Abstract
We report on the first case of a catheter-related recurrent bacteremia caused by Kocuria kristinae, a gram-positive microorganism belonging to the family Micrococcaceae, in a 51-year-old woman with ovarian cancer. This unusual pathogen may cause opportunistic infections in patients with severe underlying diseases.
CASE REPORT
In December 1999, a 51-year-old woman with abundant ascites and an irregular pelvic mass was referred to the Centro di Riferimento Oncologico of Aviano. Histologic examination of the pelvic mass revealed the presence of a poorly differentiated ovarian carcinoma. In January 2000, a permanent central venous catheter (CVC) was implanted and the patient began chemotherapy. In June 2000, after five courses of chemotherapy, the patient was readmitted for febrile neutropenia and sepsis. Complete clinical and microbiological diagnostic procedures were performed, and four pairs of blood specimens were collected from peripheral blood and the CVC for culture (both aerobic and anaerobic). A gram-positive microorganism identified as Kocuria kristinae grew from three samples cultured aerobically. The patient was empirically treated with meropenem; after the isolation of gram-positive cocci from the bloodstream, a glycopeptide was added, resulting in prompt clinical improvement. No further bacterial isolates were obtained from blood or other clinical samples. The patient was discharged from the hospital, but 1 month later she was readmitted to the Centro di Riferimento Oncologico of Aviano for another febrile episode. Among 11 blood specimens drawn from peripheral blood and the CVC, 7 specimens cultured aerobically again revealed K. kristinae; antibiotic therapy with ciprofloxacin and clindamycin was started, with resolution of fever and negativity of blood cultures. A third febrile episode occurred 1 month later. One of the two blood samples collected from the CVC and cultured aerobically yielded K. kristinae. The CVC was removed and sent to the microbiology laboratory; a gram-positive coccus, which was identified as K. kristinae, was isolated from the culture of the catheter tip. Soon after catheter removal the patient became afebrile and her clinical status improved. After 6 months of monitoring, the patient is alive and well and is in complete remission of the primary disease.
Microbiology.
Cultures of blood from peripheral veins and the CVC were performed with a BACTEC system (BACTEC 9210; Becton Dickinson) with BACTEC PLUS Aerobic/F and PLUS Anaerobic/F culture vials. The CVC tip was cultured by the quantitative technique of Cleri et al. (5). Briefly, after catheter removal, a 5-cm segment of the tip was vortexed for 30 to 60 s in 5 ml of tryptic soy broth. Three aliquots of the broth (100, 10, and 1 μl) were spread onto three sheep blood agar plates. The plates were incubated at 37°C for 18 h. More than 1,000 CFU was counted; this value is considered representative of a catheter infection. The microorganisms isolated from the peripheral blood and the CVC during the different septic episodes and from the catheter tip were gram-positive cocci occurring in tetrads. They were nonhemolytic, catalase positive, strictly aerobic, nonmotile, and unable to reduce nitrate to nitrites or to hydrolyze gelatin, arginine, and esculin. The bacterial isolates produced acid from trehalose, glucose, fructose, mannose, glycerol, and saccharose but not from mannitol, raffinose, arabinose, lactose, or ribose. On the basis of Bergey’s Manual of Systematic Bacteriology (11), they were identified as K. kristinae. This identification was confirmed with the commercially available ID32 Staph ATB system (Biomerieux), with a probability of identification of 99.9% and a T index of 1.00. The T index estimates how closely the profile corresponds to the most typical set of reactions for each taxon. Its value varies between 0 and 1 and is inversely proportional to the number of atypical tests. The cellular fatty acid profile showed that the isolates contained the cellular fatty acid components typical of this species.
A partial sequence of the 16S rRNA gene (rDNA; 642 bp) was generated in order to confirm the identification of our strains. Briefly, after extraction of genomic DNA by standard methods, we amplified the 16S rDNA by PCR with universal primers 27F and 1492R (3). The PCR products obtained were then sequenced by use of the ABI technology with the same universal primers. The nucleotide sequence was analyzed with BLAST programs, revealing the higher degree of homology (99.8%) with K. kristinae (GenBank/EMBL data library accession number AF323746).
Susceptibility tests were performed with the Vitek system (Biomerieux) and by the disk diffusion method on Mueller-Hinton agar with 24 h of incubation at 35°C. The results were expressed as susceptible, intermediate, or resistant according to the criteria of the National Committee for Clinical Laboratory Standards for the modified Kirby-Bauer method (9a). Our isolates were susceptible to clindamycin, erythromycin, ciprofloxacin, penicillin, oxacillin, cefalothin, and vancomycin.
