Abstract
Distinctive international clones of penicillin-nonsusceptible and multidrug-resistant Streptococcus pneumoniae are increasingly being reported. We investigated the spread of these clones in Canada through an active surveillance that was carried out at 11 Canadian pediatric tertiary care centers from 1991 to 1998. All penicillin-nonsusceptible isolates were serotyped, tested for antibiotic susceptibility, and genotyped by pulsed-field gel electrophoresis (PFGE) and random amplified polymorphic DNA (RAPD). Forty-five penicillin-nonsusceptible S. pneumoniae isolates were evaluated. Eleven serotype 9V isolates and six serotype 14 isolates displayed identical RAPD and PFGE fingerprint profiles. Twelve (70%) of these isolates were encountered in Quebec. The 9V/14 clone and the Spanish-French clone had similar PFGE fingerprint patterns. Eight isolates of serotype 23F and two isolates of serogroup 14 had the same fingerprint profiles and displayed resistance to three or more antibiotic drug classes. This clone was first detected in Calgary (Alberta) and in 1996 appeared simultaneously in various regions of Canada. This clone showed a PFGE fingerprint pattern similar to that of the Spanish-U.S. 23F clone. Our data show the emergence across Canada of two international clones of penicillin-nonsusceptible S. pneumoniae: (i) serotypes 9V and 14 related to the Spanish-French clone and (ii) the 23F Spanish-U.S. clone. The source of the first clone was in Quebec and the second international clone was probably originated from the United States. The exact reasons for the successful spread of these clones within Canada and their contribution to increased resistance to antibiotics have yet to be explored.
Streptococcus pneumoniae is the most common cause of acute otitis media, bacteremia, community-acquired pneumonia, and bacterial meningitis in children (25). Resistance to penicillin and other antibiotics has emerged rapidly (3, 14). Treatment of penicillin-nonsusceptible (PNS) S. pneumoniae (PNSSP) has become a challenge, and reports of treatment failure, especially with invasive multidrug-resistant (MDR) S. pneumoniae (MDRSP) infections, have increased (34). These treatment failures led to changes in the recommendations for antibiotic regimens for PNS SP infections, such as the addition of vancomycin for meningitis and increased dosages for the treatment of acute otitis media (1, 4, 16, 30).
The epidemiology of S. pneumoniae in various parts of Canada has been described in several reports in the last decade. Among children attending day care centers in 1995 to 1996, 44% were colonized with S. pneumoniae; 17% of these isolates had reduced susceptibility to penicillin, and 3% were highly resistant to penicillin (PHRSP), with MICs of ≥2 μg/ml (13). Active surveillance at 11 Canadian pediatric tertiary care centers from 1991 to 1998 by IMPACT (Immunization Monitoring Program, Active) of the Canadian Paediatric Society revealed a mean annual case load of invasive S. pneumoniae infections of 270 (range, 231 to 317). PNSSP isolates increased from 2.5% in 1991 to 11.3% in 1998. PHRSP comprised 32% of the PNSSP isolates in 1997 (3% of all isolates) (28). Among invasive isolates collected from laboratories across Canada between 1992 and 1995, the overall rate of reduced susceptibility was 8%, but a trend upward occurred from 0.6% in 1992 to 5.8% in 1995 (19). From 1996 to 1997, 10.2% of invasive isolates across Canada were PNSSP, and in Alberta, the rate was 16.4% (18).
Distinctive S. pneumoniae clones possessing resistance to penicillin and MDRSP are increasingly being reported from various parts of the world. The most common clones are of serotypes 23F, 6B, and 9V/14. Isolates of these clones share identical antibiotic susceptibility patterns; for instance, the 23F international clone displays resistance to penicillin, chloramphenicol, tetracycline, and co-trimoxazole, but regional selective pressure in different parts of the world adds resistance to erythromycin and extended-spectrum cephalosporins (24).
Phenotypic and genotypic methods have been developed to assist in epidemiological investigations. The same isolates are considered clones when they have the same antibiotic susceptibility pattern and similar profiles produced by molecular methods (24, 38). The use of molecular typing has helped in identifying the international clonal spread of PNSSP as well as MDR clones to different parts of the world (2, 5, 6, 27, 38). A common approach is to use two different molecular typing methods for looking at the whole genome and at specific genes such as the penicillin binding protein genes (8, 17, 22, 35). Since the first reports of penicillin-resistant S. pneumoniae there have been several reports of global distribution of PNSSP and MDRSP, using these methods to detect specific clones (6, 24).
The aim of this study was to identify clones of PNSSP and MDRSP among pediatric invasive isolates in Canada. In addition to serotyping and antibiogram profiles, two different molecular typing methods were used: (i) a PCR-based method, random amplified polymorphic DNA (RAPD), and (ii) pulsed-field gel electrophoresis (PFGE). A limited number of PNSSP and MDRSP international clones have been reported (23). A second aim was to compare these clones to international clones in order to track their spread in Canada.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Clinical isolates were collected through IMPACT of the Canadian Paediatric Society, which is an active surveillance of 11 Canadian pediatric tertiary care centers. All sterile-site isolates of PNSSP from patients under 18 years of age were included.
