Abstract
The adenovirus type 7 (Ad7) isolates from the 1995 nationwide outbreak in Japan were genetically and seroepidemiologically analyzed in comparison with Japanese Ad7 strains isolated before 1995 to determine their genome type and to speculate on their origin and causative factors of the outbreak. Twenty-six Ad7 isolates from the outbreak were identified by restriction enzyme analysis as the Ad7d2 genome type, while 22 Ad7 strains sporadically isolated in Japan before 1995 were identified as Ad7d. Partial nucleotide sequencing of the E3 region of Ad7d2 revealed a nucleotide substitution of G to A at position 265, resulting in the absence of the BstEII site and making Ad7d2 distinct from Ad7d. In Hiroshima City, Japan, no Ad7 was isolated from 1982 to 1994, but 43 and 50 Ad7 strains were isolated in 1995 and 1996, respectively. A seroepidemiological study of 251 serum samples collected in 1989 in Hiroshima City showed that only 2.8% of the samples were positive for Ad7. These results indicate that the 1995 outbreak of Ad7 in Japan was caused by the Ad7d2 genome type, which might have been introduced from outside Japan. The results also suggest that the low mass immunity in Japan was critical for the outbreak and that the mutation in the E3 region in Ad7d2 may have influenced transmission.
So far, 51 serotypes of human adenovirus have been identified and have been divided into six different subgenera, A to F, based on their DNA homology (4, 22, 24). Among them, adenovirus type 7 (Ad7), belonging to subgenus B, is known to cause various kinds of diseases, such as acute respiratory disease, pharyngoconjunctival fever, conjunctivitis, and gastroenteritis (22). Ad7 is one of the medically important serotypes because it causes severe or fatal lower respiratory disease in newborns and infants and causes outbreaks of respiratory disease in schools, hospitals, and military facilities (7, 22, 23). A live enteric coated Ad7 vaccine has been developed to prevent Ad7 infection (23), and molecular and epidemiological analyses of Ad7 isolates have been carried out in many countries (1, 3, 5, 7, 8, 12–14, 24–26).
In contrast with other countries (22), Ad7 was rarely isolated before 1995 in Japan (11, 30). According to the Infectious Agents Surveillance Report in Japan (11), only 30 Ad7 strains (including 15 strains isolated in 1992), 1% of all the human adenovirus isolates, were isolated during a period of 15 years, from 1980 to 1994. The low frequency of Ad7 isolations in Japan has been a unique characteristic in adenovirus epidemiology (22, 24, 25).
In 1995, a nationwide outbreak of Ad7 suddenly occurred in Japan (11, 31). The virus was first isolated in Hiroshima City, Japan, in May and was subsequently isolated in many local areas, including Akita, Yamanashi, Aichi and Nara Prefectures, and Yokohama City. Fatal cases in infants (11) and an outbreak of influenza-like illness with high fever (19), both of which were caused by Ad7, were reported in Chiba Prefecture and in a high school dormitory in Yamanashi Prefecture, respectively. A total of 75 cases of Ad7 infection were reported in 1995, and Ad7 has been isolated constantly since then (11, 32).
In this study, we genetically analyzed Ad7 isolates from the 1995 outbreak in comparison with Japanese Ad7 isolates from previous years to try to determine their origin and factors that caused the nationwide outbreak. We also studied Ad7 infection in Hiroshima City seroepidemiologically and clinically.
MATERIALS AND METHODS
Viruses.
Twelve of the 43 Ad7 strains isolated in Hiroshima City in 1995 were used for genetic analysis. Another set of 40 Ad7 strains was obtained from prefectural or municipal public health institutes in Aichi, Nara, Yamanashi, Kyoto and Akita Prefectures, and Yokohama City (Table 1). Prototypes of Ad7, Ad3, and Ad11 were obtained from the National Institute of Infectious Diseases, Tokyo, Japan. Ad7a (20) was obtained from the Aichi Prefectural Institute of Public Health. Two Ad7 isolates from Beijing, China, in 1990 were obtained from Aoki, Aoki Eye Clinic, Sapporo City, Japan. All of the viruses were passaged several times in HEp-2 cells in our laboratory.
TABLE 1.
List of Ad7 strains used in this study
| Strain code in this study | Origin
|
No. of strains used | Genome type reported bya:
|
||
|---|---|---|---|---|---|
| Place | Year | Wadell et al., Li et al. | Adrian et al. | ||
| Ad7p | United States | 1954 | 1 | Ad7p | D1 |
| Ad7a | United States | 1958 | 1 | Ad7a | D2 |
| CH/90 | Beijing, China | 1990 | 2 | Ad7d | |
| NR/87 | Nara Prefecture | 1987 | 2 | ||
| KY/87 | Kyoto Prefecture | 1987 | 1 | ||
| AI/92 | Aichi Prefecture | 1992 | 19 | ||
| HR/95 | Hiroshima City | 1995 | 12 | ||
| AK/95 | Akita Prefecture | 1995 | 1 | ||
| YM/95 | Yamanashi Prefecture | 1995 | 4 | ||
| YK/95 | Yokohama City | 1995 | 1 | ||
| NR/95 | Nara Prefecture | 1995 | 8 | ||
Extraction of viral DNA from infected cells.
