Abstract
Purpose.
To assess the frequency of shedding of herpes simplex virus type 1 (HSV-1) DNA in tears and saliva of asymptomatic individuals.
Methods
Fifty subjects without signs of ocular herpetic disease participated. Serum samples from all subjects were tested for HSV IgG antibodies by enzyme-linked immunosorbent assay (ELISA) and for HSV-1 by neutralization assay. HSV-1 DNA copy number and frequency of shedding were determined by real-time polymerase chain reaction (PCR) analysis of tear and saliva samples collected twice daily for 30 consecutive days.
Results
Thirty-seven (74%) of the 50 subjects were positive for HSV IgG by ELISA. The percentages of positive eye and mouth swabs were approximately equivalent: 33.5% (941/2806) and 37.5% (1020/2723), respectively. However, the percentage of samples with high HSV-1 genome copy numbers was greater in saliva than in tears, which may have been a result of the sample volume collected. Shedding frequency in tears was nearly the same in men (347/1003; 34.6%) and women (594/1705; 34.8%); in saliva, men had a higher frequency of shedding (457/1009; 45.3% vs. 563/1703; 33.1%, men versus women). Overall, 49 (98%) of 50 subjects shed HSV-1 DNA at least once during the course of the 30-day study.
Conclusions
The percentage of asymptomatic subjects who intermittently shed HSV-1 DNA in tears or saliva was higher than the percentage of subjects with positive ELISA or neutralization antibodies to HSV. Because most HSV transmission occurs during asymptomatic shedding, further knowledge of the prevalence of HSV-1 DNA in tears and saliva is warranted to control its spread. Shedding is simple to study, and its suppression may be an efficient way to evaluate new antivirals in humans.
Healthy individuals are known to shed herpesviruses. Humans are reservoirs, and asymptomatic shedding is a major factor in the spread of the virus. In a recent 14-month study, Epstein-Barr virus (EBV) was present in all 30 individuals tested, and 4 (13%) of 30 shed cytomegalovirus (CMV) in urine.1 In a cross-sectional study in Eritrea, Africa, with the exception of children <5 years of age, all the population groups evaluated were >80% herpes simplex virus type 1 (HSV-1) seropositive, and >90% CMV and varicella zoster virus (VZV) seropositive.2 Antibodies for HSV-1 have also been detected in tears (73%) and saliva (2.5%) of 40 healthy individuals.3
HSV-1 and, to a lesser extent, HSV-2 are known to be the leading causes of virus-induced blindness in the Western world, with approximately 500,000 individuals having herpetic eye disease in the United States.4 More than 62% of the U.S. population >12 years of age is positive for HSV-1, HSV-2, or both.4 Worldwide, 60% to 90% of the adult population is HSV-1 antibody positive.5,6 In one study, 100% of individuals older than 60 years were found to be HSV-1 seropositive.7 Despite the prevalence of HSV infections, however, only a small number of latently infected humans experience symptomatic disease. Only 1% to 6% of primary infections are clinically recognized.8 Consequently, asymptomatic shedding of HSV is considered the major form of transmission.
Thus far, studies have focused on quantifying HSV-1 and -2 antibody titers and/or frequency of shedding in patients with active herpesvirus lesions or immediately after the time of active lesions.9 Although HSV-1 seropositivity in healthy individuals has also been evaluated,3,10 asymptomatic shedding of HSV-1, especially in tears, has received less attention. To the best of our knowledge, only three studies have evaluated HSV-1 shedding in tears and saliva of healthy individuals, using relatively insensitive culturing techniques.11–13
Detection of HSV shedding is dependent on the population surveyed, as well as on the diagnostic technique used. Before the advent of real-time PCR, the presence of HSV could be determined only by relatively insensitive culturing of infectious virus.14–21 Today we are able to detect HSV DNA with PCR, the new gold standard for HSV detection in clinical samples. Hence, the percentage of individuals detected as positive for HSV-1 DNA is expected to increase significantly, as has the percentage of individuals shedding EBV in saliva.1
To the best of our knowledge, the present study is the largest cross-sectional assessment performed to date of the presence of HSV-1 DNA in the eyes and mouths of healthy individuals, in terms of population size and total samples collected. We used real-time PCR to quantify the presence and frequency of asymptomatic viral shedding in a combined total of 5529 tear and saliva specimens. Samples consisted of mouth and eye swabs collected twice daily from 50 volunteers over the course of 30 days. The subjects were instructed to swab the mouth and one eye morning and night. In addition, serum HSV IgG antibody titers were determined. We found that 98% (49/50) of the subjects in our study were positive at least once for HSV-1 DNA in tears and/or saliva.
Materials and Methods
Subjects
Participants consisted of 50 volunteers of either sex and any race, over the age of 18, with no signs of active ocular herpetic disease. Table 1 shows the cohort demographics. Volunteers were excluded if they met any of the following criteria: (1) had an active ocular herpetic lesion, or had had one in the past 30 days; (2) were taking systemic or topical antiviral drugs, or had taken antiviral drugs in the past 30 days; (3) had dry eyes; or (4) had participated in any clinical trial in the past 30 days. Before initiation of specimen collection, a history of HSV infections and of usage of ocular and/or systemic medication was taken and a 3-mL blood sample was collected for HSV antibody analysis. An external slit lamp examination of both eyes was conducted before the first swabbing; follow-up ocular examinations were performed on days 15 and 30. All subjects provided written informed consent in accordance with a protocol approved by the LSU Health Sciences Center Institutional Review Board and in agreement with the tenets of the Declaration of Helsinki.