All available isolates were sent from the IMPACT hospitals to the National Centre for Streptococcus, Edmonton, Alberta, Canada, where they were serotyped using Danish antisera and tested for penicillin resistance (6). The bacterial stocks were stored at −70°C for further analysis. Isolates that were identified as PNSSP were obtained from the national center for additional studies.
All isolates were inoculated onto Columbia blood agar base plates with 5% horse blood and were grown in a 5% CO2 incubator at 35°C overnight. Alpha-hemolytic colonies exhibiting morphologic characteristics suggestive of S. pneumoniae were isolated. Identification of these isolates as S. pneumoniae was confirmed by inhibition with optochin disks and by the bile solubility test.
Antibiotic susceptibility.
All isolates with inhibition zones of <20 mm were considered potentially PNS. Susceptibility of isolates to erythromycin, clarithromycin, trimethoprim-sulfamethoxazole (SXT), chloramphenicol, tetracycline, ofloxacin, rifampin, and clindamycin was determined by the disk diffusion methods of Bauer and Kirby, following the recommendations of the National Committee for Clinical Laboratory Standards (NCCLS) (26). The MICs of penicillin amoxicillin, cefotaxime, cefuroxime, meropenem, SXT, and erythromycin were determined by a broth microdilution assay, using Mueller-Hinton broth with 3% laked horse blood. Isolates were classified as susceptible, intermediate, or resistant according to NCCLS guidelines.
PNSSP isolates were defined as those for which the MIC was greater than 0.1 μg/ml, and PHRSP isolates were defined as those for which the MIC was greater than 1 μg/ml (5). An MDR isolate was defined by reduced susceptibility (intermediate or highly resistant) to three or more drug classes.
Genetic molecular analyses. (i) RAPD analysis.
Three loops of culture were incubated in 250 μl of Tween 20–Tris-EDTA buffer at 94°C for 20 min. After addition of 250 μl of chloroform and centrifugation (14,000 × g, 4 min), the supernatant was removed to a fresh tube and the DNA was quantitated by A260. RAPD primers (10-mers) of 213 sequences (5′ to 3′, CAGCGAACTA) were obtained from the Nucleic Acid and Protein Service Unit, University of British Columbia, Vancouver, British Columbia, Canada. For RAPD PCR, each reaction mixture (25 μl) contained 15 ng of genomic DNA, 40 pmol of oligonucleotide, 1 U of Taq polymerase (GIBCO-BRL, Gaithersburg, Md.), 250 μM deoxynucleoside triphosphate (Ultra-Pure; Pharmacia, Laval, Quebec, Canada), 10 nM Tris-Cl (pH 8), 50 mM KCl, 0.001% gelatin, and 3 mM MgCl2. Each reaction mixture was overlaid with 25 μl of mineral oil and amplified with a Perkin-Elmer Cetus DNA thermal cycler (model TC-1) as follows: (i) 4 cycles, with 1 cycle consisting of 5 min at 94°C, 5 min at 36°C, and 5 min at 72°C, and (ii) 30 cycles, with 1 cycle consisting of 1 min at 94°C, 1 min at 36°C, and 2 min at 72°C, followed by a final extension step for 10 min at 72°C. RAPD products (9 μl) were separated by electrophoresis in 1.5% agarose gels and stained with ethidium bromide (20, 21). RAPD fingerprints were analyzed on two different occasions. PCR fingerprints were defined as concordant if PCR products yielded similar patterns correlated by Molecular Analyst Fingerprinting (MAF) software (Bio-Rad Laboratories) with the Pearson product-moment correlation coefficient, unweighted pair group method using arithmetic averages.
(ii) PFGE analysis.
Genomic DNA was prepared in situ in agarose blocks and was digested with SmaI as described previously (37). PFGE was carried out with a contour-clamped homogenous electric field apparatus (DR 33; BioRad). The following parameters were used for electrophoresis: running time of 23 h at 5.3 V/cm, initial pulse time of 5 s, and final time of 35 s.
(iii) RAPD or PFGE group.
The DNA fragment patterns generated by RAPD and by PFGE were interpreted by MAF software (Bio-Rad Laboratories) and by visualization of the fingerprints, according to criteria suggested by Tenover et al. (37). All isolates with RAPD and PFGE banding patterns with a similarity index of >70% by MAF or which appeared similar visually were run again, side by side on the same gel, to confirm the identity and were judged once more by the software and visually.
(iv) Multilocus sequence typing.
Isolates FC0542 (serotype 9V), FC0539 (serotype 14), and FC0752 (serotype 9V) were kindly analyzed by multilocus sequence typing by B. Spratt and T. C. Coffey of the Wellcome Trust Centre for Epidemiology of Infectious Disease, University of Oxford, Oxford, United Kingdom.
RESULTS
Demographic data.
During the study period, a total of 1,538 isolates were tested at the National Reference Center. One hundred four isolates with reduced susceptibility to penicillin were identified. Of these, 45 PNSSP invasive isolates, collected between 1991 and 1998 and obtained from the IMPACT study, were available for analysis and constituted our study group. Thirteen (29%) PNSSP strains were isolated between 1991 and 1995, while 32 (71%) were isolated between 1996 and 1998. Thirty-three patients (72%) were under 2 years of age, and the mean patient age was 2.4 years (range, <1 year to 15 years). Twenty-two isolates (49%) were from Quebec, nine (20%) were from Alberta, and five (11%), four (9%), two (4%), and two (4%) were from Ontario, British Columbia, Nova Scotia, and Manitoba, respectively. The most common diagnoses were bacteremia (15 [33%]), pneumonia (14 [31%]), and meningitis (9 [20%]). Other diagnoses were cellulitis, otitis media, and septic arthritis.