Extraction of viral DNA from infected cells was carried out as described previously (18). Briefly, viral DNA was extracted from virus-infected HEp-2 cells by phenol-chloroform treatment with vigorous mixing after proteinase K digestion and then precipitated by ethanol and resuspended in a small amount of distilled water. The extracted DNAs were stocked at −70°C until use.
PCR.
A 1:100-diluted solution of viral DNA was used as a template DNA for PCR. The sequences of primers used are listed in Table 2. All of the primers were designed in our laboratory to amplify the hexon and fiber genes as well as the 5′-end, middle, and 3′-end parts of the E3 region. PCR was carried out in a PC9600 apparatus (PE Biosystems Japan, Tokyo, Japan) using Takara EX Taq (Takara Shuzo Co., Ltd., Kyoto, Japan) as a thermostable DNA polymerase. The conditions of PCR were as follows: denaturing at 94°C for 1 min, annealing at 50°C for 1 min, and extension at 72°C for 1 min, and the steps were repeated 35 times. Preheating (94°C, 5 min) and final extension (72°C, 10 min) were carried out before and after the reaction, respectively.
TABLE 2.
List of primers used in this study
| Primer code | Target region | Size of PCR products (bp) | Sequence of primer | Positiona |
|---|---|---|---|---|
| BH-S | Hexon | 788 | 5′-TACCTCTCCGCCGCAAACAT-3′ | 1891–1910 |
| BH-A | 5′-TCCACCTCAAAAGTCATCTC-3′ | 2678–2659 | ||
| BF-S | Fiber | 1,255 | 5′-CACTTACTTGAAATCAGCAATA-3′ | 4354–4375 |
| BF-A | 5′-AGCTGTGCGTGGGGAGAGATTG-3′ | 5608–5587 | ||
| BE3-1S | 5′ End part of E3 | 1,146 | 5′-TCGAGACGCCCAGGCCGAAG-3′ | 219–238 |
| BE3-1A | 5′-AAGCGAAAGTAAAGCAAGCACGAC-3′ | 1364–1341 | ||
| BE3-2S | Middle part of E3 | 1,380 | 5′-CTGGGTGGTAGCGGGTTTTGTA-3′ | 1206–1227 |
| BE3-2A | 5′-CTCTAGCGAACCCTCCATTATC-3′ | 2585–2564 | ||
| BE3-3S | 3′ End part of E3 | 1,912 | 5′-TACAAGGACCCCAA(G/C)AAGGCTA-3′ | 2520–2541 |
| BE3-3A | 5′-TACCACAGTTGGGAAGAGGG-3′ | 4431–4412 | ||
| BE3-3AIN | 3′ End part of E3 | 477 | 5′-CAGGCTATGCTACCA-3′ | 3508–3522 |
| BE3-3SIN | 5′-ATGAGTCGCTGTTCTGAGG-3′ | 3984–3966 |
Digestion of DNA by restriction endonuclease.
Digestion of DNA by restriction endonuclease was carried out as described previously (18). Briefly, whole viral DNAs or PCR-amplified DNA fragments were digested with 5 to 10 U of restriction endonuclease according to the manufacturer’s recommendation. The restriction enzymes used in this study were BamHI, BglI, BglII, BstEII, HindIII, PstI, PvuII, SacI, and SmaI for genome typing and AfaI, DdeI, HaeIII, HhaI, and MaeIII for restriction fragment length polymorphism (RFLP) of PCR products. All of the restriction enzymes were purchased from Toyobo Co., Ltd (Osaka, Japan), Takara Shuzo Co., Ltd., or Boehringer Mannheim Co., Ltd. (Tokyo, Japan).
Electrophoresis of digested DNA.
For genome typing, viral DNA was digested by a restriction endonuclease recognizing 6-bp sequences, separated by agarose gel electrophoresis with ethidium bromide, visualized under UV light, and photographed as previously described (18). For RFLP analysis, PCR products were digested by a restriction endonuclease, separated in a 2% agarose gel in TBE running buffer (89 mM Tris-borate and 2 mM EDTA) with 0.5 μg of ethidium bromide/ml, visualized under UV light, and photographed. λ fDNA/HindIII digests or 100 Base-Pair Ladder (Amersham Pharmacia Bioteck Ltd., Tokyo, Japan) were used as molecular weight markers.
Nucleotide sequencing.
PCR-amplified DNA was purified by agarose gel electrophoresis and spin column centrifugation. The purified DNA was sequenced using the primer that was used in the PCR amplification and a cycle-sequencing kit (Big Dye Terminator Cycle Sequencing FS Ready Reaction Kit; PE Biosystems Japan, Tokyo, Japan) according to the manufacturer’s protocol. Single-stranded DNA obtained by terminating reaction was analyzed using a Genetic Analyzer Prism 310 (PE Biosystems Japan).
Virus isolation.