Table 1.
Cohort Demographics of 50 Subjects
| n(%) | |
|---|---|
| Gender | |
| Male | 19 (38) |
| Female | 31 (62) |
| Race | |
| African American (n = 39; 78%) | |
| Male | 12 (24) |
| Female | 27 (54) |
| White (n = 10; 20%) | |
| Male | 7 (14) |
| Female | 3 (6) |
| Other (n = 1; 2%) | |
| Male | 0 |
| Female | 1 (2) |
| Age (y)* | |
| Male | 43.2 (19–71) |
| Female | 37.9 (19–70) |
| History of herpetic disease (n = 23) | |
| Ocular | 2 |
| Cutaneous | 19 |
| Genital | 2 |
Mean age, with range in parentheses.
Specimens and Collection Procedure
Each subject was asked to identify which eye would be swabbed during the course of the study. Subjects were instructed to collect tear specimens by touching the inner surface of the lower eyelid with an individually wrapped, sterile cotton swab (Fisher Scientific, Hanover Park, IL) and to place the swab immediately into a labeled sterile tube (Falcon; Becton Dickinson Labware, Franklin Lakes, NJ). Saliva specimens were obtained by vigorously swabbing the buccal mucosa. The first specimens of the day (tears and saliva) were collected soon after arising and before performing any oral hygiene. The second tear and saliva specimens were collected approximately 12 hours later. Immediately after sample collection, the tubes containing the swabs were stored in the subject’s home refrigerator. Oral and ocular samples were collected daily for 30 consecutive days and were brought to the investigator at the time of the scheduled eye examinations (days 15 and 30). Swabs were stored by the investigator at 4°C until processed.
Serum HSV-1 and -2 Detection by ELISA
Blood was obtained from the 50 subjects and centrifuged. The serum was clarified and stored at −80°C until analysis. HSV-1 and -2 IgG antibodies in the sera were detected by an enzyme-linked immunosor-bent assay (ELISA) kit, according to the manufacturer’s instructions (Diamedix, Miami, FL). Briefly, triplicate aliquots of serum serially diluted in phosphate buffer containing 0.2% NaN3 were incubated in microtiter plates in a total volume of 100 μL per well for 60 minutes at 37°C. The wells were washed three times with phosphate-buffered saline (PBS) followed by the addition of 100 μL of horseradish peroxidase (HRP)–conjugated goat anti-human IgG per well. After incubation at 37°C for 60 minutes, the wells were washed as just described. Bound antigen-antibody complexes were detected by the addition of 100 μL of 3,3′,5,5′-tetramethylbenzidine (TMB) per well. After 20 minutes at 37°C, the reaction was quenched by adding 100 μL of 1 N H3PO4 and 1 N HCl per well. The color that developed was read against a blank standard at 450 nm. Known HSV-1– and -2–positive sera were run as positive controls. The extent of reactivity in sera was measured using the following formula: enzyme units (EU)/mL/absorbance of positive control × absorbance of sample = EU/mL of sample.
Samples were considered positive for HSV-1 and/or -2 when the EU/mL exceeded 20.0 or the index value was greater than 1. If the test sample had been prediluted 1:5 before testing, the resultant EU/mL or index value was multiplied by 5.
Neutralization Assay
The antiviral neutralizing titers of the previously clarified serum samples were assayed. Samples were heat-inactivated for 30 minutes at 56°C. Serial dilutions of the heat-inactivated test serum were mixed with equal volumes of HSV-1 strain McKrae and incubated for 60 minutes at 37°C in a water bath, with gentle agitation every 15 minutes. Confluent CV-1 monolayer cells were grown in 96-well plates. After the medium was removed from the monolayers, twofold serial dilutions of 200 μL of the serum-virus mix were overlaid on CV-1 cells and cultured for 48 hours at 37°C with 5% CO2. The plates were stained with cresyl violet to assess for viral plaque formation. The dilution that was negative was considered the end point.
DNA Elution
The specimen tubes containing swabs were heated to 95°C for 5 minutes to destroy bacteria and prevent replication. All extraction procedures were performed in a designated room that was cleaned twice daily. Each swab was removed from its storage tube and placed into a 0.5-mL centrifuge tube. Then 50 μL of distilled, DNase-free, RNase-free water was added directly to the tip, with care taken to cover the entire surface. The same procedure was repeated for the negative controls, which consisted of two new sterile swabs. For the positive controls, 10 μL of a serial dilution of the HSV-1 DNA polymerase plasmid previously used to generate the standard curve was added to two additional new swabs, followed by 50 μL of distilled, DNase-free, RNase-free water. Positive and negative controls were assayed to determine the efficiency of extraction. DNA was eluted from the swabs by centrifugation at 11,000 rpm (Tabletop Centrifuge 5517R; Eppendorf, Hamburg, Germany) for 4 minutes at 4°C. DNA extracts were stored at 4°C.
