions among mycoplasmas and ureaplasmas, because a single sequencing reaction enabled us to identify the nucleotide sequences for phylogenetic analysis. The 250-bp nucleotide sequences from urine were also divergent enough to allow us to segregate the isolates from heterologous prototypes. Moreover, the five sequences from urine that showed the highest identity (99.6%) with M. genitalium and the second-highest identity (98.0%) with M. pneumoniae, were classified into M. genitalium by the phylogenetic analysis. Jensen et al. reported the heterogeneity of M. genitalium in a study using PCR-restriction fragment length polymorphism on an adhesin-coding gene (14). These data suggest that M. genitalium may comprise several genotypes. U. parvum (previously U. urealyticum, biovar 1) was detected more frequently in asymptomatic patients; however, U. urealyticum (previously U. urealyticum, biovar 2) was detected more often in symptomatic patients and those with urethritis, pelvic inflammatory disease, spontaneous abortion, etc. (1). The assay described in this report may serve to elucidate differences in the pathogenicity of human mycoplasmas and ureaplasmas.
In conclusion, we have developed a new, rapid, and simple method for identifying human mycoplasmas and ureaplasmas based on PCR and phylogenetic analysis of a partial 16S rRNA gene. A single set of seminested PCR was sufficient to amplify all prototype strains of human mycoplasmas and ureaplasmas. The PCR amplified 42 of the 148 urine specimens from urethritis patients and 15 of 42 urine specimens from asymptomatic patients. The PCR products took advantage of the heterogeneity in variable regions of the 16S rRNA gene among species, enabling us to rapidly identify the 57 DNA amplicons based on phylogenetic analysis. We believe that the method described in this communication, in conjunction with the accumulation of the nucleotide sequences in the database, will be of use not only for the rapid diagnosis of human mycoplasmas and ureaplasmas but also for etiological and epidemiological studies of these contagions.
Acknowledgments
We thank Tsuguo Sasaki at the National Institute of Infectious Diseases laboratory, Tokyo, Japan, for providing prototype strains of mycoplasmas and ureaplasmas, Kaoru Yoshihara for her invaluable precious help in sequencing, and Tadashi Narisawa for the AMPLICOR STD-1 assays.
REFERENCES
- 1.Abele-Horn, M., C. Wolff, P. Dressel, F. Pfaff, and A. Zimmermann. 1997. Association of Ureaplasma urealyticum biovars with clinical outcome for neonates, obstetric patients, and gynecological patients with pelvic inflammatory disease. J. Clin. Microbiol. 35:1199–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Artiushin, S., M. Duvall, and F. C. Minion. 1995. Phylogenetic analysis of mycoplasma strain ISM1499 and its assignment to the Acholeplasma oculi strain cluster. Int. J. Syst. Bacteriol. 45:104–109. [DOI] [PubMed] [Google Scholar]
- 3.Behbahani, N., A. Blanchard, G. H. Cassell, and L. Montagnier. 1993. Phylogenetic analysis of Mycoplasma penetrans, isolated from HIV-infected patients. FEMS Microbiol. Lett. 109:63–66. [DOI] [PubMed] [Google Scholar]
- 4.Christiansen, C., F. T. Black, and E. A. Freundt. 1981. Hybridization experiments with deoxyribonucleic acid from Ureaplasma urealyticum serovars I to III. Int. J. Syst. Bacteriol. 31:259–262. [Google Scholar]
- 5.Crotchfelt, K. A., L. E. Welsh, D. DeBonville, M. Rosenstraus, and T. C. Quinn. 1997. Detection of Neisseria gonorrhoeae and Chlamydia trachomatis in genitourinary specimens from men and women by a coamplification PCR assay. J. Clin. Microbiol. 35:1536–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deguchi, T., C. B. Gilroy, and D. Taylor-Robinson. 1995. Comparison of two PCR-based assays for detecting Mycoplasma genitalium in clinical specimens. Eur. J. Clin. Microbiol. Infect. Dis. 14:629–631. [DOI] [PubMed] [Google Scholar]
- 7.Felsenstein, J. 1985. Confidence limits on phylogenies: an aproach using the bootstrap. Evolution 39:783–791. [DOI] [PubMed] [Google Scholar]
- 8.Gambini, D., I. Decleva, L. Lupica, M. Ghislanzoni, M. Cusini, and E. Alessi. 2000. Mycoplasma genitalium in males with nongonococcal urethritis: prevalence and clinical efficacy of eradication. Sex. Transm. Dis. 27:226–229. [DOI] [PubMed] [Google Scholar]
- 9.Hillier, S. L., R. P. Nugent, D. A. Eschenbach, M. A. Krohn, R. S. Gibbs, D. H. Martin, M. F. Cotch, R. Edelman, J. G. Pastorek, II, A. V. Rao, et al. 1995. Association between bacterial vaginosis and preterm delivery of a low-birth-weight infant. The Vaginal Infections and Prematurity Study Group. N. Engl. J. Med. 333:1737–1742. [DOI] [PubMed] [Google Scholar]