Serotyping and antibiotic susceptibility.
Six serotypes were represented among these isolates. The most common serotypes were 9V, 14, and 23F, at 11 (24%) each (Table 1). Twenty-two of the 45 isolates (49%) were PHRSP (MIC, >2 μg/ml). The most common serotypes in the PHRSP group were 9V and 23F (seven isolates [32%] and nine isolates [41%], respectively). Resistance was mainly to the following antibiotics: SXT, 42 isolates (93%); cefuroxime, 32 (71%); and chloramphenicol, 20 (44%). Forty-three isolates (95%) were resistant to at least two drug classes, and 25 isolates (54%) were MDR (resistant to three drug classes or more). All serotype 23F isolates were MDR.
TABLE 1.
Antibiotic resistance of serotypes of PNSSP IMPACT study isolates
| Drug | No. of resistant isolates of serotype:
|
||||||
|---|---|---|---|---|---|---|---|
| 6B (n = 7, 16%) | 9V (n = 11, 24%) | 14 (n = 11, 24%) | 19A (n = 1, 2%) | 19F (n = 4, 10%) | 23F (n = 11, 24%) | Total (n = 45, 100%) | |
| Penicillin | 0 (7 Ia) | 7 (4 I) | 4 (7 I) | 1 | 1 (3 I) | 9 (2 I) | 22 (49%) (23 I) |
| Erythromycin | 3 | 0 | 2 | 1 | 1 | 6 | 13 (29%) |
| SXT | 6 | 11 | 10 | 1 | 3 | 1 | 42 (93%) |
| Chloramphenicol | 1 | 0 | 9 | 0 | 0 | 10 | 20 (44%) |
| Tetracycline | 1 (1 I) | 0 | 1 | 0 | 3 | 8 | 13 (29%) (1 I) |
| Clindamycin | 1 | 0 | 1 | 1 | 1 | 5 | 9 (20%) |
| Rifampin | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ofloxacin | 0 | 0 | 0 | 0 (1 I) | 0 (1 I) | 1 (1 I) | 1 (3 I) |
| Meropenem | 0 | 0 (8 I) | 0 (3 I) | 0 | 0 | 0 (6 I) | 0 (17 I) |
| Cefuroxime | 4 | 8 | 5 | 1 | 3 (1 I) | 11 | 32 (71%) (1 I) |
| Cefotaxime | 0 (4 I) | 0 (7 I) | 0 (3 I) | 0 (1 I) | 0 | 0 (6 I) | 0 (21 I) |
| 2 drugs | 6 | 11 | 11 | 1 | 3 | 12 | 43 (95%) |
| ≥3 drugs | 4 | 3 | 3 | 1 | 3 | 11 | 25 (55%) |
I, intermediate susceptibility.
General molecular typing data.
Among the 45 isolates and six serotypes, there were 21 distinct RAPD patterns and 20 different PFGE fingerprint patterns. Combination of the RAPD and PFGE groups revealed two major clones.
9V/14 clone.
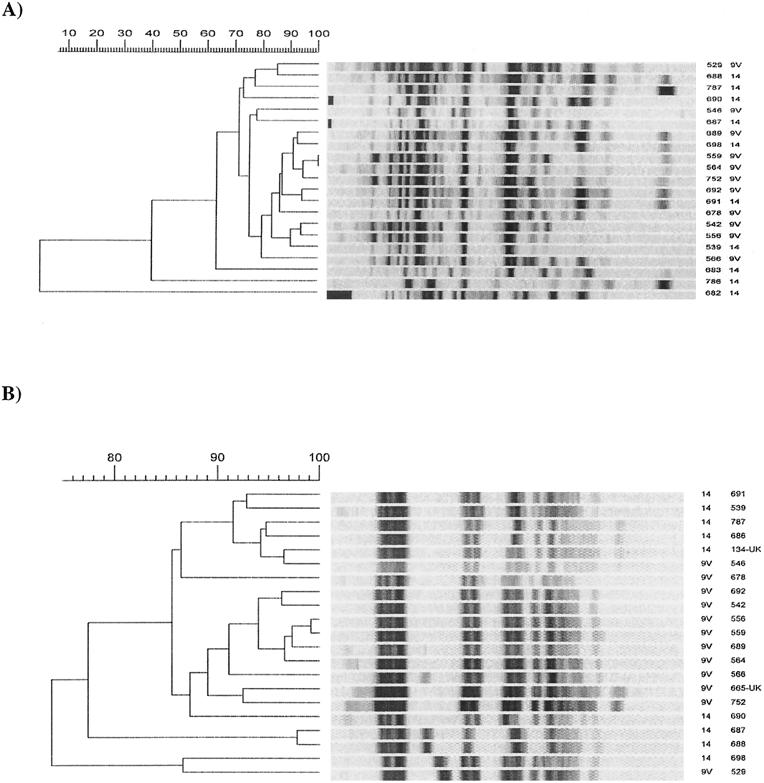
Twenty-two serotype 9V and serogroup 14 isolates were evaluated. All serotype 9V isolates were closely related, and serotype 14 produced four different fingerprint patterns by RAPD and three by PFGE.