Real-Time Quantitative PCR
Primers (Invitrogen, Carlsbad, CA) and a fluorescent probe (IDT, Coralville, IA) for the HSV-1 DNA polymerase gene were used. The forward and reverse primer sequences for HSV were 5′-CATCACCGACCCG-GAGAGGGAC-3′ and 5′-GGGCCAGGCGCTTGTTGGTGTA-3′, respectively. The fluorescent probe sequence was 5′-6FAM-CCGCCGAACT-GAGCAG-ACACCCGCGC-BHQ-1-3′. All reactions had a final volume of 50 μL, which consisted of 10× PCR buffer (Invitrogen), 50 mM MgCl2, 2 μM of each primer, 10 mM dNTP (dATP, dCTP, dGTP, dTTP), 2 μM of the probe, 1 U/μL DNA polymerase (Platinum Taq; Invitrogen), and 10 μL DNA extract. The reactions were performed in 96-well plates (Bio-Rad, Hercules, CA), which were centrifuged for <1 minute at 1000g at room temperature in a swing-out rotor (CRU 5000 centrifuge; Damon/IEC, Needham, MA) to remove any air bubbles. Amplification and detection were performed by a PCR detection system (iCycler iQ; Bio-Rad) using the following protocol: one cycle at 95°C for 3 minutes and 45 cycles at 95°C for 30 seconds, at 55°C for 30 seconds, and at 72°C for 30 seconds. Real-time fluorescence data were collected during the annealing step. In the PCR assays, three or four negative controls were included. Three or four positive controls were included in triplicate. These controls were also run at different serial dilutions (2- or 10-fold). The PCR assay was performed in a designated room separate from the extraction room.
Real-Time Data Analysis
Nine serial dilutions of a known concentration of an HSV-1 DNA polymerase plasmid were assayed in triplicate, and the baseline threshold was determined by the thermocycler system software (iCycler; Bio-Rad) to be 30. Logs of the concentrations were plotted against their corresponding threshold cycle (Ct), the fractional cycle at which the fluorescence reaches 10 times the standard deviation of the baseline, generating the following equation: Ct − 35.432/−3.133 = Y.
Hence, the threshold was manually adjusted to 30 for every subsequent assay containing unknown samples, and the HSV-1 genome copy numbers were calculated. The limit of detection was 10 copies/μL.
Result Confirmation
To assess the possibility of false positives, randomly selected PCR products of both eye and mouth swab extracts were run on 2% agarose gels. In addition, all stock components of the master mix were used as templates and assayed by nested PCR, using the DNA polymerase primer pair previously described, as well as primers for the HSV-1 ribonucleotide reductase gene. The primary forward and reverse primer sequences were 5′-ATGCCAGACCTGTTTTACAA-3′ and 5′-GTCTTTGAACATGACGAAGG-3′, respectively. The nested forward and reverse primer sequences were 5′-GAGAAGAACGTCACATG-GAC-3′ and 5′-CACTGCGCACAATGCCATAG-3′, respectively. The thermal profile used was 35 cycles of 30 seconds at 94°C, 30 seconds at 58°C, and 30 seconds at 72°C. The plasmid used to generate the standard curve was also assayed by regular PCR using our HSV-1 DNA polymerase primers, and the product was sequenced.
Statistical Analysis
Statistical evaluation of the occurrence frequencies of positive and negative PCR reactions and copy numbers was conducted in the following manner. The data from each subject individually, as well as from all subjects in aggregate, were treated as time series of concentrations of viral copy numbers occurring in the AM and PM samples over the 30-day period of the study. The resultant time series of frequencies of positives and negatives were evaluated by an exact χ2 test conducted on the aggregate eye and mouth data to detect departures from random distributions over time. The time series of aggregate data and the data for each subject were evaluated for periodic behavior (as would be seen with circadian rhythms) with the Fisher κ and Bartlett-Kolmogorov-Smirnov white noise tests.22
Results
HSV Antibody Titer
Thirty-seven (74%) of the 50 subjects were positive for HSV-1 or -2 IgG by ELISA. The neutralization assay, which tested sera only for HSV-1, yielded similar results (Table 2). Ten of the 13 subjects who were negative by ELISA were also negative by the neutralization assay (value < 1/80). Three subjects (10, 17, 45) were negative by ELISA and positive by neutralization assay. A value of < 1/80 was arbitrarily selected as the cutoff to eliminate potential false positives. Low levels of neutralizing antibodies may suggest latent HSV-1. The primary purpose of this study was to document the frequency of shedding of HSV-1 in tears and saliva. For a more thorough future study of HSV-1 antibodies, IgM analysis will be done.
Table 2.
Comparison of ELISA with HSV-1 and/or -2 IgG Antibodies and Neutralization Assay (HSV-1) in 50 Subjects
| Subject | ELISA | NA | Subject | ELISA | NA |
|---|---|---|---|---|---|
| 1 | 0.0 | 1/10 | 26 | 82.4 | 1/1280 |
| 2 | 98.2 | 1/640 | 27† | 98.6 | 1/640 |
| 3 | 103.3 | 1/1280 | 28 | 98.9 | 1/1280 |
| 4 | 105.3 | 1/640 | 29 | 56.9 | 1/40 |
| 5 | 95.7 | 1/640 | 30 | 0.0 | 1/20 |
| 6 | 87.9 | 1/640 | 31 | 0.1 | 1/20 |
| 7* | 0.2 | 1/10 | 32 | 52.4 | 1/40 |
| 8 | 0.9 | 1/20 | 33 | 85.1 | 1/1280 |
| 9 | 82.9 | 1/640 | 34‡ | 96.3 | 1/640 |
| 10 | 0.0 | 1/640 | 35 | 106 | 1/1280 |
| 11 | 0.1 | 1/10 | 36 | 93.9 | 1/640 |
| 12† | 79.2 | 1/80 | 37 | 94.5 | 1/320 |
| 13 | 102.2 | 1/640 | 38 | 0.3 | 1/20 |
| 14 | 88.9 | 1/640 | 39 | 0.9 | 1/20 |
| 15 | 88.1 | 1/5120 | 40 | 96 | 1/640 |
| 16 | 96.4 | 1/320 | 41 | 97.3 | 1/2560 |
| 17‡ | 0.0 | 1/1280 | 42 | 90.4 | 1/1280 |
| 18 | 117.4 | 1/1280 | 43 | 0.5 | 1/20 |
| 19 | 5.7 | 1/10 | 44 | 0.0 | 1/40 |
| 20 | 104.8 | 1/80 | 45 | 0.0 | 1/320 |
| 21 | 93.8 | 1/320 | 46 | 90.6 | 1/1280 |
| 22 | 109.6 | 1/640 | 47† | 91.1 | 1/2560 |
| 23 | 107.3 | 1/1280 | 48 | 98.9 | 1/640 |
| 24 | 96.1 | 1/640 | 49 | 74.5 | 1/20 |
| 25 | 99.3 | 1/640 | 50 | 102.2 | 1/640 |
NA, Neutralization assay.
