Abstract
Of 96 Helicobacter pylori isolates from patients in Shanghai and Guangzhou, China, 5 had the A2143G 23S rRNA mutation as determined by primer-mismatch PCR and were resistant to clarithromycin by the E-test. The remaining isolates were primer-mismatch PCR negative and susceptible to clarithromycin. The conclusion is that the prevalence of clarithromycin-resistant H. pylori isolates among these Chinese patients is 5%.
Helicobacter pylori causes chronic gastritis (7) and peptic ulcer disease (PUD) (15) and is involved in the pathogenesis of gastric adenocarcinoma (5). Therefore, therapy aimed at eradication of the bacterium is given to H. pylori-infected patients. Metronidazole and clarithromycin (CLR) are used in most Helicobacter eradication regimens (3). Resistance to one or both of these antimicrobials substantially reduces the efficacies of therapies containing these antibiotics (4). The mechanism of CLR resistance of H. pylori is well established. Seven point mutations in the peptidyltransferase region of domain V of the 23S rRNA gene have been found to be associated with CLR resistance (6, 8, 12, 13, 17). In a recent large multicenter study, only the A2142G, A2143G, and A2142C mutations were found, while the A2115G, G2141A, A2143C, and A2142T mutations were not observed at all (16). On the basis of this knowledge, we aimed to develop a PCR with primers mismatched at the 3′ end to detect the three prevalent types of mutations (A2142G, A2143G, and A2142C) in 23S rRNA associated with CLR resistance among H. pylori isolates from 96 patients in China. The results were confirmed by sequencing and correlated with the CLR MICs obtained by the E-test.
H. pylori isolates were cultured from gastric biopsy specimens of 96 patients who were referred for an upper gastrointestinal endoscopy in 1995 in Shanghai (46 of 81 patients had PUD and 35 of 81 patients had gastritis) and Guangzhou (15 patients had gastritis or PUD) because of dyspeptic symptoms (9). Patients who had taken antimicrobial agents within the 2 weeks prior to endoscopy, who were receiving steroids, who had an active infection requiring current antimicrobial therapy, or who had active gastrointestinal bleeding were excluded.
Reference H. pylori strains 5898 (A2142G), 5908 (A2142G), 5883 (A2143G), and 6144 (A2142C) (kindly supplied by Gregory G. Stone, Abbott Laboratories, Abbott Park, Ill.) were used to establish the primer-mismatch PCR technique. Their susceptibilities to CLR and the nucleotide sequences at domain V of their 23S rRNAs were reported previously (11).
DNAs were isolated from the H. pylori strains by phenol-chloroform-isoamyl alcohol extraction and ethanol precipitation (14) or with a QIAamp tissue kit (Qiagen Inc., Chatsworth, Calif.), according to the manufacturer’s instructions.
Primer sets (Table 1) 5g-DP5, 5g-DP4, and 5g-DP6 were used for detection of the A2143G, A2142G, and A2142C mutations, respectively. For the control PCR, primer set 5g-ZGE23 (13) was used. The volumes of the PCR mixtures were 25 μl; the reaction mixtures contained 10 ng of template DNA, 0.75 U of Taq DNA polymerase (Promega, Madison, Wis.), 0.4 μmol of primer, 2.5 μl of 10× Taq buffer (Promega), 250 μmol of each deoxynucleoside triphosphate, and 1.5 mM MgCl2. The reaction mixtures were incubated in a programmable heat block (model 480; Perkin-Elmer, Norwalk, Conn.) for 5 min at 95°C, followed by 35 cycles of 1 min at 94°C, 20 min at the annealing temperature listed in Table 1 for each primer set, and 1 min at 72°C. The PCR products were subjected to electrophoresis on a 1% agarose gel along with a 1-kb DNA marker (Gibco Life Technologies, Rockville, Md.).
TABLE 1.