Eleven serotype 9V and six serotype 14 isolates yielded closely related fingerprint patterns by PFGE and RAPD, constituting a clone. All 17 isolates of the 9V/14 clone were PNS, with MICs of more than 0.5 μg/ml, and SXT resistant but were susceptible to other antibiotic drug classes (Table 2).
TABLE 2.
Demographic and antibiotic resistance characterizations of the Canadian isolates of the 9V/14 clone
| Isolate | Serotype | Place | Date | Patient age (yr) | Diagnosis | Penicillin MIC (μg/ml) | Resistance toa:
|
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SXT | ERY | CHL | CLI | TET | |||||||
| FC0542 | 9V | Montreal | 1992 | 6 | Pneumonia | 2 | r | s | s | s | s |
| FC0539 | 14 | Ottawa | 1994 | 1 | Pneumonia | 2 | r | s | s | s | s |
| FC0556 | 9V | St. Justine (Montreal) | 1994 | <1 | Pneumonia | 2 | r | s | s | s | s |
| FC0546 | 9V | Montreal | 1994 | 4 | Pneumonia | 2 | r | s | s | s | s |
| FC0559 | 9V | St. Justine (Montreal) | 1994 | 3 | Pneumonia and OMb | 2 | r | s | s | s | s |
| FC0529 | 9V | Quebec | 1994 | 2 | Pneumonia and OM | 1 | r | s | s | s | s |
| FC0691 | 14 | St. Justine (Montreal) | 1996 | <1 | Pneumonia and OM | 2 | r | s | s | s | s |
| FC0692 | 9V | St. Justine (Montreal) | 1996 | 13 | Pneumonia | 2 | r | s | s | s | s |
| FC0564 | 9V | Edmonton | 1996 | <1 | Meningitis | 1 | r | s | s | s | s |
| FC0678 | 9V | Ottawa | 1996 | <1 | Meningitis | 1 | r | s | s | s | s |
| FC0686 | 14 | Montreal | 1997 | 7 | Meningitis | 1 | r | s | s | s | s |
| FC0787 | 14 | St. Justine (Montreal) | 1997 | 1 | Bacteremia | 1 | r | s | s | s | s |
| FC0687 | 14 | Montreal | 1997 | 1 | Pneumonia | 0.5 | r | s | s | s | s |
| FC0688 | 14 | Montreal | 1997 | 2 | Fever and OM | 1 | r | s | s | s | s |
| FC0689 | 9V | Montreal | 1997 | 1 | Cellulitis | 1 | r | s | s | s | s |
| FC0566 | 9V | Edmonton | 1997 | 6 | Pneumonia | 2 | r | s | s | s | s |
| FC0752 | 9V | Vancouver | 1998 | 5 | Bacteremia | 2 | r | s | s | s | s |
r, resistant; s, susceptible; SXT, trimethoprimsulfamethoxazole; ERY, erythromycin; CHL, chloramphenicol; CLI, clindamycin; TET, tetracycline.
OM, otitis media.
Fourteen isolates of the 9V/14 clone (82%) were recovered between 1996 and 1998. The first isolates were obtained from Montreal in 1992. In total, 12 isolates of the 9V/14 clone (70%) were encountered in Quebec. This clone spread to Ontario in 1994 and continued to spread westward, reaching Alberta in 1996 and British Columbia in 1998.
Comparison of the Canadian serotype 9V/14 clone with two representative isolates of the Spanish-French 9V/14 clone revealed identical fingerprint patterns by PFGE and similar antibiotic susceptibility patterns (Fig. 1). Furthermore, three representatives of this clone, including the first serotype 9V isolate from Montreal (isolated in 1992), a serogroup 14 isolate from Ontario (isolated) in 1994, and a 9V isolate from Vancouver, British Columbia, (isolated in 1998), were analyzed by the multilocus sequence typing method and found to be identical to the allelic profile 7-11-10-1-6-8-1, sequence type 156, of the Spanish-French clone.
FIG. 1.
Molecular typing of serotypes 9V and 14. (A) RAPD. The bottom three isolates of panel A are not related to a clone. (B) PFGE. Isolates 134-UK and 665-UK were obtained from B. Spratt and T. C. Coffey of the Wellcome Trust Centre for Epidemiology of Infectious Disease, University of Oxford, Oxford, United Kingdom, and served as representative isolates of the Spanish-French clone serotypes 9V/14.
The average age of the patients with this clone was 3.3 years (Table 2). All patients recovered from their invasive infections.
23F clone.