More than 10,000 clinical samples, including throat and conjunctival swabs, feces, urine, and cerebrospinal fluid, were collected from patients with various kinds of infectious disease in pediatric clinics or hospitals in Hiroshima City between 1982 and 1996, as part of the program of National Epidemiological Surveillance of Infectious Diseases. Virus isolation was mainly performed with HEp-2 cells, human embryonic fibroblast cells, RD-18S cells, and Vero cells as described previously (17). Isolated adenoviruses were serotyped by neutralization tests with antisera provided by the National Institute of Infectious Diseases, Tokyo, Japan, or with antisera purchased from Denka Seiken Co., Ltd. (Tokyo, Japan) or Ismunit Co. (Rome, Italy).
Sera.
A total of 251 serum samples that had been collected from 252 individuals, aged 0 to 87 years, in Hiroshima City in 1989 were used for a seroepidemiological study of Ad7 infection before the outbreak in 1995.
Neutralization and hemagglutination inhibition tests.
Neutralization (NT) tests were carried out as previously described (17). Briefly, a challenge virus was used at a dilution that showed a complete cytopathic effect (CPE) 4 days after inoculation. Sera were inactivated at 56°C for 30 min before use. A mixture of the challenge virus and diluted serum was incubated at 37°C for 2 h and then at 4°C overnight, and it was inoculated onto HEp-2 cells. The NT titer was expressed as a reciprocal of the highest dilution of serum that inhibited CPE completely 4 days after inoculation.
Hemagglutination inhibition (HI) tests were performed by a conventional method in U-type microtiter plates in phosphate-buffered saline at 37°C with 0.5% monkey erythrocytes. Antisera were treated at a final dilution of 1:4 with 25% kaolin and packed monkey erythrocytes before use (17).
Analysis of clinical data.
Data obtained from 93 Ad7 patients in Hiroshima City between 1995 and 1996 were used for analysis of clinical features. The clinical data used in this study were data obtained for clinical items ordered for examination at our laboratory for virus isolation by each of the doctors who had made diagnoses.
RESULTS
Genome types of Ad7 isolates in Japan.
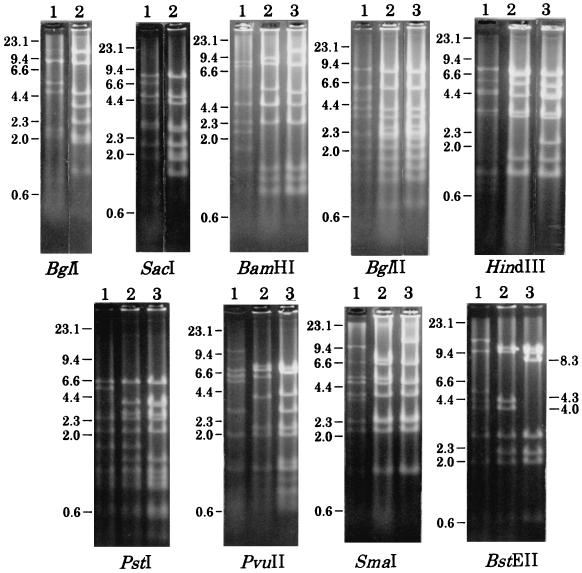
We analyzed a total of 52 Ad7 strains, listed in Table 1, using nine different restriction enzymes, BamHI, BglI, BglII, BstEII, HindIII, PstI, PvuII, SacI, and SmaI. To confirm the genome types of the reference strains, Ad7 prototype, Ad7a, and two Chinese Ad7 strains isolated in 1990 (CH/90), we compared our restriction patterns with those reported previously. Our patterns of the Ad7 prototype, Ad7a, and the two CH/90 strains, shown in Fig. 1 and Table 3, corresponded to those of Ad7p, Ad7a, and Ad7d reported by Wadell et al. (25) and Li et al. (13), and the patterns of the former two strains corresponded to those of D1 and D2 reported by Adrian et al. (1). The genome types of the Ad7 prototype, Ad7a, and the two CH/90 strains are designated here as Ad7p, Ad7a, and Ad7d, respectively (Table 3).
FIG. 1.
Restriction patterns of viral DNA of Ad7 strains digested with BglI, SacI, BamHI, BglII, HindIII, PstI, PvuII, SmaI, or BstEII. The lane numbers correspond with the codes of restriction pattern by each enzyme listed in Table 3. Molecular sizes are expressed as kilobase pairs.
TABLE 3.
Genome types of Ad7 isolates and reference strains
| Strain code (no. of strains tested) | Restriction pattern codes in panel ofa:
|
Genome type in this study | ||
|---|---|---|---|---|
| BgII, SacI | BamHI and othersa | BstEII | ||
| Ad7p (1) | 1 | 1 | 1 | Ad7p |
| Ad7a (1) | 2 | 2 | 2 | Ad7a |
| CH/90 (2) | 2 | 3 | 2 | Ad7d |
| NR/87 (2) | 2 | 3 | 2 | Ad7d |
| KY/87 (1) | 2 | 3 | 2 | Ad7d |
| AI/92 (19) | 2 | 3 | 2 | Ad7d |
| HR/95 (12) | 2 | 3 | 3 | Ad7d2 |
| AK/95 (1) | 2 | 3 | 3 | Ad7d2 |
| YM/95 (4) | 2 | 3 | 3 | Ad7d2 |
| YK/95 (1) | 2 | 3 | 3 | Ad7d2 |
| NR/95 (8) | 2 | 3 | 3 | Ad7d2 |
Restriction pattern codes correspond with the lane number in each panel in Fig. 1.
