Abstract
Annual consumption of amantadine increased abruptly after its approval for the treatment of influenza A virus infections in Japan in 1998, and the emergence of amantadine-resistant viruses is now a matter of concern. To detect resistant influenza A virus strains, we have developed a PCR-restriction fragment length polymorphism (PCR-RFLP) analysis for nasopharyngeal swabs. Three different primer sets for nested PCR were designed to incorporate restriction sites into the amplicon to differentiate single-amino-acid substitutions at positions 27, 30, and 31 that confer resistance in the transmembrane domain of the M2 protein. Each PCR product was digested with respective endonucleases (BspLU11I for amino acid change at position 27, HhaI for position 30, and ScaI for position 31), and the polymorphisms were determined by electrophoresis. Thirty-four (24.1%) of 141 PCR-positive samples had resistance patterns in eight nursing homes in the 1998–1999 season. Thirty-one viruses (91.2%) showed a change at position 31 (serine to asparagine), three viruses (8.8%) showed a change at position 30 (alanine to threonine), and none showed a change at position 27. The incidence of resistant viruses did not show any significant difference between four facilities where amantadine was used mainly for influenza treatment and four other facilities where it was used only for Parkinson’s disease, values being 27.6 and 16.3%, respectively. We have confirmed that the PCR-RFLP method is useful for detecting amantadine-resistant strains directly from nasopharyngeal swabs and that resistant viruses were circulating in nursing homes where the drug was used not only for influenza virus but also for Parkinson’s disease.
The antiviral agent amantadine has been shown to be effective for treatment and prevention of human influenza A virus infections, although treated individuals may excrete resistant viruses (7–9). Single-amino-acid changes at four positions, 26, 27, 30, and 31, within the transmembrane domain of the M2 protein can confer resistance (11, 16, 17). Resistant strains have been reported to account for one-third of viruses recovered from nursing home and household settings after persons were treated with amantadine, and they apparently can be transmitted (4, 7–9, 12, 14).
Amantadine was approved for influenza virus treatment in Japan in 1998, and sales then increased suddenly. In our earlier study, we found a high frequency of resistant strains in nursing homes using the 50% tissue culture infective dose (TCID50)/0.2-ml titration method with isolated viruses and showed predominant amino acid substitutions at position 31 (serine to asparagine [Ser-31-Asn]) in the M2 protein of resistant viruses (15). However, the number of viruses examined was limited, and the correlation between indications of use and extent of resistant virus appearance was not clear.
In this report, we present a method to detect the resistant strains with substitutions at three positions (amino acids 27, 30, and 31), using PCR-restriction fragment length polymorphism (PCR-RFLP) analysis, which enables direct analysis of nasopharyngeal swabs from patients. Furthermore, we present data on incidences of amantadine-resistant strains in eight nursing homes, with a correlation between frequency and indications for therapy, namely, Parkinson’s disease or influenza A virus infections.
MATERIALS AND METHODS
Patients and sample collections.
Institutionalized elderly (over 65 years old) with influenza-like illness (ILI) in eight nursing homes in Niigata Prefecture were enrolled from November 1998 to March 1999. The average influenza vaccination rate among elderly was 47.3%, with values ranging from 7.4 to 78.6%. Patients with ILI were defined by having a sudden onset of fever over 37°C for more than 1 day and one or more of the following signs or symptoms: coughing, sore throat, or coryza. Body temperature was routinely measured and recorded for any resident with the clinical manifestations. Amantadine was administered for influenza therapy (at a dosage of 50 to 100 mg/day in two divided doses for 3 to 5 days) within 48 h of onset and not for prophylaxis or Parkinson’s disease (at a dosage of 100 to 150 mg/day throughout the study period). Nasopharyngeal swabs collected from ILI patients were stored in transport medium. The samples were stored at 4°C for a few days, until viral culture, and an aliquot was kept at −80°C. An outbreak was defined on the basis of an overall attack rate of at least 10% in the nursing homes within any 7-day period.