Saliva and tears negative for HSV-1 DNA.
Saliva negative for HSV-1 DNA.
Tears negative for HSV-1 DNA.
HSV Genome Copy Number
Of the 5529 specimens assayed (2806 eye swabs and 2723 mouth swabs), 1961 (35.5%) contained HSV-1 DNA (Fig. 1). However, the HSV-1 genome copy numbers in the saliva samples were greater than those in the tear samples (Fig. 2), which could be a result of the sample volume collected.
Figure 1.
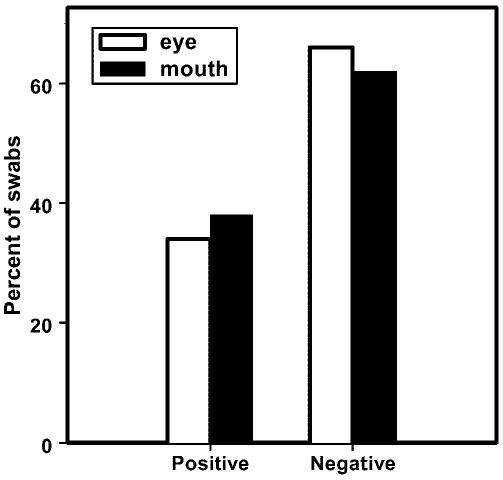
Distribution of 2806 eye and 2723 mouth swab results obtained with HSV-1 DNA polymerase real-time PCR. Of the total swabs assayed, 941 (33.5%) eye and 1020 (37.5%) mouth swabs were positive.
Figure 2.
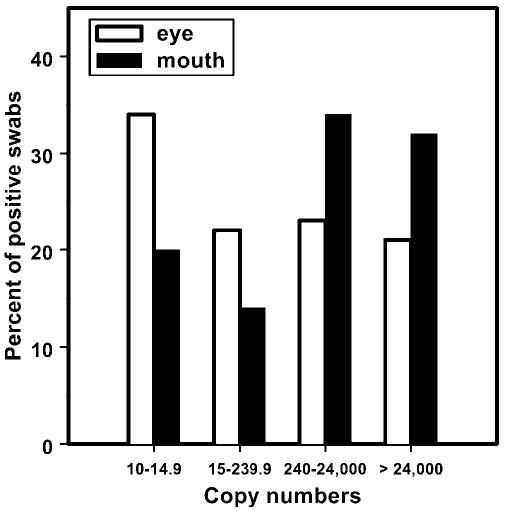
Distribution of HSV-1 DNA copy numbers per 10 μL for 941 positive eye and 1020 positive mouth swabs.
Only 21% (195/941) of the positive eye swabs contained >24,000 copies/10 μL, compared with 32% (325/1020) of the mouth swabs. The largest group of positive eye swabs (34%; 323/941) contained <15 copies/10 μL, whereas the largest group of mouth swabs (34%; 348/1020) contained 240 to 24,000 copies/10 μL (Fig. 2). Overall, when eye and mouth swabs were combined, the <15 copies/10 μL group and the 240 to 24,000 copies/10 μL group each contained 28% of the total positive specimens (552/1961 and 556/1961, respectively).
There was no correlation between a subject’s HSV antibody titer and detected HSV-1 DNA shedding pattern. Ten subjects were HSV antibody-negative by both ELISA and the neutralization assay. Only one subject (7) was antibody negative by both assays and had no HSV-1 DNA detected in tears or saliva by real-time PCR.
Shedding Frequency
The frequency of HSV-1 DNA shedding (positive swabs/total swabs) for all 50 subjects over the course of the 30-day study is shown in Figure 3. As mentioned, only one subject (7) did not shed HSV-1 DNA in tears or saliva at any time during the study. Three subjects (12, 27, 47) shed HSV-1 DNA in their tears but not their saliva, and two (17, 34) had only positive saliva swabs. Forty-four (88%) subjects had both positive tears and saliva. Forty-six (92%) excreted virus DNA in tears at some time during the study, and 45 (90%) excreted virus DNA in saliva at some time during the study. Figure 4 shows examples of the patterns of ocular and oral HSV-1 DNA shedding in three individual subjects (3, 16, 28).
Figure 3.
