Abstract
Caldesmon inhibits the binding of skeletal muscle subfragment-1 (S-1)·ATP to actin but enhances the binding of smooth muscle heavy meromyosin (HMM)·ATP to actin. This effect results from the direct binding of caldesmon to myosin in the order of affinity: smooth muscle HMM > skeletal muscle HMM > smooth muscle S-1> skeletal muscle S-1 Hemric, M. E., and Chalovich, J. M. (1988) J. Biol. Chem. 263, 1878–1885). We now show that the difference between skeletal muscle HMM and S-1 is due to the presence of the S-2 region in HMM and is unrelated to light chain composition or to two-headed versus single-headed binding. Differences between the binding of smooth and skeletal muscle myosin subfragments to actin do not result from the lack of light chain 2 in skeletal muscle S-1. In the presence of ATP, caldesmon binds to smooth muscle myosin filaments with a stoichiometry of 1:1 (K = 1 × 106 m−1). Similar results were obtained for the binding of caldesmon to smooth muscle rod as well as the binding of the purified myosin-binding fragment of caldesmon to smooth muscle myosin. The binding of caldesmon to intact myosin is ATP sensitive. The interaction of caldesmon with myosin is apparently specific and sensitive to the structure of both proteins.
Caldesmon was first isolated as an actin- and calmodulin-binding protein which inhibits actomyosin superprecipitation (2). Caldesmon was later shown to inhibit actin-activated ATP hydrolysis of both smooth (3, 4) and skeletal muscle myosins (5, 6), and their subfragments (7–9). Despite the fact that caldesmon inhibits the rate of ATP hydrolysis of all myosins, the effect of caldesmon on the binding of the myosin·ATP complexes to actin-tropomyosin depends on the source of the myosin and on the type of myosin subfragment. Thus, caldesmon inhibits the binding of skeletal muscle S-1·1 ATP to actin in parallel with the inhibition of ATPase activity (7). In contrast, caldesmon causes an increase in the apparent binding of smooth muscle HMM·ATP to actin (8, 9), and it has been suggested that the inhibition of ATPase activity of smooth muscle acto-HMM is due to a decrease in the rate of release of either Pi (1) or ADP (9). Rather than postulating a different mechanism for caldesmon function with skeletal muscle S-1, we had proposed a model in which caldesmon inhibits the binding of all myosin·ATP complexes to actin resulting in inhibition of ATP hydrolysis. The apparent increase in binding of smooth muscle HMM·ATP to actin was proposed to be due to a direct binding of caldesmon to smooth muscle myosin which formed an inactive actin-caldesmon-myosin complex (8).
Support for this model, involving an actin-caldesmon-myosin complex, came from observations that the effect of caldesmon on actin binding had no relationship to the inhibition of ATPase activity (8). Thus caldesmon caused an increase in the apparent binding of myosin·ATP to actin in the order: smooth muscle HMM > skeletal muscle HMM > smooth muscle S-1 although the effect of caldesmon on the ATPase activities of these species was similar (8). Furthermore, we and others have observed a direct interaction between myosin and caldesmon (11–15).
Further support for our competitive binding model of caldesmon came from the demonstration that the actin-binding fragment of caldesmon inhibits hydrolysis of ATP in parallel with inhibition of binding of smooth muscle HMM·ATP to actin (15, 16). This occurs because the actin-binding fragment of caldesmon cannot cross-link actin to myosin. Furthermore, both caldesmon and the actin-binding fragment of caldesmon cause detachment of weak binding cross-bridges in skinned skeletal muscle fibers (17).
We have now further characterized the binding of caldesmon to myosin. In particular, we now show that the different effects of caldesmon on S-1 and HMM, from the same myosin, are due to the presence of the S-2 region and not to differences in light chain composition or in the number of active sites. The S-1 regions of smooth and skeletal muscle myosin are very different in their interaction with caldesmon. The source of these differences is not known. Using direct binding studies, we have shown that the binding of caldesmon to smooth muscle myosin is saturable and specifically reversible. The strength of binding agrees well with the value predicted from our model (8). The strength of binding decreases with increasing ionic strength although under physiological conditions the interaction of caldesmon with smooth muscle myosin is expected to be stronger than the interaction of myosin·ATP to actin. Therefore, the interaction of caldesmon with myosin may have a physiological function. In the presence of ATP, the binding constant of caldesmon to smooth muscle myosin increases 22-fold and the stoichiometry changes from 2 or 3 mol of caldesmon/mol of myosin to 1:1. In the absence of ATP, the binding of caldesmon to smooth muscle myosin rod, as well as the binding of the isolated myosin-binding fragment of caldesmon to myosin, also occur with a stoichiometry of 1:1, suggesting that the interaction of caldesmon with myosin is sensitive to the conformation of both proteins.