Viruses.
As reference viruses, we used amantadine-resistant viruses which have been produced through serial (three to five) passages of sensitive strains in the presence of amantadine (2.0 μg/ml) in vitro and also verified by partial nucleotide sequence analysis of the viral M2 gene (15). The amino acid changes are valine to alanine at position 27 (Val-27-Ala), alanine to threonine at position 30 (Ala-30-Thr), and serine to asparagine at position 31 (Ser-31-Asn).
One hundred microliters of supernatant of nasopharyngeal swabs from patients with ILI was inoculated into Madin-Darby canine kidney (MDCK) cells for virus culture. An amantadine susceptibility test was done with two series of 10-fold dilution of virus from a cytopathic effect-positive culture, plated in triplicate in a 96-well microplate on MDCK cells, with one dilution series containing amantadine (2.0 μg/ml) in the medium (15). Amantadine-resistant strains were identified when a <2.0-fold difference in log TCID50/0.2-ml titer was observed with and without the drug after 48 h of inoculation. Subtyping of influenza viruses using hemagglutinin type-specific antisera was also performed.
Extraction of viral RNA.
Viral RNA was extracted from patients’ nasopharyngeal swabs or supernatants of culture medium where viruses were inoculated, and 100-μl samples were mixed with 500 μl of TRIzol (GIBCO BRL, Life Technologies, Rockville, Md.) and 100 μl of chloroform. After incubation for 5 min at room temperature, the mixtures were centrifuged for separation of RNA from the upper aqueous phase. RNA was precipitated with 100% isopropanol at room temperature for 10 min and then purified by ether extraction.
Reverse transcription and cDNA synthesis.
RNA pellets were resuspended in 11 μl of RNase-free sterile distilled water; mixed with 5 μl of 5× first-strand buffer (GIBCO BRL, Life Technologies), 1 μl (each) of 2.5 mM deoxynucleoside triphosphate (Promega, Madison, Wis.), 2 μl of 0.1 mM dithiothreitol (GIBCO BRL, Life Technologies), 1 μl of Random Primers (Promega), 1 μl of RNase inhibitor (GIBCO BRL, Life Technologies), and 1 μl of Moloney murine leukemia virus reverse transcriptase (GIBCO BRL, Life Technologies); and incubated at 37°C for 1 h for cDNA synthesis.
PCR-RFLP analysis.
We developed a method to detect single-amino-acid changes at three sites (positions 27, 30, and 31) in the 27 amino acids spanning the transmembrane domain in the M2 protein, directly from nasopharyngeal swabs, using PCR-RFLP analysis.
Oligonucleotide primers.
The primers (GIBCO BRL, Life Technologies) were selected for the highly conserved M2 protein region of the known influenza virus genomes, using available primer-designing computer programs (Primer 3; Whitehead Institute for Biomedical Research). The product amplified by the forward primer, M2-For3 (5′-CTAGTCAGGCCAGGCAAATG-3′), and the reverse primer, M2-Rev (5′-ACTGTCGTCAGCATCCACAG-3′), in the first PCR was 339 nucleotides. We designed three specific nested PCR primer sets and selected corresponding endonucleases for three single-amino-acid changes in M2 (Table 1). GenBank analysis showed that the restriction site for nuclease BspLu11I, which cleaves within the region encoding amino acid 27, amplified by sense M2-27For (incorporating two mismatched bases at positions 25 and 26) and antisense M2-Rev2, is present in all registered M2 segments of drug-sensitive epidemic viruses. The same is also the case for the restriction site for endonuclease HhaI, which cleaves within the region encoding amino acid 30, amplified by sense M2-For4 and antisense M2-30Rev (incorporating one mismatched nucleotide at position 31), and for endonuclease ScaI, which cleaves within the region encoding amino acid 31, amplified by sense M2-For5 and antisense M2-31 (incorporating two mismatched nucleotides at position 32). If single nucleotide changes which confer resistance appear in the triplet coding for amino acids 27 (from GTT to GCT) or 30 (from GCG to ACG or GTG) or 31 (from AGT to AAT), the respective cleavage sites disappear and double-stranded DNA becomes insensitive to the endonucleases.