- 10.Hoffman, R. W., F. X. O’Sullivan, K. R. Schafermeyer, T. L. Moore, D. Roussell, R. Watson-McKown, M. F. Kim, and K. S. Wise. 1997. Mycoplasma infection and rheumatoid arthritis: analysis of their relationship using immunoblotting and an ultrasensitive polymerase chain reaction detection method. Arthritis Rheum. 40:1219–1228. [DOI] [PubMed] [Google Scholar]
- 11.Horner, P. J., C. B. Gilroy, B. J. Thomas, R. O. Naidoo, and D. Taylor-Robinson. 1993. Association of Mycoplasma genitalium with acute non-gonococcal urethritis. Lancet 342:582–585. [DOI] [PubMed] [Google Scholar]
- 12.Hosoya, M., M. Sato, K. Honzumi, M. Katayose, Y. Kawasaki, H. Sakuma, K. Kato, Y. Shimada, H. Ishiko, and H. Suzuki. 2001. Association of nonpolio enteroviral infection in the central nervous system of children with febrile seizures. Pediatrics 107:E12. [DOI] [PubMed] [Google Scholar]
- 13.Jensen, J. S., H. T. Hansen, and K. Lind. 1996. Isolation of Mycoplasma genitalium strains from the male urethra. J. Clin. Microbiol. 34:286–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen, J. S., S. A. Uldum, J. Sondergard-Andersen, J. Vuust, and K. Lind. 1991. Polymerase chain reaction for detection of Mycoplasma genitalium in clinical samples. J. Clin. Microbiol. 29:46–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katoh, N. 1985. Significance of Ureaplasma urealyticum and Clostridium difficile in nongonococcal urethritis. Kansenshogaku Zasshi 59:687–700. [DOI] [PubMed] [Google Scholar]
- 16.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111–120. [DOI] [PubMed] [Google Scholar]
- 17.kong, F., G. James, Z. Ma, S. Gordon, W. Bin, and G. L. Gilbert. 1999. Phylogenetic analysis of Ureaplasma urealyticum–support for the establishment of a new species, Ureaplasma parvum. Int. J. Syst. Bacteriol. 49:1879–1889. [DOI] [PubMed] [Google Scholar]
- 18.Maeda, S., M. Tamaki, M. Nakano, M. Uno, T. Deguchi, and Y. Kawada. 1998. Detection of Mycoplasma genitalium in patients with urethritis. J. Urol. 159:405–407. [DOI] [PubMed] [Google Scholar]
- 19.Maniatis, T., J. Sambrook, and E. F. Fritsch. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Palmer, H. M., C. B. Gilroy, P. M. Furr, and D. Taylor-Robinson. 1991. Development and evaluation of the polymerase chain reaction to detect Mycoplasma genitalium. FEMS Microbiol. Lett. 61:199–203. [DOI] [PubMed] [Google Scholar]
- 21.Rawadi, G., A. Dujeancourt-Henry, B. Lemercier, and D. Roulland-Dussoix. 1998. Phylogenetic position of rare human mycoplasmas, Mycoplasma faucium, M. buccale, M. primatum and M. spermatophilum, based on 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 48:305–309. [DOI] [PubMed] [Google Scholar]
- 22.Razin, S., and D. Yogev. 1986. Genetic relatedness among Ureaplasma urealyticum serotypes (serovars). Pediatr. Infect. Dis. 5:S300–S304. [DOI] [PubMed] [Google Scholar]
- 23.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robertson, J. A., and M. H. Chen. 1984. Effects of manganese on the growth and morphology of Ureaplasma urealyticum. J. Clin. Microbiol. 19:857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425. [DOI] [PubMed] [Google Scholar]
- 26.Schwartz, M. A., and T. M. Hooton. 1998. Etiology of nongonococcal nonchlamydial urethritis. Dermatol. Clin. 16:727–733, xi. [DOI] [PubMed] [Google Scholar]
- 27.Shimada, M., T. Kotani, S. Ohtaki, H. Sameshima, T. Ikenoue, T. Sasaki, and T. Kenri. 1999. Clinicobacteriological studies on the nine cases with upper genital tract Mycoplasma hominis infection. Kansenshogaku Zasshi 73:646–651. [DOI] [PubMed] [Google Scholar]
- 28.Smith, D. B., F. Davidson, and P. Simmonds. 1995. Hepatitis C virus variants and the role of genotyping. J. Hepatol. 23:26–31. [PubMed] [Google Scholar]
- 29.Taylor-Robinson, D. 1995. The Harrison Lecture. The history and role of Mycoplasma genitalium in sexually transmitted diseases. Genitourin. Med. 71:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor-Robinson, D., and P. M. Furr. 1997. Genital mycoplasma infections. Wien. Klin. Wochenschr. 109:578–583. [PubMed] [Google Scholar]
- 31.Tully, J. G., D. Taylor-Robinson, R. M. Cole, and D. L. Rose. 1981. A newly discovered mycoplasma in the human urogenital tract. Lancet i:1288–1291. [DOI] [PubMed] [Google Scholar]
- 32.Tully, J. G., D. Taylor-Robinson, D. L. Rose, P. M. Furr, C. E. Graham, and M. F. Barile. 1986. Urogenital challenge of primate species with Mycoplasma genitalium and characteristics of infection induced in chimpanzees. J. Infect. Dis. 153:1046–1054. [DOI] [PubMed] [Google Scholar]
- 33.van Kuppeveld, F. J., J. T. van der Logt, A. F. Angulo, M. J. van Zoest, W. G. Quint, H. G. Niesters, J. M. Galama, and W. J. Melchers. 1992. Genus- and species-specific identification of mycoplasmas by 16S rRNA amplification. Appl. Environ. Microbiol. 58:2606–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]