MATERIALS AND METHODS
Smooth muscle myosin was prepared from chicken gizzards as described by Persechini and Hartshorne (18) except that 0.1% Triton X-100 was included in the first wash and myosin was extracted with 40 mm imidazole-HCl (pH 7.2), 8 mm ATP, 60 mm KC1, 1 mm EDTA, 100 mg/liter streptomycin sulfate, and 0.5 mm dithiothreitol. Skeletal muscle myosin was isolated from the back muscles of rabbits by a standard procedure (19). Smooth muscle HMM was prepared by the method of Seidel (20). Smooth muscle S-1 and skeletal muscle papain (Mg2+) S-1 were prepared as described by Green et al. (21). Skeletal muscle S-1 was made by a standard procedure (22). Skeletal muscle single-headed HMM, smooth muscle rod, and smooth muscle LMM were prepared as described by Margossian and Lowey (23). Actin was isolated from rabbit skeletal muscle by a modification of the method of Spudich and Watt (24, 25). Smooth muscle tropomyosin was isolated from turkey gizzards by the method of Bretscher (26), and caldesmon was isolated from the same source by a modification of the method of Bretscher (26). The myosin-binding fragment of caldesmon (15) and [14C]iodoacetamide-labeled caldesmon (27) were prepared as described previously. Protein concentrations were determined by absorbance at 280 nm except for caldesmon, smooth muscle tropomyosin, skeletal muscle single-headed HMM, smooth muscle rod, and smooth muscle LMM, whose concentrations were determined by the Lowry (28) method using bovine serum albumin as a standard. The smooth muscle myosin rod concentration was confirmed by reaction of acid-hydrolyzed smooth muscle rod with o-phthalaldehyde using alanine as a standard (29). The amino acid composition data of smooth muscle myosin was obtained from Yanagisawa et al. (30). Similar results were obtained from both the Lowry assay and the o-phthalaldehyde assay. The molecular weights used for calculations of protein concentrations are: myosin, 532,000; HMM, 350,000; rod, 263,000; single headed HMM, 230,000; LMM, 130,000; S-1, 120,000; caldesmon, 87,000 (31); actin, 42,000; tropomyosin, 68,000; and caldesmon myosin-binding fragment, 19,050 (15).
ATPase Assays
Rates of ATP hydrolysis were measured at 25 °C as the rate of liberation of 32Pi from [32P]ATP (32). Each assay consisted of a minimum of three time points taken during the initial 20% of the reaction. The conditions of the assays are given in the figure legends.
Binding Assays
The binding of myosin subfragments to actin was measured by sedimentating 1.0 ml of an actin/myosin subfragment mixture in an ultracentrifuge and determining the free S-1 concentration by an NH4+/EDTA/ATPase assay (32). The control, zero actin point, contained the same concentration of smooth muscle tropomyosin as the experimental points. In addition, 0.25 mg/ml of smooth muscle tropomyosin was added to the NH4+/EDTA/ATPase mixture to avoid denaturation of the myosin subfragments at low concentrations. The mixing procedure (7) and design of control experiments (8) were described previously.
The binding of radioactively labeled caldesmon to smooth muscle myosin was determined by a low speed sedimentation assay. Depolymerized smooth muscle myosin and [14C]iodoacetamide-caldesmon were mixed and dialyzed against low ionic strength binding buffer. The amount of caldesmon bound was calculated from the depletion of radioactivity from the supernatant following centrifugation in a low speed Eppendorf centrifuge at 25 °C for 5 min. The data were corrected for the fraction of total caldesmon which did not bind to myosin even at saturating myosin concentrations. This fraction of inactive protein was about 26% for [14C]iodoacetamide-labeled caldesmon and 55% for the [14C]iodoacetamide-labeled myosin-binding fragment of caldesmon. This correction for inactive caldesmon affected the constant for caldesmon binding to myosin but had little effect on the stoichiometry of binding. The data were also corrected for the fraction of caldesmon that sedimented in the absence of myosin. This fraction was approximately 3% for both intact and fragmented caldesmon. This correction for nonspecific sedimentation affected the stoichiometry of binding but had little effect on the binding constant. Finally, a correction was applied for the fraction of the myosin which did not sediment. This fraction was 1% at 50 mm ionic strength and 2% at 100 mm ionic strength. This correction had essentially no effect on experimental values.