TABLE 1.
Nested primersa used for RFLP analysis in genotyping of substitutions in the transmembrane domain of the M2 protein of amantadine-resistant strains
| Substitution position | Sense primer
|
Antisense primer
|
Size (bp) | Endo- nuclease | Fragment lengths (bp) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Name | Sequence | Positionb | Name | Sequence | Position | ||||
| 27 | M2-27For | GGGGGTGCAGATGCAACGAT TCAAGTGACCCACAT | 732 | M2-Rev2 | TCCGTAGAAGGCCCTCTTTT | 885 | 154 | BspLU11I | 32, 122 |
| 30 | M2-For4 | CTATCAGAAACGAATGGGGG | 717 | M2-30Rev | CCACAATATCAAGTGCAAGAT CCCAATGATACGC | 811 | 95 | HhaI | 33, 62 |
| 31 | M2-For5 | TCCTAGCTCCAGTGCTGGTC | 666 | M2-31 | GAAGAACCACAATATCAAGTG CAAGATCCCAATAGT | 818 | 153 | ScaI | 37, 116 |
Sense and antisense are defined in terms of the sequence complementary to the RNA genome sequence.
Position number represents the primer’s 5′-end nucleotide position in the matrix protein of the published sequence of H3N2 A/Shiga/25/97 (accession no. AF038274).
First and nested PCR.
Complementary viral DNA (1 μl) was added to 50 μl of reaction mixtures (Promega) containing 5 μl of storage buffer A (50 mM Tris-HCl, 100 mM NaCl, 0.1 mM EDTA, 1 mM dithiothreitol, 50% glycerol, 1.0% Triton X-100), 1 μl (each) of 2.5 mM deoxynucleoside triphosphate, 3 μl (25 mM) of magnesium chloride, 2 μl (20 pmol/ml) of primers, 2 U of Taq polymerase, and 36.8 μl of sterile distilled water. Processing was done with Gene Amp PCR system 2400R (PE Corporation Applied Biosystems, Foster, Calif.). The first PCR conditions were as follows: 94°C for 5 min followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, and the final extension was run at 72°C for 7 min. Aliquots of 1 μl of the first PCR product were further amplified by nested PCR, with 50 μl of the reaction mixture detailed above except for the primer sets contained. The nested PCR conditions were as follows: 94°C for 3 min followed by 30 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s, and the final extension was run at 72°C for 7 min. In order to avoid possible influence of laboratory contamination, as a common practice both positive and negative controls were included along with the samples for every reaction.
PCR-RFLP analysis.
Each 5-μl aliquot of nested PCR product was treated with specific endonucleases. These amplified with M2-27For and M2-Rev2 were digested with 5 U of BspLu11I (Roche Diagnostics GmbH, Mannheim, Germany) for 2 h at 48°C in 1.5 μl of buffer recommended by the manufacturer and 8.0 μl of sterile distilled water. Those amplified by M2-For4 and M2-30Rev, or M2-For5 and M2-31, were digested with 5 U of HhaI (Takara Biomedicals, Ohtsu, Japan) or ScaI (New England Biolabs, Beverly, Mass.), for 2 h at 37°C, respectively, with the same mixture ratio of buffer to distilled water. The digested samples were analyzed by electrophoresis using 4% agarose X gels (Nippon Gene, Tokyo, Japan) containing ethidium bromide. The restriction fragments were separated in 0.5× Tris-borate-EDTA buffer at 100 V for 30 min and examined by transillumination before being photographed. A 50-bp DNA ladder (Promega) was used as the standard molecular size marker.