Other enzymes were BgIII, HindIII, PstI, PvuII, and SmaI.
Twenty-two Japanese Ad7 strains isolated before 1995, including two strains isolated in Nara Prefecture in 1987 (NR/87), one isolated in Kyoto Prefecture in 1987 (KY/87), and 19 isolated in Aichi Prefecture in 1992 (AI/92), showed the same restriction patterns as those of Ad7d by digestions with the nine restriction enzymes (Fig. 1 and Table 3). The results indicate that the genome type of Ad7 strains isolated in Japan before 1995 is Ad7d. On the other hand, 26 Ad7 strains isolated in 1995, including 12 strains isolated in Hiroshima City (HR/95), eight isolated in Nara Prefecture (NR/95), four isolated in Yamanashi Prefecture (YM/95), one isolated in Akita Prefecture (AK/95), and one isolated in Yokohama City (YK/95), showed a distinct pattern from those of Ad7d after BstEII digestion (Fig. 1 and Table 3). The other eight enzymes generated identical patterns. With BstEII, the Ad7 strains isolated in 1995 lacked two fragments of about 4,300 and 4,000 bp in length, which appeared in Ad7d, and had an additional fragment of about 8,300 bp (Fig. 1, BstEII). Our present restriction pattern of the 1995 Ad7 isolates seemed to be identical to that of Ad7d2 reported by Azar et al. (3), in which Ad7d2 only differed from Ad7d in BstEII digestion when using seven enzymes; an additional 8,350-bp fragment appeared with 4,350- and 4,030-bp fragments missing. Thus, we identified the Japanese Ad7 strains isolated in 1995 as Ad7d2.
Genetic analysis in E3 region of Ad7.
The restriction pattern change observed seemed to have resulted from a mutation(s) at a single BstEII site, because the sum of the length of the 4,300- and 4,000-bp fragments of Ad7d is equal to that of the 8,300-bp fragment of Ad7d2, including the Ad7 isolates obtained in 1995. The mutation site(s) of Ad7d2 appeared to be located at about 0.86 map units of the genome according to the restriction site map reported by Adrian et al. (1). This site is located near the 3′ end of the E3 region, which encodes proteins shown to modulate the host’s immune response (29). We therefore determined the partial nucleotide sequences of the E3 region, including the mutation site(s), with the PCR products of 10 Ad7 strains (Table 4). The 401-bp sequences from position 37 to position 437 on the PCR products were compared. The results for the Ad7d2 isolates, HR/95 and YM/95, showed a nucleotide substitution of G to A at position 265 in the open reading frame encoding a 14.9-kDa protein compared to Ad7d strains, including the Japanese Ad7d isolates, and the substitution resulted in the absence of the BstEII site. This nucleotide change was accompanied by a predicted amino acid substitution of Gly at position 89 to Ser. The other nucleotide substitutions found in the sequences were not associated with any predicted amino acid changes.
TABLE 4.
Comparison of nucleotide sequences near 3′ end of E3 region among Ad7 strains
| Strain code (no. of strains tested) | Accession no. | Nucleotide in PCR products at positiona:
|
|||
|---|---|---|---|---|---|
| 186 (204) | 247 (265) | 276 (294) | 342 (360) | ||
| Ad7p (1) | AB054824 | A | G | T | G |
| Ad7a (1) | AB054825 | C | G | C | G |
| CH/90 (2) | AB054818 | C | G | C | A |
| NR/87 (1) | AB054819 | C | G | C | A |
| KY/87 (1) | AB054820 | C | G | C | A |
| AI/92 (2) | AB054821 | C | G | C | A |
| HR/95 (1) | AB054822 | C | A | C | A |
| YM/95 (1) | AB054823 | C | A | C | A |
| Predicted amino acid substitution | Silent | Gly to Ser | Silent | Silent | |
Values in parentheses indicate the nucleotide positions in the 14.9-kDa protein-encoding open reading frame of the Ad7 prototype (Accession no. M23696).
To find other possible genetic differences between the E3 regions of Ad7d and Ad7d2, RFLP analysis with four restriction endonucleases recognizing 4-bp sequences, AfaI, DdeI, HaeIII, and HhaI, was performed. However, no other genetic differences were found between them in the PCR products analyzed (data not shown).
Antigenic and genetic analyses of hexons and fibers in Ad7 isolates.
Several antigenically intermediate adenoviruses in subgenus B have been reported (2, 28). We therefore analyzed the hexons and fibers in Ad7 isolates antigenically and genetically. The results of NT and HI tests using anti-Ad7, anti-Ad3, and anti-Ad11 sera showed that Ad7d and Ad7d2 had common antigenicity to that of the Ad7 prototype. No antigenic difference was found between the Ad7d and Ad7d2 (data not shown). The Ad3 prototype reacted weakly with anti-Ad7 serum in NT tests.