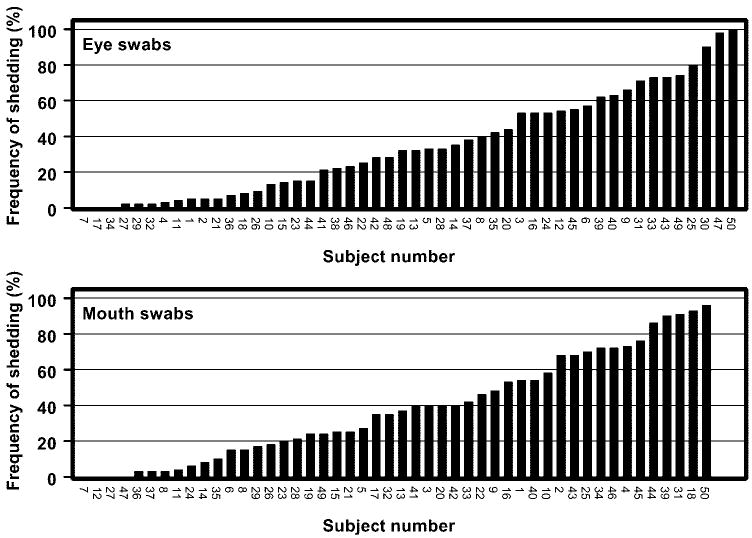
Frequency of HSV-1 DNA shedding over the course of the 30-day study (positive swabs/total swabs). Overall, 49 (98%) of the 50 subjects shed HSV DNA at least once. Only subject 7 did not shed virus in any of the samples. All 50 subjects are represented in the eye swab data; 48 subjects are represented in the mouth swab data (subjects 30, 38 not available).
Figure 4.
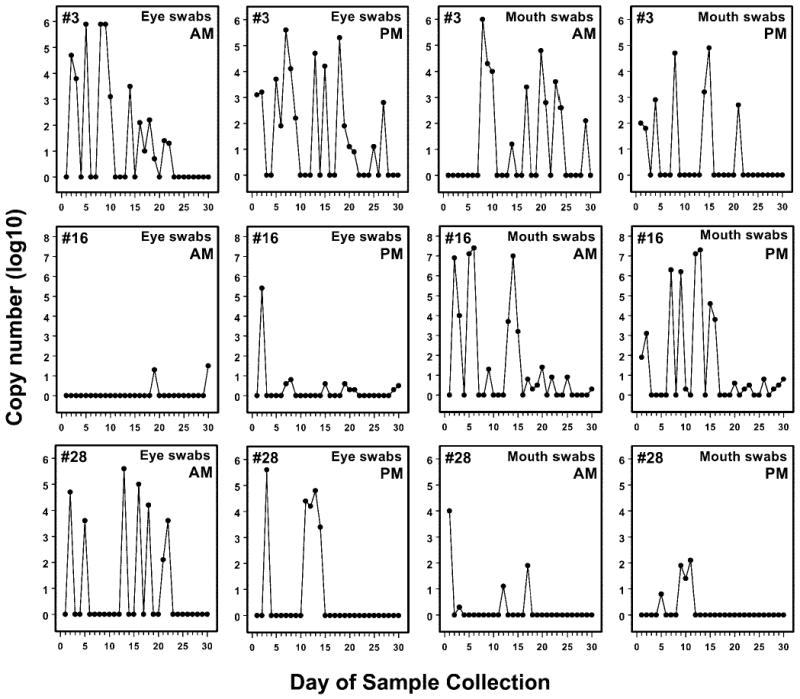
Distribution of HSV-1 DNA copy numbers in tears from subjects 3, 16, and 28 for 30 consecutive days. AM samples were obtained before oral hygiene was performed. PM samples were obtained approximately 12 hours later.
For the overall exact χ2 tests of the frequencies of positives over the 30-day study period, there were no significant departures from random distributions over time for either eye or mouth. For white noise tests, there were no significant departures from the hypothesis that the time series of copy numbers departed from the expectation that the series were broad-spectrum (white) noise. Individual analyses showed four subjects who had significant results on both Fisher κ and Bartlett-Kolmogorov-Smirnov tests for the eye data and one subject who had significant results for both eye and mouth tests (reject the white-noise hypothesis). This frequency was slightly more than would be expected to arise from testing this many individual random sequences subjects (expected, approximately three). However, as the mouth and the eye data did not appear, for the most part, as significant in the same individuals, the conservative conclusion is that some few individuals of a sample of 50 showed a hint of periodicity in HSV shedding followed over 30 days, but this was not a striking aspect of the overall sample.
Shedding Frequency and Demographics
The percentages of positive eye swabs in men and women were nearly identical (347/1003; 34.6% and 594/1709; 34.8%, respectively). However, men had a higher proportion of positive mouth swabs (457/1009; 45.3%) than did women (563/1703; 33.1%). Thirty-four percent (762/2272) of the tear samples and 35% (747/2131) of the assayed saliva samples from African Americans were positive for HSV-1 DNA. The white subjects had 37.3% (179/480) and 47.0% (273/581) positive tear and saliva samples, respectively. African-American women had significantly fewer positive saliva samples than did white women (500/1615: 31.0% and 63/88; 71.6%, respectively; P < 0.0001), but a significantly higher frequency of shedding in tears (555/1564; 35.5% and 39/145; 26.9%, respectively; P = 0.0082). By contrast, white men had a significantly higher percentage of HSV-1 shedding in tears than did African-American men (198/406; 48.8% and 150/647; 23.2%, respectively; P < 0.0001). Shedding in saliva was similar in both groups (157/357; 44.0% and 300/652; 46.0%, respectively). Familial relationships had no apparent influence on viral shedding among relatives in this study. A more detailed epidemiologic study of factors involved in HSV-1 shedding, their stability, and their correlation with serum antibodies will be conducted in the future.