RESULTS
One possible reason for the tighter binding of caldesmon to smooth muscle S-1 compared to skeletal muscle S-1 is that the preparation of smooth muscle S-1 with papain leaves light chain 2 intact. In contrast, digestion of skeletal muscle myosin with chymotrypsin results in loss of light chain 2. To determine the effect of light chain 2, binding experiments were done with skeletal muscle S-1 produced by digesting skeletal muscle myosin with papain. Because S-1 and caldesmon are both soluble proteins, the interaction of caldesmon with S-1 was determined indirectly by the affect of caldesmon on the binding of the S-1 to actin. If the binding of S-1 to caldesmon is weaker than its binding to actin, caldesmon will decrease the binding of S-1 to actin. However, if S-1 binds more tightly to caldesmon than to actin, the apparent binding of the S-1 to actin will be enhanced by caldesmon. Fig. 1 shows the effect of caldesmon on the actin-activated ATPase rate of skeletal muscle papain S-1and the binding of skeletal muscle papain S-1·ATP to skeletal muscle actin. Caldesmon inhibits the binding of the S-1, containing intact light chain 2, just as it inhibited the binding of chymotryptically prepared S-1 (7). Since light chain 2 is not responsible for the interaction of caldesmon with myosin, the binding of caldesmon to smooth muscle S-1 must reflect a difference in the heavy chains of smooth and skeletal muscle S-1.
Fig. 1. Effect of caldesmon on the actin-activated ATPase activity of skeletal muscle papain (Mg2+) S-1 and the binding of skeletal muscle papain (Mg2+) S-1·ATP to actin-tropomyosin.
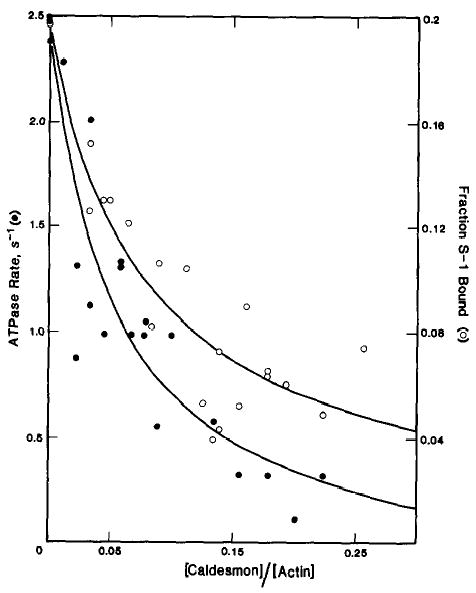
ATPase rates (•) and binding (○) were measured in the presence of 50 μm skeletal muscle actin, 10 μm smooth muscle tropomyosin, and 0.2 μm skeletal muscle papain (Mg2+) S-1. The conditions used were 42 mm NaCl, 4.2 mm MgCl2, 10 mm imidazole-HCl (pH 7), 1 mm dithiothreitol, 2 mm ATP, at 25 °C. The ATPase activity in the absence of caldesmon was 2.5 s−1. The fraction of skeletal muscle papain (Mg2+) S-1 bound in the absence of caldesmon was 0.2.
To determine whether the stronger binding of HMM relative to S-1 is due to the presence of two active S-1 head regions, the effect of caldesmon on the binding of skeletal muscle single-headed HMM to skeletal muscle actin was studied. Fig. 2 shows that caldesmon enhances the apparent binding of skeletal muscle single-headed HMM·ATP to skeletal muscle actin in a manner similar to that observed earlier with intact skeletal muscle HMM (8). Therefore, the binding of caldesmon to HMM does not require the presence of two S-1 regions. The stronger interaction of caldesmon with HMM compared with S-1 is probably due to the presence of the S-2 region of HMM.
Fig. 2. Effect of caldesmon on the binding of skeletal muscle single-headed HMM·ATP to actin-tropomyosin.
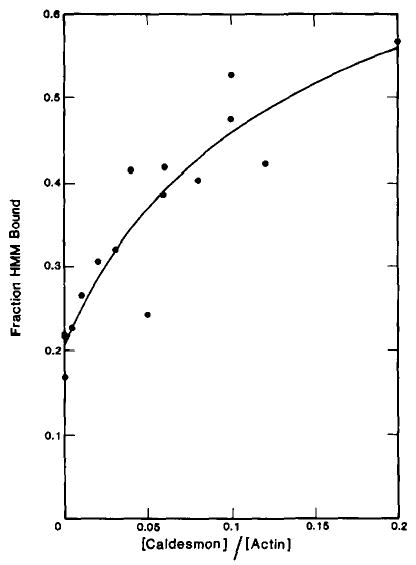
Binding was measured with the same protein concentrations and under the same conditions as described in the legend to Fig. 1. The fraction of skeletal muscle single-headed HMM·ATP bound in the absence of caldesmon was 0.21.