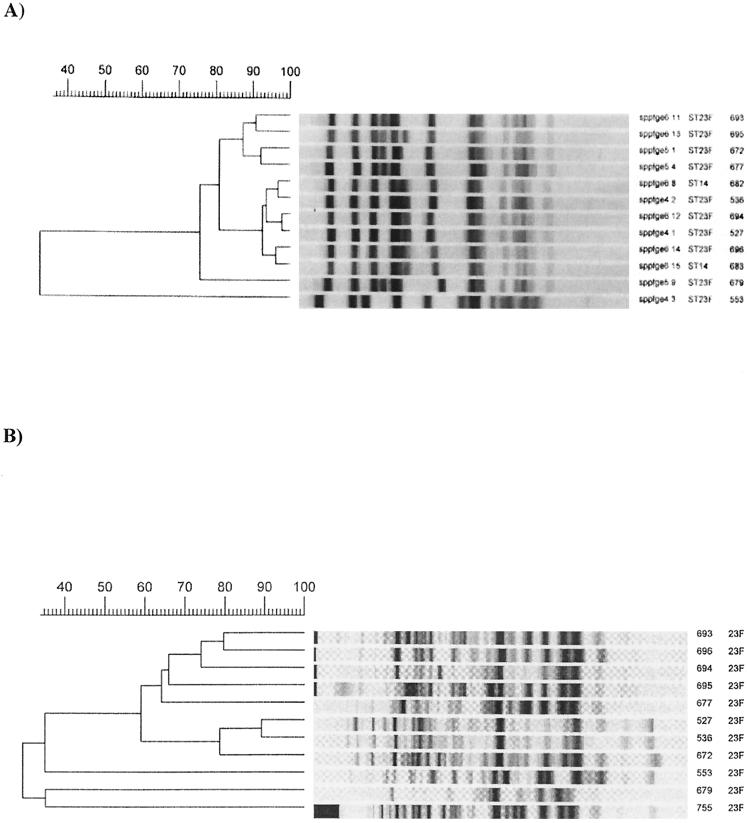
Eleven isolates of serotype 23F produced three different fingerprint pattern groups by both molecular typing methods. The largest group contained eight isolates. In addition, two isolates of serogroup 14 had a fingerprint pattern similar to that of the largest 23F subgroup by both molecular typing methods and were defined within this clone (Fig. 2).
FIG. 2.
Molecular typing of serotype 23F. (A) PFGE. (B) RAPD. The bottom two isolates of panel A and the bottom three isolates of panel B are not related to a clone.
All 10 isolates of this clone were PNS, and all isolates displayed resistance at least to three antibiotic drug classes, with the most common being penicillin, SXT, chloramphenicol, and tetracycline, with variable resistance to erythromycin. All isolates but one were clindamycin susceptible (Table 3).
TABLE 3.
Demographic and antibiotic resistance characterizations of the isolates of the 23F clone
| Isolate | Serotype | Place | Date | Patient age (yr) | Diagnosis | Penicillin MIC (μg/ml) | Resistance toa:
|
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SXT | ERY | CHL | CLI | TET | |||||||
| FC0527 | 23F | Halifax | 1997 | <1 | Pneumonia | 2 | r | s | r | s | r |
| FC0536 | 23F | Vancouver | 1996 | 8 | Bacteremia | >2 | r | r | r | s | r |
| FC0672 | 23F | Quebec | 1998 | <1 | Pneumonia | 2 | r | s | r | s | r |
| FC0677 | 23F | Winnipeg | 1997 | <1 | Bacteremia | 1 | r | s | r | s | r |
| FC0693 | 23F | St. Justine (Montreal) | 1997 | <1 | Meningitis | >2 | r | s | r | s | r |
| FC0694 | 23F | St. Justine (Montreal) | 1997 | <1 | Meningitis | 2 | r | s | r | s | r |
| FC0695 | 23F | St. Justine (Montreal) | 1996 | 2 | Pneumonia | 2 | r | r | r | r | s |
| FC0696 | 23F | Edmonton | 1996 | 6 | Pneumonia | 2 | r | s | r | s | s |
| FC0682 | 14 | Calgary | 1997 | <1 | Septic arthritis | 1 | r | r | s | s | s |
| FC0683 | 14 | Calgary | 1995 | 1 | Pneumonia | 2 | r | r | r | s | s |
SXT, trimethoprimsulfamethoxazole; Ery, erythromycin; CHL, chloramphenicol; CLI, clindamycin; TET, tetracycline; r, resistant; s, susceptible.
This clone was first detected in Calgary in 1995 and then appeared simultaneously in other regions of Canada in 1996. Eight patients (80%) were under 2 years old (Table 3). All patients recovered.
Comparison of this clone to the Spanish-U.S. 23F clone showed similar fingerprint patterns by PFGE.
DISCUSSION
The incidence of PNSSP varies from 15 to 38% in different countries. The highest incidences were found in Korea (>70%), in South Africa (45%), and in Spain (44%) (7, 9, 11, 15, 33, 39). PNSSP and MDRSP have rapidly increased in frequency in Canada over the last decade (10, 11). This study documents the spread of major clones of PNSSP and MDRSP in children under 18 years old across Canada from 1991 to 1998. Among 45 invasive PNSSP isolates, approximately half were highly penicillin resistant (MIC, >2 μg/ml), 95% were resistant to two antibacterial drug classes, and 55% were MDR. The most common drug resistance was to SXT, which was present in 93% of the isolates.
Six serotypes or serogroups were represented among PNSSP isolates. The most common were 23F, 9V, and 14, which accounted for three-quarters of all isolates. These serotypes or serogroups are well known as the most common causes of invasive S. pneumoniae infections in children and are those most often associated with antibacterial drug resistance (29). The current S. pneumoniae conjugate vaccine covers all of these isolates (32).