Real-Time PCR Result Confirmation
The real-time PCR products randomly selected and run on 2% agarose gels confirmed the presence of the target 92-bp HSV DNA polymerase, excluding the possibility of false positives. In addition, all the master mix reagents that were assayed individually by regular PCR for possible contamination were negative. Sequencing of the PCR product of the HSV-1 DNA polymerase plasmid used to generate the standard curve yielded 98% homology with HSV-1 strain 17Syn+ (GenBank accession number X14112; GenBank http://www.ncbi.nlm.nih.gov/Genbank; provided in the public domain by the National Center for Biotechnology Information, Bethesda, MD).
Discussion
Unlike HSV-1 seroprevalence, little is known about asymptomatic shedding of HSV-1 in tears. To the best of our knowledge, there have been only three studies to date in which the presence of HSV-1 was assessed in tears of healthy individuals.11–13 Kaye et al.12 found no shedding of HSV-1, and Kaufman et al.11 and Okinaga13 found 0.8% and 0.05%, respectively, but these studies used relatively insensitive culturing techniques. In comparison to those findings, our present tear data showed significantly higher HSV-DNA–positive results. However, as is the case in assessing HSV seropositivity, the present study results (33.5% and 37.5% positive eye and mouth specimens, respectively) are in part attributable to the sensitivity of the real-time PCR diagnostic technique used, in contrast to earlier studies that relied on culturing of viable HSV. In addition, shedding appears to be intermittent, and so the number of individuals with detectable virus depends on an adequate duration of the study.
In contrast, asymptomatic shedding of HSV-1 (infectious virus and DNA) in saliva has been more widely analyzed. Reported results range from 0.45% to 76.0%.11–13,23–27 Our findings are in accordance with the report of Knaup et al.,23 in which 25 (83.3%) of 30 healthy individuals were HSV seropositive by ELISA. Of the 25 seropositive individuals, 19 (76.0%) shed HSV-1 DNA at least once in 8 to 12 oral swabs collected over a period of 58 to 167 days, as shown by nested PCR. None of the five seronegative individuals had positive oral swabs at any time. In our present study, 37 (74%) of the 50 volunteers were antibody positive, and all the antibody-positive and 11 of the 13 antibody-negative subjects shed HSV-1 DNA. The discrepancy between the results obtained from serum analysis by ELISA and tear and saliva analysis by real-time PCR in our study could be due to the different sensitivities of the assays. Serum antibodies for HSV IgG may have been present in the 11 seronegative subjects that shed virus, but not in sufficiently high titers to be detected by ELISA. Real-time PCR, however, has been shown to be more sensitive, and it detects HSV DNA irrespective of the presence of infectious virus.
A report from Posavad et al.28 may offer one explanation as to how an individual could be seronegative and still shed HSV-1 DNA. In the course of testing 24 immunocompetent subjects with no history of oral, genital, or labial herpes for HSV-specific T-cell immunity, they found that 6 of the 24 individuals exhibited T-cell immunity to HSV, but were seronegative to both HSV-1 and -2. The investigators concluded, “The identification of persistent T-cell responses to HSV in seronegative subjects is novel to the herpesvirus field and suggests either undetected infection or acquired immunity in the absence of infection.” We believe that “undetected infection” is the more likely explanation. In our study, the ELISA determination of seropositivity was general for both HSV-1 and -2. That we were unable to repeat the serologies with a type-specific assay, due to a lack of serum from the subjects, is a limitation of this work.
Real-time PCR offers numerous advantages over conventional methods used to detect HSV DNA. It requires less hands-on involvement; therefore, it is faster, and, more important, there is a reduced possibility of cross-contamination. Also, real-time PCR has consistently been reported to be significantly more sensitive than viral culture and antigen detection.14–21 A 43.9% increase in HSV-1 detection and a 62.5% increase in HSV-2 detection were observed in comparison with shell vial culture.29 In addition, real-time PCR is not hindered by the quality of the sample.17,30 However, its detection capability can be affected by the sample origin.29 The most important difference between real-time PCR and other methods used for detection of HSV in clinical specimens and probably the reason that 96% of our subjects were positive is that the PCR assay does not rely on the presence of infectious virus or on the quality and presence of appropriately infected cells.31 Although the effect of storage time of the collected samples is not known, the presence of HSV-1 DNA in high concentrations in approximately one third of these samples is sufficient to be striking and important. Some, of course, may have degraded, but that does not detract from the findings in this study.
Shedding is clearly intermittent, and a longer-term study would be expected to show an overall higher number of positive subjects, as noted in this study in comparison with other shorter studies that have focused on asymptomatic HSV-1 DNA shedding in tears and saliva.11–13,23 The probability of detecting HSV DNA is directly proportional to the duration of the study and the number of samples collected. In our study, 33.5% of assayed tear samples and 37.5% of assayed saliva samples were positive, but 98% of the 50 subjects secreted viral DNA at some time over the course of 30 days. The number of episodes of intermittent shedding (defined as one or more days of shedding separated by one or more days of not shedding) ranged from 0 to 11 for the eye swabs and 0 to 10 for the mouth swabs. The averages were 3.98 (eye) and 4 (mouth).