We also performed RFLP analysis of PCR products in hexon and fiber genes of Ad7 isolates with restriction enzymes recognizing 4-bp sequences. DNA fragments of 788 bp in length for the hexon gene and 1,255 bp in length for the fiber gene were amplified. In RFLP analysis of the hexon gene, the restriction patterns of Ad7p and Ad7a were different with AfaI, DdeI, HaeIII, and HhaI (data not shown). Ad7d (NR/87 and AI/92) and Ad7d2 (HR/95) showed the same patterns as those of Ad7a with the four enzymes, indicating that the hexon gene of Ad7d and Ad7d2 is more closely related to Ad7a than to Ad7p. In RFLP analysis of the fiber gene, Ad7p, Ad7a, Ad7d (NR/87 and AI/92), and Ad7d2 (HR/95) showed the same restriction patterns as AfaI, DdeI, HaeIII, and MaeIII (data not shown). None of the PCR products of the fiber gene were digested with HhaI. Thus, no genetic difference was found between Ad7d and Ad7d2 in hexon and fiber genes.
Isolation and seroepidemiolgy of Ad7 in Hiroshima City before 1995 outbreak.
We have isolated adenoviruses associated with various kinds of infectious diseases, such as respiratory, gastrointestinal, and eye diseases, from patients in Hiroshima City. A total of 818 adenoviruses (14 serotypes) were isolated over a 13-year period, from 1982 to 1994. Of the 818 isolates, 265 (32.3%) corresponded to serotype 3, suggesting that Ad3 was the most prevalent serotype circulating during this period. In contrast, no Ad7 was isolated during the same period. Ad7 was first isolated in May 1995 from a female infant aged 1 year who was diagnosed as having acute bronchitis and tympanitis. Since then, Ad7 has been frequently isolated. The numbers of Ad7 and Ad3 isolates were 43 and 28 in 1995 and 50 and 17 in 1996, respectively. In 1996, Ad7 was the most frequently isolated serotype among all of the adenoviruses. A seroepidemiological study of Ad7 before the Ad7 outbreak in 1995 was performed using 251 serum samples collected in 1989 in Hiroshima City. Seven of the 251 serum samples (2.8%) were positive for Ad7 in a 1:4 dilution by NT tests (Table 5). In contrast, 107 (42.6%) were positive for Ad3. These results of the virus isolation and seroepidemiological study suggested that Ad7 had rarely existed in Hiroshima City before the Ad7 outbreak in 1995.
TABLE 5.
Seroepidemiology of Ad7 and Ad3 infections in Hiroshima City in 1989
| Age of person | No. of samples | No. (%) of positive cases of:
|
|
|---|---|---|---|
| Ad7 | Ad3 | ||
| 0–5 months | 36 | 1(2.8) | 17(47.2) |
| 6–11 months | 18 | 0(0.0) | 1(5.6) |
| 1 year | 19 | 0(0.0) | 1(5.3) |
| 2 years | 17 | 0(0.0) | 3(17.6) |
| 3 years | 20 | 0(0.0) | 3(15.0) |
| 4 years | 19 | 0(0.0) | 7(36.8) |
| 5 years | 20 | 1(5.0) | 12(60.0) |
| 6–9 years | 20 | 2(10.0) | 15(75.0) |
| 10–14 years | 21 | 0(0.0) | 12(57.1) |
| 14–19 years | 19 | 0(0.0) | 10(52.6) |
| ≥20 years | 42 | 3(7.1) | 26(61.9) |
| Total | 251 | 7(2.8) | 107(42.6) |
Clinical features of Ad7 patients in Hiroshima City.
We examined clinical features of 93 patients infected with Ad7 during the period from January 1995 to December 1996 in Hiroshima City. The main clinical symptoms of the Ad7 patients were fever (91.4%), upper respiratory symptoms (65.6%), lower respiratory symptoms (26.9%), diarrhea (21.5%), vomiting (18.3%), tonsillitis (12.9%), and gastroenteritis (11.8%). Among the Ad7-infected patients, 49.5% had a fever of 40°C or higher and 74.2% were less than 6 years of age.
DISCUSSION
Ad7 had rarely been isolated in Japan before 1995 (11, 30). After the first isolation of Ad7 in Hiroshima City in May 1995 (11), the virus was isolated in eight regional institutes of public health and one commercial diagnostic laboratory in various districts in Japan during the period from May to October 1995 (11, 31). A total of 75 Ad7 cases were reported in 1995, and the virus has been isolated constantly since then (11, 32). In the present study, we genetically analyzed Ad7 strains isolated from different local areas in the early period of the outbreak to examine the genetic diversity among the Ad7 isolates and to speculate on their origin and the causative factors for the nationwide Ad7 outbreak. Restriction enzyme analysis using nine 6-bp-recognizing endonucleases demonstrated that all the 26 Ad7 isolates in 1995 belonged to an identical genome type, which was closely related to Ad7d. The only difference found between the isolates and Ad7d was in the BstEII restriction pattern, which seemed to be identical to that of the Ad7d2 strain from Israel reported by Azar et al. (3), and hence we concluded that Japanese Ad7 isolates strains isolated in 1995 were the Ad7d2 genome type. Direct comparison of restriction patterns between the Japanese and Israeli strains is needed to confirm their identity. In contrast, the 22 Ad7 strains isolated from sporadic cases before 1995 were identified as being the Ad7d genome type.