Nucleotide sequencing.
We confirmed the results by direct sequencing of the nested PCR products with a Thermo Sequenase Cy5.5 Terminator sequencing kit (Amersham Pharmacia Biotech, Piscataway, N.J.), using an automated Gene Rapid sequencer (Amersham Pharmacia Biotech). Amantadine-resistant viruses with substitutions at position 27, 30, or 31 in the M2 protein were compared with the corresponding codons on the reverse complement of the sequence obtained from known sensitive and resistant isolates (15).
Statistical analysis.
All statistical analyses were performed using the Epi Info program (6.04b; Centers for Disease Control and Prevention, Atlanta, Ga.). A P of <0.05 was regarded as statistically significant.
RESULTS
Reference viruses and isolated viruses.
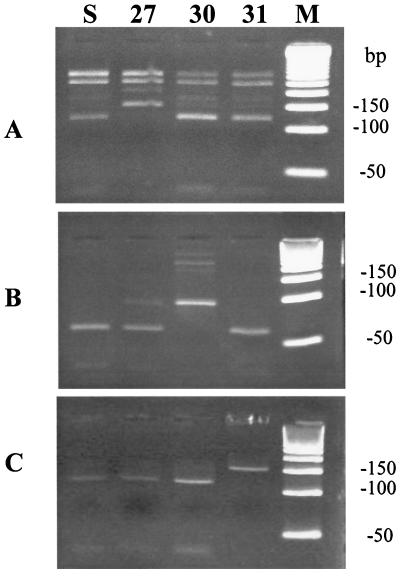
Three amantadine-resistant viruses with different substitution sites were utilized as reference viruses. They were produced in vitro and verified by partial nucleotide sequence analysis of the viral M2 gene (15). By using PCR-RFLP analysis, we could clearly differentiate the three nucleotide substitutions, namely, Val-27-Ala, Ala-30-Thr, and Ser-31-Asn (Fig. 1). The nested PCR product of Val-27-Ala was not digested with BspLU11I, in contrast to the Ala-30-Thr, Ser-31-Asn, and sensitive strains (Fig. 1A). The nested PCR product of Ala-30-Thr was not cleaved with HhaI (Fig. 1B), and that of Ser-31-Asn was not sensitive to ScaI (Fig. 1C). The results of PCR-RFLP analysis resolved by agarose gel electrophoresis were nearly identical to the predicted fragments based on the nucleotide sequence data. However, the 32-bp BspLU11I fragment, the 33-bp HhaI fragment and the 37-bp ScaI fragment were too small to be clearly visualized (Table 1; Fig. 1).
FIG. 1.
PCR-RFLP analysis of amantadine-resistant reference viruses. Each aliquot of 5 μl of reverse transcription-PCR product, amplified by specific nested primer sets, was treated with 5 U of BspLu11I (A) at 48°C for 2 h and HhaI (B) or ScaI (C) at 37°C for 2 h, respectively, and then electrophoresed in 4% agarose X gels. Lanes: S, amantadine-sensitive virus without substitution; 27, 30, and 31, and strains having amantadine resistance substitutions at amino acids 27, 30, and 31 of the M2 protein, respectively; M, 50-bp molecular size marker.
A total of 12 viruses (4.9%) from 246 patients in eight nursing homes from November 1998 to March 1999 were isolated. They were antigenically consistent with A/Sydney/95 (H3N2), a strain circulating in the community during the study period. Eight of the viruses were resistant using TCID50/0.2-ml titration, and all of them exhibited the Ser-31-Asn type in PCR-RFLP and sequencing analysis. The other four viruses sensitive by TCID50/0.2-ml titration showed sensitive patterns on RFLP analysis, and no amino acid changes at positions 27, 30, and 31 in the transmembrane domain were found by sequencing.
Clinical nasopharyngeal swabs.