Primers used for detection of 23S rRNA gene point mutations in H. pylori isolates from patients in China
| Sequence detected or purpose | Primer | Nucleotide sequence | Location (size [bp]) of PCR producta | Annealing temp (°C) |
|---|---|---|---|---|
| 23S rRNA | 5gb | 5′-TCAGGGTGATGGACTGC-3′ | 1027–2665 (1,639) | 56 |
| ZGE23b | 5′-CACAGGCCAGTTAGCTA-3′ | |||
| A2142 G | 5gb | 5′-TCAGGGTGATGGACTGC-3′ | 1027–2530 (1,504) | 60 |
| DP4b | 5′-AGGTCCACGGGGTCTTC-3′ | |||
| A2143G | 5gb | 5′-TCAGGGTGATGGACTGC-3′ | 1027–2531 (1,505) | 59 |
| DP5b | 5′-AAGGTCCACGGGGTCTC-3′ | |||
| A2142C | 5gb | 5′-TCAGGGTGATGGACTGC-3′ | 1027–2535 (1,509) | 65 |
| DP6 | 5′-AGTAAAGGTCCACGGGGTCTTG-3′ | |||
| Sequencing | 2451-F | 5′-TCAACCAGAGATTCAGT-3′ | 2451–2665 (215) | 50 |
| ZGE23b | 5′-CACAGGCCAGTTAGCTA-3′ |
Primer set 2451-F-ZGE23 was used to amplify a 215-bp fragment comprising the point mutation region in the 23S rRNA gene. The amplicons were subjected to PCR-based sequencing with BigDye terminators according to the manufacturer’s instructions (Applied Biosystems Incorporated, Foster City, Calif.). The sequences were analyzed on an automatic sequencer (model 373; Applied Biosystems).
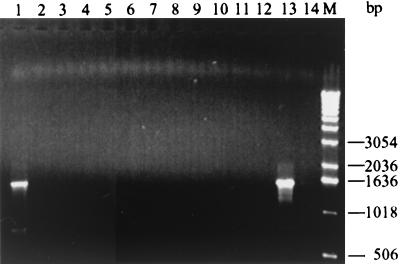
The CLR MICs for the H. pylori isolates were assessed by the E-test (AB Biodisk, Solna, Sweden) as described before (19). A strain was considered resistant if the MIC was ≥2 mg/liter (8, 10). The chromosomal DNAs of the 96 Chinese H. pylori isolates yielded amplicons of about 1,600 bp in the control PCR with primer set 5g-ZGE23, indicating no inhibition in this PCR. In contrast, the PCRs with primer set 5g-DP4 or primer set 5g-DP6 gave negative results, indicating that the A2142G and A2142C 23S rRNA mutations did not occur among these 96 isolates. Of 96 Chinese isolates, 5 (4 of 81 isolates from patients in Shanghai and 1 of 15 isolates from patients in Guangzhou) yielded an amplicon of about 1,500 bp with primer set 5g-DP5, indicating that these strains have the 23S rRNA A2143G mutation. An example of the results obtained for the 12 H. pylori isolates are presented in Fig. 1 for the isolate with the 23S rRNA A2143G mutation. Sequencing results confirmed the presence of the A2143G mutation in these five isolates. The CLR MICs for these isolates ranged between 2 and 24 mg/liter CLR by the E-test.
FIG. 1.
PCR amplification of 12 representative H. pylori isolates from patients in China with primers 5g and DP5. Lanes: 1, H. pylori strain R-50 with the A2143G mutation (CLR resistant); 2 to 12, wild-type H. pylori strains; 13, CLR-resistant reference H. pylori strain 5883 (A2143G); 14, CLR-sensitive wild-type H. pylori strain 26695; M, DNA molecular size marker.
The remaining 91 H. pylori isolates, all of which were negative by all three primer-mismatch PCRs, were susceptible to CLR (MICs, ≤0.016 mg/liter). Of these, 10 randomly chosen isolates had the wild-type 23S rRNA genotype by sequencing.