As mentioned previously, population demographics play a fundamental role in the prevalence of HSV infections. Consequently, the percentage of individuals shedding HSV-1 DNA noted in the present study may be in part a function of our population sample and indicative only of the incidence of HSV-1 in this particular region of the United States. The predominantly African-American female (54%) cohort is representative of the demographics of this southeastern city, the population of which comprises 67.3% African Americans and 53.1% women—two significant predictors of HSV-1 infection.32 Other documented significant predictors are age, stress, socioeconomic status, level of education, age of first intercourse, and total years of sexual activity.
Also, natural stress factors, such as sunlight exposure, may have been a contributing factor to HSV-1 DNA shedding. The present study was conducted during the summer, and although there is no consensus on whether seasonal changes affect HSV-1 shedding in humans, in animals, UV exposure is a known method for reactivation of latent HSV. Viral DNA shedding, as well as antibody titers, may have also been affected by the subject’s age. Several studies have noted an increase in HSV-1 and -2 antibody titers and frequency of herpetic disease with increased age. This is probably a result of repeated infection and/or reactivation of the primary HSV infection.
Control of virus excretion could well limit transmission, especially of more virulent strains of virus. More important, if such a high proportion of adults excrete virus, the reduction and/or prevention of virus excretion may be a simple, cost-effective way to evaluate new antiviral drugs.
Acknowledgments
The authors thank Susan Massare and Burnell Scales for excellent technical assistance.
Footnotes
Supported in part by National Eye Institute Grants R01EY002672 (HEK), R01EY006311 (JMH), and P30EY002377 (LSU Eye Center Core grant), and an unrestricted departmental grant (LSU Eye Center) and a Senior Scientific Investigator award (JMH) from Research to Prevent Blindness, Inc.
Disclosure: H.E. Kaufman, None; A.M. Azcuy, None; E.D. Varnell, None; G.D. Sloop, None; H.W. Thompson, None; J.M. Hill, None
References
- 1.Ling PD, Lednicky JA, Keitel WA, et al. The dynamics of herpesvirus and polyomavirus reactivation and shedding in healthy adults: a 14-month longitudinal study. J Infect Dis. 2003;187:1571–1580. doi: 10.1086/374739. [DOI] [PubMed] [Google Scholar]
- 2.Ghebrekidan H, Ruden U, Cox S, Wahren B, Grandien M. Prevalence of herpes simplex virus types 1 and 2, cytomegalovirus, and varicella-zoster virus infections in Eritrea. J Clin Virol. 1999;12:53–64. doi: 10.1016/s0928-0197(98)00064-6. [DOI] [PubMed] [Google Scholar]
- 3.Coyle PK, Sibony PA. Viral antibodies in normal tears. Invest Ophthalmol Vis Sci. 1988;29:1552–1558. [PubMed] [Google Scholar]
- 4.Xu F, Schillinger J, Sternberg M, et al. Seroprevalence and coinfection with herpes simplex virus type 1 and type 2 in the United States, 1988–1994. J Infect Dis. 2002;185:1019–1024. doi: 10.1086/340041. [DOI] [PubMed] [Google Scholar]
- 5.Carr DJJ, Härle P, Gebhardt BM. The immune response to ocular herpes simplex virus type 1 infection. Exp Biol Med. 2001;226:353–366. doi: 10.1177/153537020122600501. [DOI] [PubMed] [Google Scholar]
- 6.Streilein JW, Dana MR, Ksander BR. Immunity causing blindness: five different paths to herpes stromal keratitis. Immunol Today. 1997;18:443–449. doi: 10.1016/s0167-5699(97)01114-6. [DOI] [PubMed] [Google Scholar]
- 7.Becker Y. HSV-1 brain infection by the olfactory nerve route and virus latency and reactivation may cause learning and behavioral deficiencies and violence in children and adults: a point of view. Virus Genes. 1995;10:217–226. doi: 10.1007/BF01701811. [DOI] [PubMed] [Google Scholar]
- 8.Liesegang TJ. Herpes simplex virus epidemiology and ocular importance. Cornea. 2001;20:1–13. doi: 10.1097/00003226-200101000-00001. [DOI] [PubMed] [Google Scholar]
- 9.Smith JS, Robinson NJ. Age-specific prevalence of infection with herpes simplex virus types 2 and 1: a global review. J Infect Dis. 2002;186:S3–S28. doi: 10.1086/343739. [DOI] [PubMed] [Google Scholar]
- 10.Miranda-Saksena M, Boadle RA, Armati P, Cunningham AL. In rat dorsal root ganglion neurons, herpes simplex virus type 1 tegument forms in the cytoplasm of the cell body. J Virol. 2002;76:9934–9951. doi: 10.1128/JVI.76.19.9934-9951.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaufman HE, Brown DC, Ellison EM. Recurrent herpes in the rabbit and man. Science. 1967;156:1628–1629. doi: 10.1126/science.156.3782.1628. [DOI] [PubMed] [Google Scholar]
- 12.Kaye SB, Madan N, Dowd TC, Hart CA, McCarthy K, Patterson A. Ocular shedding of herpes simplex virus. Br J Ophthalmol. 1990;74:114–116. doi: 10.1136/bjo.74.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okinaga S. Shedding of herpes simplex virus type 1 into tears and saliva in healthy Japanese adults. Kurume Med J. 2000;47:273–277. doi: 10.2739/kurumemedj.47.273. [DOI] [PubMed] [Google Scholar]