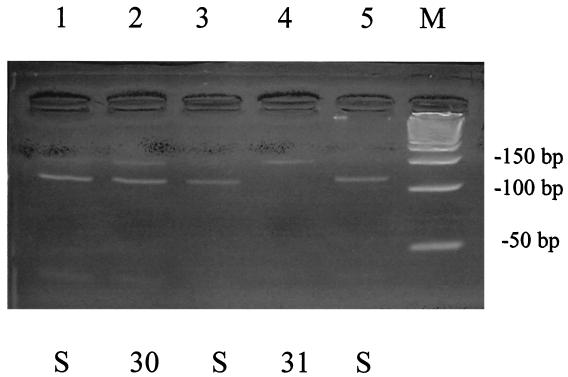
With regard to the above-mentioned 12 samples, we compared PCR-RFLP patterns of samples from isolated viruses and directly from nasopharyngeal swabs (Fig. 2). Eight resistant strains all showed the same Ser-31-Asn pattern, and four sensitive strains showed sensitive patterns by both methods.
FIG. 2.
Representative result of PCR-RFLP analysis of amantadine-resistant strains directly from nasopharyngeal swabs. Aliquots of 5 μl of nested PCR product were treated with 5 U of ScaI at 37°C for 2 h and then electrophoresed in 4% agarose X gel. Lanes: S, amantadine-sensitive virus without mutations; 30 and 31, strains having amantadine resistance substitutions at amino acids 30 and 31 of the M2 protein, respectively; M, 50-bp molecular size marker.
A total of 141 samples were PCR positive among 246 nasopharyngeal swabs collected from eight nursing homes. Thirty-four (24.1%) of them had resistant patterns by RFLP analysis; 31 viruses (91.2%) had substitutions at position 31 (Ser-31-Asn), 3 (8.8%) had substitutions at position 30 (Ala-3-Thr), and none had substitutions at position 27 (Val-27-Ala) as confirmed by partial nucleotide sequencing analysis of the M2 protein (Table 2). No samples with resistant patterns on RFLP had dual point mutations at position 27, 30, or 31 in a single allele in the M2 protein by sequencing analysis.
TABLE 2.
Frequency of resistant strains among residents in eight nursing homes in the 1998–1999 season, Niigata Prefecture, Japan
| Facility (no. of residents) | Outbreak | No. of patients receiving amantadine for:
|
No. of resistant strainsa/ no. of PCR-positive strains (%) | No. of strains with substitution in M2b at position:
|
|||
|---|---|---|---|---|---|---|---|
| Flu | Parkinson’s disease | 27 | 30 | 31 | |||
| A (95) | 5 | 1 | 3/15 (20.0) | 0 | 1 | 2 | |
| B (93) | + | 3 | 1 | 2/11 (18.2) | 0 | 0 | 2 |
| C (94) | + | 62 | 0 | 4/18 (22.2) | 0 | 0 | 3 |
| D (160) | + | 34 | 0 | 18/54 (33.3) | 0 | 1 | 18 |
| Subtotal | 104 | 2 | 27/98 (27.6)c | 0 | 2 | 25 | |
| E (88) | + | 0 | 1 | 3/26 (11.5) | 0 | 1 | 2 |
| F (112) | 0 | 5 | 3/9 (33.3) | 0 | 0 | 3 | |
| G (68) | 0 | 3 | 1/4 (25.0) | 0 | 0 | 1 | |
| H (50) | 0 | 1 | 0/4 (0.0) | 0 | 0 | 0 | |
| Subtotal | 0 | 10 | 7/43 (16.3)c | 0 | 1 | 6 | |
A sample was considered to be resistant if it showed a resistant pattern in one of three single amino acids in M2 by PCR-RFLP analysis.
Substitution position of the amino acid in the M2 protein verified by RFLP analysis and sequencing.
There was no statistical difference, with regard to frequency subtotals, between facilities where amantadine was used mainly for flu and facilities where amantadine was used only for Parkinson’s disease.