In addition to the serotyping method, we used two different molecular typing methods in order to subdivide these isolates and define major clones. Two major clones were found. The largest clone included all isolates of serotype 9V and six isolates of serogroup 14. This clone was resistant to penicillin and SXT but was susceptible to all other drug classes. The 9V/14 clone was initially detected in Quebec in 1992, and most subsequent isolates of this clone were encountered in this region. These serotypes were reported also to be the most common serotypes among invasive isolates associated with penicillin resistance in patients mainly younger than 2 years of age from Quebec (11). This clone was previously found in several European and South American countries and was known as the Spanish-French 9V/14 clone (31, 36, 39). The Canadian 9V/14 clone was found to be identical to the Spanish-French 9V/14 clone. It is possible that the 9V/14 clone was introduced into Canada from France, because of the close relations shared by the two countries (particularly with Quebec), either by tourism or immigration, and from there it spread to the west and over time was found in Alberta and afterward in British Columbia.
The second major clone was detected within serotype 23F and serogroup 14. These isolates were MDR and were uniformly resistant to SXT. This clone emerged in different regions of Canada during 2 years: 1995 to 1997. Comparison of this clone to the Spanish-U.S. 23F international MDR clone by PFGE revealed closely related patterns. Our hypothesis is that this clone was introduced into Canada across the border simultaneously on different occasions and spread independently within each region.
The demographic characteristics of the patients in each clonal group are different. With the 23F clone, most patients were under 2 years of age, while with the 9V/14 clone patients were older, with a mean age of 3 years. This age differences can possibly be explained by the fact that serotype 23F is carried mostly in the nasopharynx by young children, from where it can invade the blood (12). The 9V serotype is carried less often in the nasopharynx, and thus it becomes a more common cause of infection in older children. The 9V/14 clone was detected 3 years before the 23F clone. The 9V/14 clone was encountered mainly in Quebec, while the 23F clone was widespread across the country. Thus, it is possible to speculate that the modes of introduction and spread of these clones in Canada were independent of each other.
The strength of this study lies in the fact that all available cases were captured by IMPACT, minimizing reporting biases. The collection of cases was national in scope, with IMPACT centers accounting for nearly 90% of all tertiary pediatric beds. All isolates were assessed at a national reference laboratory for penicillin susceptibility as well as for serotyping. The limitation of the study is that the isolates were only from invasive infections and from only a subset of children, which limited the ability to detect the presence of these clones from other body sites such as ears or nasopharynx. Thus, these major clones represent invasive clones and may be different from mucosal clones, which are very important in day care center transmissions, where serotypes such as 6B and 23F are more common (40). In addition only 45 isolates (out of 104, or 43%) were available for molecular typing, but 22 of 26 (85%) of all penicillin-resistant (MIC, >1 μg/ml) S. pneumoniae isolates were analyzed.
It has been shown that there are limited international PNSSP and MDRSP clones, spreading mainly from Spain to different parts of the world. In this study, we were able to identify two major clones of PNSSP with different epidemiological backgrounds spreading in Canada over 8 years. Both international clones were related to the Spanish clones but each of them was probably introduced into Canada in a different fashion. The 9V/14 clone was introduced through a French Quebec connection, while the 23F MDR clone was introduced across the U.S. border.
PNSSP isolates increased from 2.5% in 1991 to 11.3% in 1998. In addition, among invasive isolates isolated from patients of various ages, including adults, a trend upward was observed, from 0.6% in 1992 to 5.8% in 1995 and to 10.2% in 1997 (18). This trend was parallel to the increase of the major clones observed in our study, and the 9V/14 and 23F clones might have contributed to this proclivity.
Knowledge of the clonal distribution of PNSSP and MDRSP within Canada will contribute to understanding the risk factors linked to its occurrence, such as geographic location, and will help initiation of strategies to prevent the spread of these organisms. In addition, it will help decision makers to implement newly developed vaccines strategies aimed at preventing S. pneumoniae infections in the future. A limited number of PNSSP and MDRSP international clones are known to have spread to many countries. Recently, nomenclature for these major international clones was published (23). These data will help in development of a world database of PNSSP and MDRSP. In addition, this database will contribute to detection of changes following the future use of conjugate vaccines in Canada, such as genetic changes (i.e., capsular switches), among S. pneumoniae invasive isolates.
The exact reasons for the successful spread of these clones within Canada and their contribution to increased resistance to antibiotics have yet to be explored.
REFERENCES
- 1.American Academy of Pediatrics. Report of the Committee on Infectious Diseases, p. 452 to 460. In Red book 2000, 25th ed. American Academy of Pediatrics, Elk Grove Village, Ill.