So far, many genome types of Ad7 strains have been identified (1, 3, 5, 8, 12–14, 24–26). According to reports on the genome typing of Ad7 strains isolated in many countries (13, 25), Ad7d was isolated only in China between 1958 and 1984. Li et al. (14) also reported that Ad7d was incomparably the predominant genome type from 1980 to 1990 in China. Azar et al. (3) reported that genome types other than Ad7d2 had been isolated between 1968 and 1984 in Israel and that only Ad7d2 was isolated between 1992 and 1995. Although Ad7d2 had not been found in the other countries except for Japan (1, 5, 8, 12–14, 25, 26), an outbreak of Ad7d2 was reported in the United States in 1998 (7). These results suggest that Ad7d or Ad7d-related genome types such as Ad7d2 might have been a characteristic genome type for Asian countries until 1998. Considering the present results together with the geographical distribution of Ad7d2 and Ad7d described above, we speculate that the Japanese Ad7d2 in 1995 was imported into Japan from an Asian country such as Israel just before 1995 and subsequently spread throughout Japan. A similar explanation was given for the emergence of Ad7d2 in Israel (3). Ad7d2 may have been introduced into Japan during the Asian Olympic Games held in Hiroshima City in 1994. The sporadic isolates of NR/87, KY/87, and AI/92 might also have been imported from an Asian country such as China.
Although the possibility that the Japanese Ad7d2 emerged domestically due to mutations from the sporadic Ad7d viruses before 1995 cannot be ruled out, the above view appeared to be supported by the results of our preliminary analysis of the Ad7d and Ad7d2 isolates with 4-bp-recognizing endonucleases (data not shown). NR/87 and KY/87 in the sporadic Ad7d strains showed the same restriction patterns with the four enzymes as those of the CH/90 strain, while AI/92 showed different patterns with the two enzymes from CH/90, suggesting that NR/87 and KY/87 were more closely related to CH/90 than to AI/92. Furthermore, Ad7d2 strains isolated in 1995 (HR/95 and YM/95) were distinct from those Ad7d strains in their patterns by the four enzyme digestions. Notably, AI/92 possessed a unique fragment(s) not found in NR/87, KY/87, HR/95, and YM/95, suggesting that Ad7d2 isolated in Japan in 1995 did not evolve from AI/92, the strain that appeared in Japan just before 1995. It is possible that the American Ad7d2 isolated in 1998 might also have been introduced from an outside source, but there are no available data on genome types of adenoviruses prevailing in the United States before 1998 to confirm this speculation (7).
The results of the seroepidemiological study using 251 serum samples collected in Hiroshima City in 1989 indicated that the incidence of Ad7 infection in Hiroshima City was significantly lower than that of Ad3 infection (2.8 and 42.6%, respectively). Ad7 had not been isolated in Hiroshima City during a period of 13 years from 1982 to 1994. Other seroepidemiological studies (15, 16, 19, 27) and studies on virus isolation (11, 30) have also indicated that the incidences of Ad7 infection in various areas in Japan had been low for a period of about 20 years before the nationwide outbreak, although Nishio et al. (16) showed seroepidemiologically that there had been a high incidence of Ad7 infection before 1960, and then it disappeared. Thus, Ad7 rarely existed in Japan for a few decades before the outbreak in 1995. The fact that the level of mass immunity of the Japanese population against Ad7 before 1995 was very low must have been a critical factor in the nationwide outbreak of Ad7. If and how mass immunity influences adenovirus circulation and outbreaks has been not well understood. It would be interesting to further examine the change in the seroprevalence of Ad7 in the same population following the outbreak and the continuous circulation to determine the reasons for the replacement of Ad3 by Ad7 and for the outbreak.
The nationwide outbreak of Ad7 in 1995, however, cannot be explained only by the low positive rate of Ad7 antibody in the Japanese population, because some Ad7 epidemics before 1995, for instance, in Nara and Kyoto Prefectures in 1987 and in Aichi Prefecture in 1992 (11), resulted in sporadic cases despite the similar seroepidemiological background. It is possible that Ad7d2 has a stronger transmissibility, infectivity, or pathogenicity than does Ad7d. In the first step of investigating this issue, we examined the mutation(s) at a BstEII site of Ad7d2 DNA because the mutation(s) seemed to be located near the 3′ end of the E3 region, which encodes immunomodulatory proteins (29). A comparison of the nucleotide sequence of Ad7d2 with that of Ad7d showed a mutation of G at position 265 to A in the E3 region, causing a predicted amino acid change of Gly at position 89 to Ser in a 14.9-kDa protein. The function of the 14.9-kDa protein of Ad7 is unknown, but the corresponding proteins of Ad2 and Ad5 have been shown to inhibit cytolysis, apoptosis, and inflammation induced by tumor necrosis factor (6, 9, 29). Further genetic and biochemical analyses are needed to demonstrate whether the mutation in the 14.9-kDa protein is related to the pathogenicity of Ad7d2.