Only six (17.6%) of 34 patients with resistant strains were receiving amantadine for influenza treatment at the time of sample collection, but the remainder (82.4%) had no history of amantadine therapy during the study period. Two resistant strains were detected on day 2, and one each was detected on days 3 and 4 after starting amantadine treatment. Interestingly, two patients with resistant strains were identified on the same day that the therapy started.
Influenza outbreaks occurred at four of eight nursing homes. The vaccine strain matched strains circulating during the study period, but the influenza outbreaks tended to occur in nursing homes with low vaccination rates. Amantadine was used for influenza therapy in three of these facilities, but the outbreaks did not subside (Table 2). The incidences of resistant viruses did not demonstrate significant differences between facilities with or without outbreaks: 27 of 109 (24.8%) and 7 of 31 (21.9%), respectively. There was no evidence of outbreaks solely due to resistant strains.
Amantadine was administered mainly for influenza therapy in facilities A to D, where 27 (27.6%) out of 98 PCR positives had resistant patterns (Table 2). On the other hand, the drug was used only for Parkinson’s disease in facilities E to H, where 7 (16.3%) of 43 PCR-positive samples were resistant. The frequency of resistant viruses was thus higher in the former, although without statistical significance.
DISCUSSION
In recent years, many investigators have revealed the appearance of amantadine-resistant viruses in nursing homes or household settings where prophylactic or therapeutic use of the drug occurs (8, 12, 14). Up to approximately one-third of patients may shed resistant viruses when amantadine or rimantadine is used for therapy (7, 8). Naturally occurring influenza A viruses can be viewed as mixtures of sensitive and resistant strains with a ratio of 10,000:1; the latter would be selected within 2 to 3 days of starting amantadine therapy (8). It is well-established that single nucleotide changes leading to corresponding amino acid substitutions of one of four critical sites—amino acids 26, 27, 30, and 31—in the transmembrane region of the M2 protein confer resistance (6, 11, 13, 16). In the clinical field, we can detect actual resistant viruses by several methods, such as enzyme-linked immunosorbent assay (1, 9, 12, 14, 18), plaque reduction (7), TCID50/0.2-ml titration (15), sequencing analysis (1, 9, 12–15), or PCR-RFLP methods (6, 13).
Our earlier report indicated the efficacy of the TCID50/0.2-ml titration method with isolated viruses (15), although the virus detection rate by tissue culture was less sensitive than that by PCR. In this study, we could isolate only 12 viruses (5%) from 246 samples, but we identified 141 PCR-positive samples (57%). Carman et al. also reported that virus isolation was less than half as sensitive as PCR in detecting influenza virus in the elderly (2). The difference in our study occurred in some samples because detection of influenza virus was improved in PCR when sampling was done during periods of low viral load in the upper respiratory tract and in others because viability of the virus may have been lost during transport and processing of samples.
With low virus isolation rates, we need to develop a sensitive and rapid laboratory method. The PCR-RFLP genotyping method reported by Klimov et al. is quite useful but requires virus isolation and costly endonucleases (13). We therefore developed a PCR-RFLP analysis to distinguish resistant viruses, focusing on amino acid substitutions at positions 27, 30, and 31, using inexpensive and commercially available endonucleases. The advantage of our approach is that we can detect amantadine-resistant strains directly from patient’s nasopharyngeal swabs in as little as 8 h. The outcomes demonstrated a good match between phenotyping by TCID50/0.2-ml titration and genotyping by the PCR-RFLP method and amino acid sequencing, indicating utility for screening for resistant strains in the clinical field.
Since a thorough literature search and our previous report indicated that 70 to 80% of substitutions in amantadine-resistant viruses occur at position 31 (Ser to Asn) and that around 10% each are found at positions 27 (Val to Ala) and 30 (Ala to Thr or Ala to Val) (1, 6, 8, 9, 12–15, 18), we focused on these three genotypes with our RFLP method, covering over 90% of amino acid changes in resistant viruses. The reason why these particular genotypes are frequent is unknown.