Primer-mismatch PCR has previously been used to detect the A2142G and A2143G mutations but not the A2142C mutation with transformed CLR-resistant H. pylori isolates (13). Recently, primer-mismatch PCR has been used to detect only the A2142C point mutation in H. pylori isolates from patients in Spain (2). The technique is rapid and suitable for use for epidemiological surveys of large patient populations. It is possible that such surveys could be done by assessment of biopsy specimens by this method. However, the technique should be optimized and validated in the local laboratory. In the present study, primer-mismatch PCRs for the detection of the A2142G and A2143G mutations in the 23S rRNA of H. pylori were initially performed with the primers and under the conditions (1.0 mM MgCl2 and an annealing temperature of 57°C) described previously (13). However, under these conditions these PCRs with chromosomal DNAs of reference strains 5898 (A2142G) and 5883 (A2143G) were negative. In addition, the control PCR with chromosomal DNAs of all reference strains and the DNA of strain 26695 (wild type) was also negative. Therefore, the MgCl2 concentration was adjusted to 1.5 mM and the annealing temperature for each set of primers was carefully optimized to achieve the best sensitivity and specificity.
In conclusion, the prevalence of CLR-resistant H. pylori isolates among the group of patients residing in Shanghai and Guangzhou is 5%, which is in the range of the prevalences of CLR-resistant H. pylori in other countries (1 to 10%) (1). CLR was not available in China before 1996, which might explain the lower prevalence of CLR-resistant H. pylori in China compared to the 11% prevalence of CLR-resistant H. pylori among patients from Hong Kong (18). However, the possibility of the previous use of macrolides like josamycin or erythromycin cannot be excluded.
Acknowledgments
Z.-J. Pan and W.-W. Su contributed equally to the present study.
This work was supported by grants from the Dutch Ministry of Education and Science and the Royal Dutch Academic of Science (KNAW) and from the Chinese Ministry of Public Health (to Z.-J.P.).
We thank Gregory G. Stone from Abbott Laboratories for providing the reference H. pylori strains.
REFERENCES
- 1.Alarcon, T., D. Domingo, and M. Lopez-Brea. 1999. Antibiotic resistance problems with Helicobacter pylori. Int. J. Antimicrob. Agents 12:19–26. [DOI] [PubMed] [Google Scholar]
- 2.Alarcon, T., D. Domingo, N. Prieto, and M. Lopez-Brea. 2000. PCR using 3′-mismatched primers to detect A2142C mutation in 23S rRNA conferring resistance to clarithromycin in Helicobacter pylori clinical isolates. J. Clin. Microbiol. 38:923–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graham, D. Y. 2000. Therapy of Helicobacter pylori: current status and issues. Gastroenterology 118:S2–S8. [DOI] [PubMed] [Google Scholar]
- 4.Houben, M. H., D. van der Beek, E. F. Hensen, A. J. Craen, E. A. J. Rauws, and G. N. J. Tytgat. 1999. A systematic review of Helicobacter pylori eradication therapy—the impact of resistance on eradication rates. Aliment. Pharmacol. Ther. 13:1047–1055. [DOI] [PubMed] [Google Scholar]
- 5.Huang, J. Q., S. Sridhar, Y. Chen, and R. H. Hunt. 1998. Meta-analysis of the relationship between Helicobacter pylori seropositivity and gastric cancer. Gastroenterology 114:1169–1179. [DOI] [PubMed] [Google Scholar]
- 6.Hultén, K., A. Gibreel, O. Skold, and L. Engstrand. 1997. Macrolide resistance in Helicobacter pylori: mechanism and stability in strains from clarithromycin-treated patients. Antimicrob. Agents Chemother. 41:2550–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marshall, B. J., J. A. Amstrong, D. B. McGechie, and R. J. Glancy. 1985. Attempt to fulfil Koch’s postulates for pyloric Campylobacter. Med. J. Aust. 142:436–439. [DOI] [PubMed] [Google Scholar]