Analysis of clinical features of Ad7 patients in Hiroshima City demonstrated that Ad7d2 caused a variety of mild diseases. Although five fatal cases of Ad7 infection (two in Chiba Prefecture, one in Asahikawa City, and two in Kushiro City) were reported during the 1995 nationwide outbreak (11, 21, 33), these cases corresponded to infants with basal diseases. The restriction pattern of one isolate from the fatal cases in Chiba Prefecture was shown to be the same as that of our Ad7d2 strains (10). These results suggest that Ad7d2 causes usually mild diseases but may cause severe or fatal diseases in patients with some underlying diseases. The clinical features observed in this study are similar to those observed in Ad7d2-infected patients in a pediatric chronic-care facility and tertiary-care hospital in the United States (7).
In conclusion, the Ad7 nationwide outbreak in Japan in 1995 was caused by genome type d2, which could have been introduced from another Asian country. Although the factors causing the outbreak are still not clear, the low mass immunity against Ad7 must have been a critical factor. In addition, it is possible that an amino acid-changing mutation found in the E3 region of Ad7d2, possibly encoding immunomodulatory proteins, might have caused its greater transmissibility or pathogenicity.
Acknowledgments
We thank Koki Aoki for providing Chinese Ad7 strains.
REFERENCES
- 1.Adrian, T., M. Becker, J. C. Hierholzer, and R. Wigand. 1989. Molecular epidemiology and restriction site mapping of adenovirus 7 genome types. Arch. Virol. 106:73–84. [DOI] [PubMed] [Google Scholar]
- 2.Adrian, T., and R. Wigand. 1986. Adenovirus 3-7, an intermediate strain of subgenus B. Intervirology 26:202–206. [DOI] [PubMed] [Google Scholar]
- 3.Azar, R., N. Varsano, F. Mileguir, and E. Mendelson. 1998. Molecular epidemiology of adenovirus type 7 in Israel: identification of two new genome types, Ad7k and Ad7d2. J. Med. Virol. 54:291–299. [DOI] [PubMed] [Google Scholar]
- 4.De Jong, J. C., A. G. Wermenbol, M. W. Verweij-Uijterwaal, K. W. Slaterus, P. Wertheim-Van Dillen, G. J. Van Doornum, S. H. Khoo, and J. C. Hierholzer. 1999. Adenoviruses from human immunodeficiency virus-infected individuals, including two strains that represent new candidate serotypes Ad50 and Ad51 of species B1 and D, respectively. J. Clin. Microbiol. 37:3940–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Silva, L. M., P. Colditz, and G. Wadell. 1989. Adenovirus type 7 infections in children in New South Wales, Australia. J. Med. Virol. 29:28–32. [DOI] [PubMed] [Google Scholar]
- 6.Dimitrov, T., P. Krajcsi, T. W. Hermiston, A. E. Tollefson, M. Hannink, and W. S. Wold. 1997. Adenovirus E3–10.4K/14.5K protein complex inhibits tumor necrosis factor-induced translocation of cytosolic phospholipase A2 to membranes. J. Virol. 71:2830–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerber, S. I., D. D. Erdman, S. L. Pur, P. S. Diaz, J. Segreti, A. E. Kajon, R. P. Belkengren, and R. C. Jones. 2001. Outbreak of adenovirus genome type 7d2 infection in a pediatric chronic-care facility and tertiary-care hospital. Clin. Infect. Dis. 32:694–700. [DOI] [PubMed] [Google Scholar]
- 8.Golovina, G. I., F. N. Zolotaryov, and T. I. Yurlova. 1991. Sensitive analysis of genetic heterogeneity of adenovirus types 3 and 7 in the Soviet Union. J. Clin. Microbiol. 29:2313–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gooding, L. R., T. S. Ranheim, A. E. Tollefson, L. Aquino, P. Duerksen-Hughes, T. M. Horton, and W. S. Wold. 1991. The 10,400- and 14,500-dalton proteins encoded by region E3 of adenovirus function together to protect many but not all mouse cell lines against lysis by tumor necrosis factor. J. Virol. 65:4114–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inada, T. 1998. Brief history, situation in the world, abroad, national surveillance, diagnosis and molecular epidemiology of adenovirus type 7 infection. Rinsho To Uirusu [Clin. Virol.] 26:205–215.(In Japanese.) [Google Scholar]
- 11.Infectious Agents Surveillance Center. 1996. Emergence of adenovirus type 7, Japan, 1995. Infect. Agents Surveillance Rep. 17:1–2. [Google Scholar]
- 12.Kajon, A., and G. Wadell. 1994. Genome analysis of South American adenovirus strains of serotype 7 collected over a 7-year period. J. Clin. Microbiol. 32:2321–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, Q. G., and G. Wadell. 1986. Analysis of 15 different genome types of adenovirus type 7 isolated on five continents. J. Virol. 60:331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, Q. G., Q. J. Zheng, Y. H. Liu, and G. Wadell. 1996. Molecular epidemiology of adenovirus types 3 and 7 isolated from children with pneumonia in Beijing. J. Med. Virol. 49:170–177. [DOI] [PubMed] [Google Scholar]