The overall frequency of resistant viruses in eight nursing homes in Niigata Prefecture in the season of 1998 to 1999 was 24.1%, with the predominant substitution at position 31. In a previous report, up to approximately one-third of patients shed resistant viruses when amantadine or rimantadine was used for therapy (8). We found roughly 80% of patients who shed amantadine-resistant-pattern viruses did not have a history of the drug administration. Two patients shed resistant-pattern strains on the same day that the therapy started. We could not confirm whether the samples were collected before or after the actual administration, but the duration was too short for replacement of sensitive with resistant viruses, so transmission from other patients can be assumed. These findings suggest frequent transmission of resistant viruses among nursing home residents as they stay in closed communal settings.
In the present study, resistant viruses could be recovered not only from the facilities where the drug was used for influenza therapy but also after application for Parkinson’s disease. Interestingly, the proportions of resistant viruses between the two groups did not show statistical difference. In an earlier study, we encountered resistant strains in nursing homes where the drug was used for Parkinson’s disease (15), but the number was too limited to perform statistical analysis.
We strongly support the recommendations of the Advisory Committee on Immunization Practices to prevent the potential transmission of drug-resistant virus during institutional outbreaks (3). Measures should be taken to reduce contact as much as possible between persons taking and not taking antiviral drugs for treatment or chemoprophylaxis. Furthermore, we also suggest that persons taking amantadine for neurological indications should be included when such measures are taken.
In conclusion, the present investigation provided clear evidence that resistant influenza virus strains were circulating in nursing homes at a high frequency in Japan. While there appears to be no need to change existing recommendations for the use of amantadine, we request that a nationwide monitoring system be established to survey the appearance of resistant influenza A virus in such facilities.
Acknowledgments
This investigation was supported by a Research Grant for Science and Welfare, Subcommittee of Emerging and Re-emerging Diseases, Ministry of Health and Welfare, Japan, and Yujin Memorial Grant (Alumni Grant of Niigata University School of Medicine).
We thank the research team for their hard work, K. Kamimura, I. Terada, K. Kameyama, A. Sumi, K. Saito, K. Yeda, O. Sekine, Y. Oguma, M. Ohnishi, and T. Saito; all the health care workers who took part in the studies in nursing homes in Niigata Prefecture; M. Nishikawa and M. Sasagawa for their technical and scientific advice on influenza diagnosis; the staff of Denka Seiken Co., Ltd., for scientific discussions; and A. Watanabe and S. Aida for their excellent laboratory assistance.
References
- 1.Belshe, R. B., B. Burk, F. Newman, R. L. Cerrutti, and I. S. Sim. 1989. Resistance of influenza A virus to amantadine: results of one decade of surveillance. J. Infect. Dis. 159:430–435. [DOI] [PubMed] [Google Scholar]
- 2.Carman, W. F., A. G. Elder, L. A. Wallace, K. McAulay, A. Walker, G. D. Murray, and D. J. Stott. 2000. Effects of influenza vaccination of health-care workers on mortality of elderly people in long-term care: a randomized control trial. Lancet 355:93–97. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2001. Prevention and control of influenza: recommendations of the Advisory Committee on Immunization Practices (ACIP). Morb. Mortal. Wkly. Rep. 50(RR-4):1–44. [Google Scholar]