- 2.Austrian, R. 1976. The quellung reaction, a neglected microbiologic technique. Mt. Sinai J. Med. 43:699–709. [PubMed] [Google Scholar]
- 3.Breiman, R. F., J. C. Butler, F. C. Tenover, J. A. Elliott, and R. R. Facklam. 1994. Emergence of drug-resistant pneumococcal infections in the United States. JAMA 271:1831–1835. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 1996. Defining the public health impact of drug-resistant Streptococcus pneumoniae: report of a working group. Morb. Mortal. Wkly. Rep. 45:1–26. [PubMed] [Google Scholar]
- 5.Coffey, T. J., C. G. Dowson, M. Daniels, J. Zhou, C. Martin, B. G. Spratt, and J. M. Musser. 1991. Horizontal transfer of multiple penicillin binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol. Microbiol. 5:2255–2260. [DOI] [PubMed] [Google Scholar]
- 6.Dowson, C. G., T. J. Coffey, and B. G. Spratt. 1994. Origin and molecular epidemiology of penicillin-binding-protein-mediated resistance in β-lactam antibiotics. Trends Microbiol. 2:361–366. [DOI] [PubMed] [Google Scholar]
- 7.Friedland, I. R., and K. P. Klugman. 1992. Antibiotic-resistant pneumococcal disease in South African children. Am. J. Dis. Child. 146:920–923. [DOI] [PubMed] [Google Scholar]
- 8.Hall, L. M. C. 1998. Application of molecular typing of the epidemiology of Streptococcus pneumoniae. J. Clin. Pathol. 51:270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hermans, P. W., M. Sluijter, T. Hoogenboezem, H. Heersma, A. Van Belkum, and R. Groot. 1995. Comparative study of five different DNA fingerprint techniques for molecular typing of Streptococcus pneumoniae strains. J. Clin. Microbiol. 33:1606–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jette, L. P., F. Lamothe, and the Pneumococcus Study Group. 1989. Surveillance of invasive Streptococcus pneumoniae infection in Quebec, Canada, from 1984 to 1986: serotype distribution, antimicrobial susceptibility, and clinical characteristics. J. Clin. Microbiol. 27:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jette, L. P., G. Delage, L. Ringuette, R. Allard, P. De Weals, F. Lamothe, V. Loo, and the Pneumococcal Study Group. 2001. Surveillance of invasive Streptococcus pneumoniae infection in the province of Quebec, Canada, from 1996 to 1998: serotype distribution, antimicrobial susceptibility, and clinical characteristics. J. Clin. Microbiol. 39:733–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kellner, J. D., and E. L. Ford-Jones. 1999. Streptococcus pneumoniae carriage in children attending 59 Canadian child care centers. Toronto Child Care Centre Study Group. Arch. Pediatr. Adolesc. Med. 153:495–502. [DOI] [PubMed] [Google Scholar]
- 13.Kellner, J. D., A. McGeer, M. S. Cetron, D. E. Low, J. C. Butler, A. Matlow, J. Talbot, and E. L. Ford-Jones. 1998. The use of Streptococcus pneumoniae nasopharyngeal isolates from healthy children to predict features of invasive disease. Pediatr. Infect. Dis. J. 17:279–286. [DOI] [PubMed] [Google Scholar]
- 14.Klugman, K. P., and I. R. Friedland. 1995. Antibiotic-resistant pneumococci in pediatric disease. Microb. Drug Resist. 1:5–8. [DOI] [PubMed] [Google Scholar]
- 15.Lee, H. J., J. Y. Park, S. H. Jang, J. H. Kim, E. C. Kim, and K. W. Choi. 1995. High incidence of resistance to multiple antimicrobials in clinical isolates of Streptococcus pneumonae from university hospital in Korea. Clin. Infect. Dis. 20:826–835. [DOI] [PubMed] [Google Scholar]
- 16.Leibovitz, E., and R. Dagan. 2000. Antibiotic treatment for acute otitis media. Int. J. Antimicrob. Agents 15:169–177. [DOI] [PubMed] [Google Scholar]
- 17.Lipuma, J. J. 1998. Molecular tools for epidemiologic study of infectious diseases. Pediatr. Infect. Dis. J. 17:667–675. [DOI] [PubMed] [Google Scholar]
- 18.Lovgren, M., and J. A. Talbot. 1999. Antimicrobial-resistant Streptococcus pneumoniae. Can. J. Infect. Dis. 10:27A–29A. [DOI] [PubMed] [Google Scholar]
- 19.Lovgren, M., J. S. Spika, and J. A. Talbot. 1998. Invasive Streptococcus pneumoniae infections: serotype distribution and antimicrobial resistance in Canada, 1992 to 1995. Can. Med. Assoc. J. 158:327–331. [PMC free article] [PubMed] [Google Scholar]