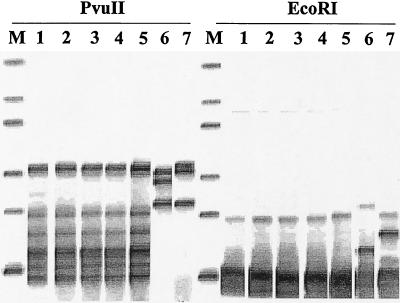
To type the isolates, we used the automated RiboPrinter microbial characterization system (Qualicon, Wilmington, Del.), which allows the acquisition and normalization of ribotype patterns from bacteria (4, 6). These genetic fingerprints, or RiboPrint patterns, are normalized digital representations of the genetic data for the isolated organism. Patterns are grouped within a specific similarity range to form RiboGroups, which define the genetic relatedness of samples. This process has previously been described in detail (2, 4). Identification is obtained by matching the given pattern against a set of reference patterns for specific organisms; the patterns have been given the classical taxonomic names of the organisms and have been confirmed by extensive phenotypic testing. The database for EcoRI-digested fragments contains more than 1,100 reference patterns for genera such as Bacillus, Enterococcus, Escherichia, Lactobacillus, Listeria, Pseudomonas, Salmonella, Staphylococcus, and Vibrio (4). A library for PvuII-digested fragments, which contains some patterns including one for a K. kristinae strain, is also available. Our samples were prepared according to the manufacturer’s protocol. Briefly, isolated bacterial colonies grown overnight at 37°C on blood agar were collected with a plastic stick and suspended in the sample buffer supplied with the system. The sample suspension was treated for 10 min at 80°C to reduce viability and to inactivate nucleases. After addition of the two lytic enzymes supplied with the system, the samples were then loaded into the RiboPrinter system. The standard process includes restriction enzyme digestion with EcoRI and hybridization with an rRNA gene probe (Escherichia coli region encoding the 16S, 23S, and 5S rRNA genes and the spacer region including Glu-tRNA). By automated ribotyping we analyzed five K. kristinae strains isolated from the CVC and the bloodstream of our patient using EcoRI, which is highly discriminatory for coagulase-negative staphylococci (7), and PvuII to compare the patterns obtained with those in the internal database. K. kristinae strains ATCC 27570 and ATCC 27572 were used as internal controls. The five strains had the same pattern, which was different from those obtained from the reference strains (Fig. 1).
FIG. 1.
Automated ribotyping obtained with EcoRI and PvuII restriction enzymes (modified from an image generated with RiboPrinter). Lanes M, molecular mass marker (from the top, 1, 2.2, 3.3, 6.3, 9.6, and 50 kbp); lanes 1, Kocuria strain isolated from the CVC; lanes 2 to 5, Kocuria strains isolated from different blood samples; lanes 6, reference strain K. kristinae ATCC 27572; lanes 7, reference strain K. kristinae ATCC 27570.
Micrococcus spp. are gram-positive, strictly aerobic microorganisms that often occur in tetrads or irregular clusters and that usually grow on simple media. In humans, they may colonize the skin, mucosae, and oropharynx; they are also isolated from a wide variety of animal sources and soil. Recently, Stackebrandt and colleagues (8, 12) made a taxonomic revision of this class of microorganisms; some strains were reclassified in the new genus Kocuria spp. (Kocuria rosea, previously described as Deinococcus erythromyxa or Micrococcus roseus; K. kristinae, formerly Micrococcus kristinae; Kocuria varians, basonym: Micrococcus varians; Kocuria palustris; and Kocuria rhizophila sp. nov.). Micrococcus strains have been isolated from different clinical specimens, although their clinical relevance is questionable. The identification of micrococci isolated from clinical specimens is often cumbersome for the laboratory, and the clinical interpretation of positive results may be misleading. In fact, these microorganisms are often not correctly identified or are hastily discarded as contaminants. It has been reported that Micrococcus luteus may be associated with bacteremia, intracranial abscesses, septic arthritis, and meningitis in immunosuppressed or immunocompetent hosts (1, 10, 13). To review the spectrum of clinical diseases caused by K. kristinae in humans, we performed a Medline search using the terms “Micrococcus kristinae” and “Kocuria.” Only Kocuria sedentarius has been associated with prosthetic valve endocarditis, whereas we were unable to find published studies on systemic infections caused by K. kristinae. We have isolated from repeated cultures of blood from a patient with clinical signs of a septic syndrome a gram-positive coccus that was unequivocally identified as K. kristinae by biochemical and molecular biology-based assays.
Automated ribotyping with EcoRI and PvuII as restriction enzymes seems to be effective for characterization of these microorganisms. In particular, this method allows one to distinguish unrelated strains: it works on a DNA fragment that is highly conserved within species, so that major variations in patterns obtained by analysis of this region indicate different strains. In our experiments all the strains isolated from our patient were identical and were remarkably different from those obtained by analysis of reference strains from the American Type Culture Collection. Although pulsed-field gel electrophoresis is more discriminatory than ribotyping for epidemiological purposes, the advantages of automated ribotyping are multiple, including increased standardization, speed, and accuracy for between-run comparisons and studies with small numbers of nontypeable isolates. The strictly epidemiological relationship among our strains and the identities of the ribotype patterns that we obtained with two different restriction enzymes allowed us to demonstrate that the microorganisms analyzed were probably clonal, suggesting a recurrent infection rather than reinfection caused by K. kristinae.