- 4.Degelau, J., S. K. Somani, S. L. Cooper, D. R. P. Guay, and K. B. Crossley. 1992. Amantadine-resistant influenza A in a nursing facility. Arch. Intern. Med. 152:390–392. [PubMed] [Google Scholar]
- 5.Douglas, R. G., Jr. 1990. Prophylaxis and treatment of influenza. N. Engl. J. Med. 322:443–450. [DOI] [PubMed] [Google Scholar]
- 6.Englund, J. A., R. E. Champlin, P. R. Wyde, H. Kantarjian, R. L. Atmar, J. Tarrand, H. Yousuf, H. Regnery A. J. Klimov, N. J. Cox, and E. Whimbey. 1998. Common emergence of amantadine- and rimantadine-resistant influenza A viruses in symptomatic immunocompromised adults. Clin. Infect. Dis. 26:1418–1424. [DOI] [PubMed] [Google Scholar]
- 7.Hall, C. B., R. Dolin, C. L. Gala, D. M. Markovitz, Y. Q. Zhang, P. H. Madore, et al. 1987. Children with influenza A infection: treatment with rimantadine. Pediatrics 80:275–282. [PubMed] [Google Scholar]
- 8.Hayden, F. G., R. B. Belshe, R. D. Clover, A. J. Hay, M. G. Oakes, and W. Soo. 1989. Emergence and apparent transmission of rimantadine-resistant influenza A virus in families. N. Engl. J. Med. 321:1696–1702. [DOI] [PubMed] [Google Scholar]
- 9.Hayden, F. G., S. J. Sperber, R. B. Belshe, R. D. Clover, A. J. Hay, and S. Pyke. 1991. Recovery of drug-resistant influenza A virus during therapeutic use of rimantadine. Antimicrob. Agents Chemother. 35:1741–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayden, F. G. 1997. Antiviral for pandemic influenza. J. Infect. Dis. 176 (Suppl.1):S56–S61. [DOI] [PubMed] [Google Scholar]
- 11.Holsinger, L. I., D. Nichani, L. H. Pinto, and R. A. Lamb. 1994. Influenza A virus M2 ion channel protein: a structure-function analysis. J. Virol. 68:1551–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Houck, P., M. Hemphill, S. LaCroix, D. Hirsh, and N. Cox. 1995. Amantadine-resistant influenza in nursing homes. Arch. Intern. Med. 155:533–537. [PubMed] [Google Scholar]
- 13.Klimov, A. J., E. Rocha, F. G. Hayden, P. S. Shult, L. F. Roumillat, and N. J. Cox. 1995. Prolonged shedding of amantadine-resistant influenza A viruses by immunodeficient patients: detection by polymerase chain reaction-restriction analysis. J. Infect. Dis. 172:1352–1355. [DOI] [PubMed] [Google Scholar]
- 14.Mast, E. E., M. W. Harmon, S. Gravenstein, S. P. Wu, N. H. Arden, R. Circo, G. Tyszka, A. P. Kendal, and J. P. Davis. 1991. Emergence and possible transmission of amantadine-resistant viruses during nursing home outbreaks of influenza A (H3N2). Am. J. Epidemiol. 134:988–997. [DOI] [PubMed] [Google Scholar]
- 15.Masuda, H., H. Suzuki, H. Oshitani, R. Saito, S. Kawasaki, M. Nishikawa, and H. Satoh. 2000. Incidence of amantadine-resistant influenza A viruses in sentinel surveillance sites and nursing homes in Niigata, Japan. Microbiol. Immunol. 44:833–839. [DOI] [PubMed] [Google Scholar]
- 16.Pinto, L. H., L. J. Holsinger, and R. A. Lamb. 1992. Influenza virus M2 protein has ion channel activity. Cell 69:517–528. [DOI] [PubMed] [Google Scholar]
- 17.Wang, C., K. Takeuchi, L. H. Pinto, and R. A. Lamb. 1993. Ion channel activity of influenza A virus M2 protein: characterization of the amantadine block. J. Virol. 67:5585–5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ziegler, T., M. L. Hemphill, M. L. Ziegler, G. Perez-Oronoz, A. I. Klimov, W. Hampson, H. L. Regnery, and N. J. Cox. 1999. Low incidence of rimantadine resistance in field isolates of influenza A viruses. J. Infect. Dis. 180:935–939. [DOI] [PubMed] [Google Scholar]