- 15.Miyamoto, Y., M. Honda, T. Katsuki, K. Kajiwara, Y. Tsutsumi, and Y. Maeda. 1995. Serological survey for virus antibodies of the Fukuoka citizens. 1. Adenovirus. Fukuoka-shi Eiseishikensyo Nenpo 20:82–86.(In Japanese.) [Google Scholar]
- 16.Nishio, O., K. Matsui, M. Akiyama, M. Nanbu, T. Oka, Y. Matsunaga, and S. Inouye. 1998. Change in antibody prevalence to adenovirus type 7 during the past 20 years in Japan. Rinsho To Uirusu [Clin. Virol.] 26:255–259.(In Japanese.) [Google Scholar]
- 17.Noda, M., Y. Miyamoto, Y. Ikeda, T. Matsuishi, and T. Ogino. 1991. Intermediate human adenovirus type 22/H10,19,37 as a new etiological agent of conjunctivitis. J. Clin. Microbiol. 29:1286–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noda, M., Y. Otagaki, Y. Ikeda, T. Matsuishi, and T. Ogino. 1988. Genome types of adenovirus types 19 and 37 from patients with conjuctivitis in Hiroshima City. J. Med. Virol. 26:15–22. [DOI] [PubMed] [Google Scholar]
- 19.Ozawa, S., T. Yamagami, Y. Watanabe, A. Machida, H. Yokoyama, and O. Nishio. 1998. An outbreak of adenovirus type 7 in a dormitory of a high school. Rinsho To Uirusu [Clin. Virol.] 26:226–237.(In Japanese.) [Google Scholar]
- 20.Rowe, W. P., J. W. Hartley, and R. J. Huebner. 1958. Serotype composition of the adenovirus group. Proc. Soc. Exp. Biol. Med. 97:465–470. [DOI] [PubMed] [Google Scholar]
- 21.Sakata, H. 1998. Clinical study of severe illness due to adenovirus type 7. Rinsho To Uirusu [Clin. Virol.] 26:238–243.(In Japanese.) [Google Scholar]
- 22.Sharp, I. R., and G. Wadell. 1995. Adenoviruses, p.287–308. In A. J. Zuckerman, J. E. Banatvala, and J. R. Pattison (ed.), Principles and practice of clinical virology, 3rd ed. John Wiley & Sons Ltd., New York, N.Y.
- 23.Top, F. H., Jr. 1975. Control of adenovirus acute respiratory disease in U.S. Army trainees. Yale J. Biol. Med. 48:185–195. [PMC free article] [PubMed] [Google Scholar]
- 24.Wadell, G. 1984. Molecular epidemiology of human adenoviruses. Curr. Top. Microbiol. Immunol. 110:191–220. [DOI] [PubMed] [Google Scholar]
- 25.Wadell, G., M. K. Cooney, A. da Costa Linhares, L. de Silvia, M. L. Kennett, R. Kono, R. Gui-Fang, K. Lindman, J. P. Nascimento, B. D. Schoub, and C. D. Smith. 1985. Molecular epidemiology of adenoviruses: global distribution of adenovirus 7 genome types. J. Clin. Microbiol. 21:403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wadell, G., J. C. de Jong, and S. Wolontis. 1981. Molecular epidemiology of adenoviruses: alternating appearance of two different genome types of adenovirus 7 during epidemic outbreaks in Europe from 1958 to 1980. Infect. Immun. 34:368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe, M. 1968. Adenovirus antibodies in Gifu prefecture. Nippon Kosyueiseigaku Zassi 15:797–802.(In Japanese.) [Google Scholar]
- 28.Wigand, R, N. Sehn, J. C. Hierholzer, J. C. de Jong, and T. Adrian. 1985. Immunological and biochemical characterization of human adenoviruses from subgenus B. I. Antigenic relationships. Arch. Virol. 84:63–78. [DOI] [PubMed] [Google Scholar]
- 29.Wold, W. S., and L. R. Gooding. 1991. Region E3 of adenovirus: a cassette of genes involved in host immunosurveillance and virus-cell interactions. Virology 184:1–8. [DOI] [PubMed] [Google Scholar]
- 30.Yamadera, S., K. Yamashita, M. Akatsuka, N. Kato, M. Hashido, S. Inouye, and S. Yamazaki. 1995. Adenovirus surveillance, 1982–1993, Japan. A report of the National Epidemiological Surveillance of Infectious Agents in Japan. Jpn. J. Med. Sci. Biol. 48:199–210. [PubMed] [Google Scholar]
- 31.Yamadera, S., K. Yamashita, M. Akatsuka, N. Kato, and S. Inouye. 1995. Trend of adenovirus type 7 infection, an emerging disease in Japan. A report of the National Epidemiological Surveillance of Infectious Agents in Japan. Jpn. J. Med. Sci. Biol. 51:43–51. [DOI] [PubMed] [Google Scholar]
- 32.Yamadera, S., K. Yamashita, M. Akatsuka, N. Kato, M. Tokunaga, and S. Inouye. 2000. Adenovirus type 7 outbreaks in Japan in 1998. Jpn. J. Infect. Dis. 53:22–23. [PubMed] [Google Scholar]
- 33.Yokozawa, M., and R. Endoh. 1998. Adenovirus type 7 pneumonia in Kushiro City; a study of 8 hospital cases. Rinsho To Uirusu [Clin. Virol.] 26:244–249.(In Japanese.) [Google Scholar]