- 14.Chichili GR, Athmanathan S, Farhatullah S, et al. Multiplex polymerase chain reaction for the detection of herpes simplex virus, varicella-zoster virus and cytomegalovirus in ocular specimens. Curr Eye Res. 2003;27:85–90. doi: 10.1076/ceyr.27.2.85.15947. [DOI] [PubMed] [Google Scholar]
- 15.Stocher M, Holzl G, Stekel H, Berg J. Automated detection of five human herpes virus DNAs by a set of LightCycler PCRs complemented with a single multiple internal control. J Clin Virol. 2004;29:171–178. doi: 10.1016/S1386-6532(03)00121-5. [DOI] [PubMed] [Google Scholar]
- 16.Schmutzhard J, Merete Riedel H, Zweygberg Wirgart B, Grillner L. Detection of herpes simplex virus type 1, herpes simplex virus type 2 and varicella-zoster virus in skin lesions: comparison of real-time PCR, nested PCR and virus isolation. J Clin Virol. 2004;29:120–126. doi: 10.1016/s1386-6532(03)00113-6. [DOI] [PubMed] [Google Scholar]
- 17.Thouvenot D, Morfin F. Management of mucocutaneous herpes simplex virus infections in immunocompetent patients: signification and limits of antigen detection culture methods and antibody detection. Ann Dermatol Venereol. 2002;129:609–619. [PubMed] [Google Scholar]
- 18.Breinig MK, Kingsley LA, Armstrong JA, Freeman DJ, Ho M. Epidemiology of genital herpes in Pittsburgh: serologic, sexual, and racial correlates of apparent and inapparent herpes simplex infections. J Infect Dis. 1990;162:299–305. doi: 10.1093/infdis/162.2.299. [DOI] [PubMed] [Google Scholar]
- 19.Espy MJ, Uhl JR, Mitchell PS, et al. Diagnosis of herpes simplex virus infections in the clinical laboratory by LightCycler PCR. J Clin Microbiol. 2000;38:795–799. doi: 10.1128/jcm.38.2.795-799.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Espy MJ, Ross TK, Teo R, et al. Evaluation of LightCycler PCR for implementation of laboratory diagnosis of herpes simplex virus infections. J Clin Microbiol. 2000;38:3116–3118. doi: 10.1128/jcm.38.8.3116-3118.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robert P-Y, Adenis J-P, Denis F, Alain S, Ranger-Rogez S. Herpes simplex virus DNA in corneal transplants: prospective study of 38 recipients. J Med Virol. 2003;71:69–74. doi: 10.1002/jmv.10454. [DOI] [PubMed] [Google Scholar]
- 22.Fuller WA. Introduction to Statistical Time Series. New York: John Wiley & Sons; 1976.
- 23.Knaup B, Schunemann S, Wolff MH. Subclinical reactivation of herpes simplex virus type 1 in the oral cavity. Oral Microbiol Immunol. 2000;15:281–283. doi: 10.1034/j.1399-302x.2000.150502.x. [DOI] [PubMed] [Google Scholar]
- 24.Douglas RG, Jr, Couch RB. A prospective study of chronic herpes simplex virus infection and recurrent herpes labialis in humans. J Immunol. 1970;104:289–295. [PubMed] [Google Scholar]
- 25.Kameyama T, Sajaku C, Yamamoto S, Hwang CBC, Shillitoe EJ. Shedding of herpes simplex virus type 1 into saliva. J Oral Pathol. 1988;17:478–481. doi: 10.1111/j.1600-0714.1988.tb01320.x. [DOI] [PubMed] [Google Scholar]
- 26.Scott DA, Coulter WA, Lamey PJ. Oral shedding of herpes simplex virus type 1: a review. J Oral Pathol Med. 1997;26:441–447. doi: 10.1111/j.1600-0714.1997.tb00012.x. [DOI] [PubMed] [Google Scholar]
- 27.Välimaa H, Waris M, Hukkanen V, et al. Salivary defense factors in herpes simplex virus infection. J Dent Res. 2002;81:416–421. doi: 10.1177/154405910208100612. [DOI] [PubMed] [Google Scholar]
- 28.Posavad CM, Wald A, Hosken N, et al. T cell immunity to herpes simplex viruses in seronegative subjects: silent infection or acquired immunity? J Immunol. 2003;170:4380–4388. doi: 10.4049/jimmunol.170.8.4380. [DOI] [PubMed] [Google Scholar]
- 29.Stranska R, Schuurman R, De Vos M, van Loon AM. Routine use of a highly automated and internally controlled real time PCR assay for the diagnosis of herpes simplex and varicella-zoster virus infections. J Clin Virol. 2004;30:39–44. doi: 10.1016/j.jcv.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 30.Garweg JG, Boehnke M. Low rate shedding of HSV-1 DNA, but not of infectious virus from human donor cornea into culture media. J Med Virol. 1997;52:320–325. doi: 10.1002/(sici)1096-9071(199707)52:3<320::aid-jmv14>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 31.Whiley DM, Syrmis MW, Mackay IM, Sloots TP. Preliminary comparison of three LightCycler PCR assays for the detection of herpes simplex virus in swab specimens. Eur J Clin Microbiol Infect Dis. 2003;22:764–767. doi: 10.1007/s10096-003-1031-2. [DOI] [PubMed] [Google Scholar]
- 32.Gibson JJ, Hornung CA, Alexander GR, Lee FK, Potts A, Nahmias AJ. A cross-sectional study of herpes simplex virus types 1 and 2 in college students: occurrence and determinants of infection. J Infect Dis. 1990;162:306–312. doi: 10.1093/infdis/162.2.306. [DOI] [PubMed] [Google Scholar]