- 8.Occhialini, A., M. Urdaci, F. Doucet-Populaire, C. M. Bebear, H. Lamouliatte, and F. Megraud. 1997. Macrolide resistance in Helicobacter pylori: rapid detection of point mutations and assays of macrolide binding to ribosomes. Antimicrob. Agents Chemother. 41:2724–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan, Z. J., D. E. Berg, R. W. M. van der Hulst, W. W. Su, A. Raudonikiene, S. D. Xiao, J. Dankert, G. N. J. Tytgat, and A. van der Ende. 1998. Prevalence of vacuolating cytotoxin production and distribution of distinct vacA alleles in Heliobacter pylori from China. J. Infect. Dis. 178:220–226. [DOI] [PubMed] [Google Scholar]
- 10.Pina, M., A. Occhialini, L. Monteiro, H. P. Doermann, and F. Megraud. 1998. Detection of point mutations associated with resistance of Helicobacter pylori to clarithromycin by hybridization in liquid phase. J. Clin. Microbiol. 36:3285–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stone, G. G., D. Shortrdige, J. Versalovic, J. Beyer, R. Flamm, D. Y. Graham, A. T. Ghoneim, and S. K. Tanaka. 1997. A PCR-oligonucleotide ligation assay to determine the prevalence of 23S rRNA gene mutations in clarithromycin resistant Helicobacter pylori. Antimicrob. Agents Chemother. 41:712–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stone, G. G., D. Shortridge, R. K. Flamm, J. Versalovic, J. Beyer, K. Idler, L. Zulawinski, and S. K. Tanaka. 1996. Identification of a 23S rRNA gene mutation in clarithromycin-resistant Helicobacter pylori. Helicobacter 1:227–228. [DOI] [PubMed] [Google Scholar]
- 13.Taylor, D. E., Z. Ge, D. Purych, T. Lo, and K. Hiratsuka. 1997. Cloning and sequence analysis of two copies of a 23S rRNA gene from Helicobacter pylori and association of clarithromycin resistance with 23S rRNA mutations. Antimicrob. Agents Chemother. 41:2621–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Ende, A., E. A. J. Rauws, M. Feller, C. J. Mulder, G. N. J. Tytgat, and J. Dankert. 1996. Heterogeneous Helicobacter pylori isolates from members of a family with a history of peptic ulcer disease. Gastroenterology 111:638–647. [DOI] [PubMed] [Google Scholar]
- 15.van der Hulst, R. W. M., E. A. J. Rauws, B. Köycü, J. J. Keller, J. G. P. Tyssen, M. Bruno, and G. N. J. Tytgat. 1997. Prevention of ulcer recurrence after successful eradication of Helicobacter pylori infection: a prospective long term follow-up study. Gastroenterology 113:1082–1086. [DOI] [PubMed] [Google Scholar]
- 16.van Doorn, L.-J., Y. Glupczynski, J. G. Kusters, F. Mégraud, P. Midolo, N. Maggi-Solcà, D. M. M. Queiroz, N. Nouhan, E. Stet, and W. G. V. Quint. Accurate prediction of macrolide resistance of Helicobacter pylori by a PCR line probe assay for the detection of mutations in the 23S rRNA gene: a multicenter validation study. Antimicrob. Agents Chemother. 45:1500–1504. [DOI] [PMC free article] [PubMed]
- 17.Versalovic, J., D. Shortridge, K. Kibler, M. V. Griffy, J. Beyer, R. K. Flamm, S. K. Tanaka, D. Y. Graham, and M. F. Go. 1996. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 40:477–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, W. H., B. C. Wong, A. K. Mukhopadhyay, D. E. Berg, C. H. Cho, L. C. Lai, W. H. Hu, F. M. Fung, W. M. Hui, and S. K. Lam. 2001. High prevalence of Helicobacter pylori infection with dual resistance to metronidazole and clarithromycin in Hong Kong. Aliment. Pharmacol. Ther. 14:901–910. [DOI] [PubMed] [Google Scholar]
- 19.Weel, J. F., R. W. M. van der Hulst, Y. Gerrits, G. N. J. Tytgat, A. van der Ende, and J. Dankert. 1996. Heterogeneity in susceptibility to metronidazole among Helicobacter pylori isolates from patients with gastritis or peptic ulcer disease. J. Clin. Microbiol. 34:2158–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]