- 20.Mahenthiralingam, E., M. E. Campbell, D. A. Henry, and D. P. Speert. 1996. Epidemiology of Burkholderia cepacia infection in patients with cystic fibrosis: analysis by random amplified polymorphic DNA fingerprinting. J. Clin. Microbiol. 34:2914–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahenthiralingam, E., M. Campbell, D. Henry, and D. P. Speert. 1996. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J Clin Microbiol. 34:1129–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDugal, L. K., R. Facklam, M. Reeves, S. Hunter, J. M. Swenson, B. C. Hill, and F. C. Tenover. 1992. Analysis of multiply antimicrobial-resistant isolates of Streptococcus pneumoniae from the United States. Antimicrob. Agents Chemother. 36:2176–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefevre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J. Clin. Microbiol. 7:2565–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munoz, R., T. Coffey, M. Daniels, C. G. Dowson, G. Laible, J. Casal, R. Hakenbeck, M. Jacobs, J. M. Musser, B. G. Spratt, and A. Tomasz. 1991. International spread of a multi resistant clone of serotype 23F Streptococcus pneumoniae. J. Infect. Dis. 164:302–306. [DOI] [PubMed] [Google Scholar]
- 25.Musher, D. M. 1992. Infections cause by Streptococcus pneumoniae: clinical spectrum, pathogenesis, immunity, and treatment. Clin. Infect. Dis. 14:801–809. [DOI] [PubMed] [Google Scholar]
- 26.National Committee for Clinical Laboratory Standards. 1997. Performance standards for antimicrobial susceptibility testing. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 27.Reichmann, P., A. Koning, J. Linnares, F. Alcaide, F. D. Tenover, L. McDougal, S. Swidsinski, and R. A. Hakenbeck. 1997. Global gene pool for high-level cephalosporin resistance in comensal Streptococcus species and Streptococcus pneumoniae. J. Infect. Dis. 176:1001–1012. [DOI] [PubMed] [Google Scholar]
- 28.Scheifele, D., S. Halperin, L. Pelletier, and J. Talbot. 2000. Invasive pneumococcal infections in Canadian children, 1991–1998: implications for new vaccination strategies. Clin. Infect. Dis. 31:58–64. [DOI] [PubMed] [Google Scholar]
- 29.Scott, J. A., A. J. Hall, R. Dagan, J. M. Dixon, S. J. Eykyn, A. Fenoll, M. Hortal, L. P. Jetté, J. H. Jorgensen, F. Lamothe, C. Latore, J. T. Macfarlane, D. M. Shlaes, L. E. Smart, and A. Taunay. 1996. Serogroup-specific epidemiology of Streptococcus pneumoniae: associations with age, sex and geography in 7,000 episodes of invasive disease. Clin. Infect. Dis. 22:973–981. [DOI] [PubMed] [Google Scholar]
- 30.Seikel, K., S. Shelton, and G. H. McCracken, Jr.. 1998. Middle ear fluid concentrations of amoxicillin after large dosages in children with acute otitis media. Pediatr. Infect. Dis. J. 10:969–970. [DOI] [PubMed] [Google Scholar]
- 31.Setchanova, L., and A. Tomasz. 1999. Molecular characterization of penicillin-resistant Streptococcus pneumoniae isolates from Bulgaria. J. Clin. Microbiol. 37:638–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shinefield, H. R., S. Black, P. Ray, I. Chang, N. Lewis, B. Fireman, J. Hackell, P. R. Paradiso, G. Siber, R. Kohberger, D. V. Madore, F. J. Malinowski, A. Kimura, C. Le, I. Landaw, J. Aguilar, and J. Hansen. 1999. Safety and immunogenicity of heptavalent pneumococcal CRM197 conjugate vaccine in infants and toddlers. Pediatr. Infect. Dis. J. 18:757–763. [DOI] [PubMed] [Google Scholar]
- 33.Simor, A. E., M. Louie, and D. E. Low. 1996. Canadian national survey of prevalence of antimicrobial resistance among clinical isolates of Streptococcus pneumoniae. Canadian Bacterial Surveillance Network. Antimicrob. Agents Chemother. 40:2190–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sloas, M. M., F. F. Barrett, and P. J. Chesney. 1992. Cephalosporin treatment failure in penicillin- and cephalosporin-resistant Streptococcus pneumoniae meningitis. Pediatr. Infect. Dis. J. 11:662–666. [PubMed] [Google Scholar]
- 35.Smith A. M., and K. P. Klugman. Methods in molecular medicine, vol. 15. Molecular bacteriology: protocols and clinical applications. Humana Press Inc., Totowa, N.J.
- 36.Talon, D., M. Blandine, M. J. Dupont, A. M. Chareton, and M. Thouverez. 1998. Epidemology of penicillin resistance in Streptococcus pneumoniae isolates in eastern France. Clin. Microbiol. Infect. 4:11–17. [DOI] [PubMed] [Google Scholar]
- 37.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomasz, A. 1994. Benefit and risk in the β-lactam antibiotic-resistance strategies of Streptococcus pneumoniae and Staphylococcus aureus. Trends Microbiol. 2:380–385. [DOI] [PubMed] [Google Scholar]
- 39.Tomasz, A., A. Coraso, and Members of the PAHO/Rockefeller University Workshop (G. Echaniz-Aviles, M. C. de C. Brandileone, T. Cmou, E. Castaneda, O. Figueroa, A. Rossi, and E. P. Severina). 1998. Molecular epidemiological characterization of penicillin-resistant Streptococcus pneumoniae invasive pediatric isolates recovered in six South American countries. Microb. Drug Resist. 4:195–207. [DOI] [PubMed] [Google Scholar]
- 40.Yagupsky, P., N. Porat, F. Prajgrod, D. Fraser, M. Merires, L. McGee, K. P. Klugman, and R. Dagan. 1998. Acquisition, carriage, and transmission of pneumococci with decreased antibiotic susceptibility in young children attending a day care facility in southern Israel. J. Infect. Dis. 177:1003–1012. [DOI] [PubMed] [Google Scholar]