The repeated isolation of K. kristinae from different blood cultures in the absence of other microorganisms, together with isolation of the organism from the catheter, strongly suggests that K. kristinae may cause catheter-related bacteremias. Because infections remain a major complication related to the use of permanent catheters for long-term therapy for oncology patients (9), the importance of the repeated isolation of a Kocuria strain from blood cultures should not be underestimated by the clinician. Although uncommon, the possibility that a central venous line may be the portal of entry should be evaluated.
Nucleotide sequence accession number.
The partial sequence of the rDNA sequence of the K. kristiniae isolate from the present study has been deposited in the GenBank/EMBL data library under accession number AJ316579.
Acknowledgments
This work was partially supported by a grant from FSN Ricerca Finalizzata 1998 (to P. De Paoli) and by grants from IRCCS Policlinico San Matteo, Pavia (Ricerca Corrente 1998) (to P. Marone).
We thank Susanne Verbarg and Jola Swiderski of the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH for helpful assistance with the verification of strain identification and Claudio Bandi for expert assistance with sequencing of the 16S rDNA.
REFERENCES
- 1.Adang, R., H. C. Schouten, F. van Tiel, and G. H. Blijham. 1992. Pneumonia due to Micrococcus spp. in a patient with acute myeloid leukemia. Leukemia 6:224–226. [PubMed] [Google Scholar]
- 2.Allerberger, F., and S. J. Fritschel. 1999. Epidemiological typing of Austrian Listeria monocytogenes isolates by automated ribotyping. J. Microbiol. Methods 35:237–244. [DOI] [PubMed] [Google Scholar]
- 3.Bandi, C., G. Damiani, L. Magrassi, A. Gigolo, R. Fani, and L. Sacchi. 1994. Flavobacteria as intracellular symbionts in cockroaches. Proc. R. Soc. London Ser. B 257:43–48. [DOI] [PubMed] [Google Scholar]
- 4.Bruce, J. 1996. Automated system rapidly identifies and characterizes microorganisms in food. Food Technol. 50:77–81. [Google Scholar]
- 5.Cleri, D. J., M. L. Corrado, and S. J. Seligman. 1980. Quantitative culture of intravenous catheters and other intravascular inserts. J. Infect. Dis. 141:781–786. [DOI] [PubMed] [Google Scholar]
- 6.Hubner, R. J., E. M. Cole, J. L. Bruce, C. I. McDowell, and J. A. Webster. 1995. Predicted types of Listeria monocytogenes created by the positions of EcoRI cleavage sites relative to rRNA sequences. Proc. Natl. Acad. Sci. USA 92:5234–5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Izard, N. C., H. Hachler, M. Grehn, and F. H. Kayser. 1992. Ribotyping of coagulase-negative staphylococci with special emphasis on intraspecific typing of Staphylococcus epidermidis. J. Clin. Microbiol. 30:817–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kovács, G., J. Burghardt, S. Pradella, P. Schumann, E. Stackebrandt, and K. Màrialigeti. 1999. Kocuria palustris sp. nov. and Kocuria rhizophila sp. nov., isolated from the rhizoplane of the narrow-leaved cattail (Typha angustifolia). Int. J. Syst. Bacteriol. 49:167–173. [DOI] [PubMed] [Google Scholar]
- 9.Mueller, B., J. Skelton, P. E. Callender, D. Marshall, J. Gress, D. Longo, J. Norton, M. Rubin, D. Venzon, and P. A. Pizzo. 1992. A prospective randomized clinical trial comparing the infectious and noninfectious complications of an externalized catheter versus a subcutaneously implanted device in cancer patients. J. Clin. Oncol. 10:1943–1948. [DOI] [PubMed] [Google Scholar]
- 9a.National Committee for Clinical Laboratory Standards. 1997. Performance standards for antimicrobial disk susceptibility tests. Approved standard M2-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 10.Peces, R., E. Gago, F. Tejada, A. S. Laures, and J. Alvarez-Grande. 1997. Relapsing bacteraemia due to Micrococcus luteus in a haemodialysis patient with a Perm-Cath cathether. Nephrol. Dial. Transplant. 12:2428–2429. [DOI] [PubMed] [Google Scholar]
- 11.Schleifer, K. H. 1986. Gram-positive cocci, p.999–1103. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey’ manual of systematic bacteriology, vol. 2. The Williams & Wilkins Co., Baltimore, Md.
- 12.Stackebrandt, E., C. Koch, O. Gvozdiak, and P. Schumann. 1995. Taxonomic dissection of the genus Micrococcus: Kocuria gen. nov., Nesterenkonia gen. nov. Kytococcus gen. nov., Dermacoccus gen. nov., and Micrococcus Cohn 1872 gen. emend. Int. J. Syst. Bacteriol. 45:682–692. [DOI] [PubMed] [Google Scholar]
- 13.von Eiff, C., N. Kuhn, M. Herrman, S. Weber, and G. Peters. 1996. Micrococcus luteus as a cause of recurrent bacteremia. Pediatr. J. Infect. Dis. 15:711–713. [DOI] [PubMed] [Google Scholar]