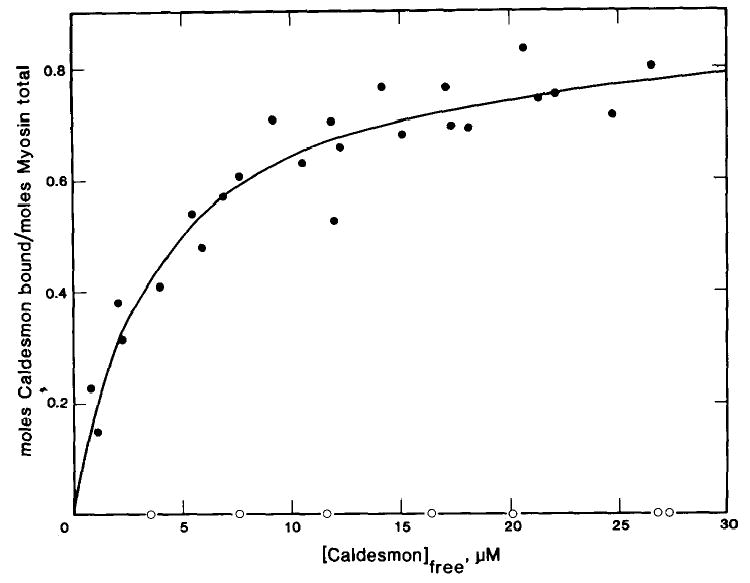
To determine the stoichiometry of binding of caldesmon to myosin and to directly show the importance of the S-2 region in the binding, a more direct binding assay was developed using filaments of myosin or myosin fragments. The binding of [14C]iodoacetamide-labeled caldesmon to smooth muscle myosin filaments was measured by a co-sedimentation assay which allowed an estimation of both the binding constant and stoichiometry to be made. Because of the instability of smooth muscle myosin filaments in the presence of ATP at ionic strengths greater than 100 mm (33), binding was measured in the presence of 1 mm ATP at 50 mm ionic strength. Fig. 3 is a plot of the molar ratio of caldesmon bound per total myosin as a function of the free caldesmon concentration. These data are from three experiments using different preparations of [14C]iodoacetamide-labeled caldesmon and smooth muscle myosin. The data are adequately described by a theoretical curve for a single class of binding sites with an association constant of 9.9 × 105 m−1 and a stoichiometry of 0.92 caldesmon molecules/myosin molecule.
Fig. 3. Binding of caldesmon to smooth muscle myosin filaments in the presence of ATP.
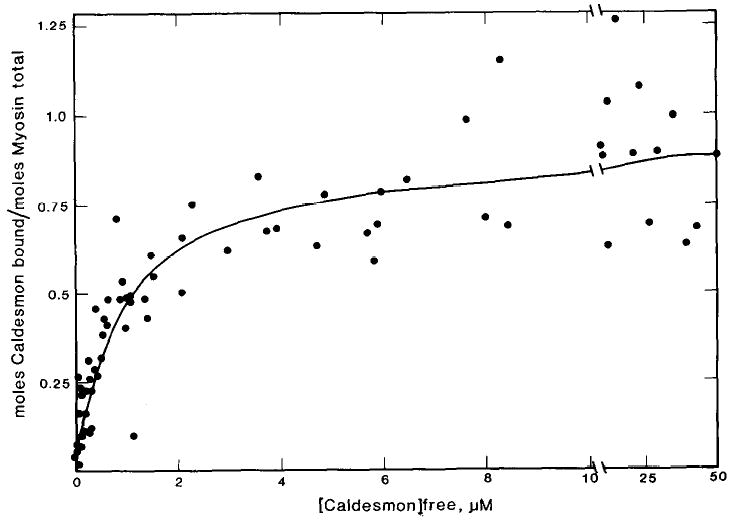
Binding was measured in the presence of 2.5 μm smooth muscle myosin and varied concentrations of [14C]iodoacetamide-labeled caldesmon. The conditions used were: 30 mm NaCl, 5 mM MgCl2, 10 mm imidazole-HCl (pH 7), 1 mm ATP, 1 mm dithiothreitol, at 25 °C. The theoretical curve describes a single class of binding sites with an association constant of 9.9 × 105 m−1 and a stoichiometry of 1 caldesmon molecule/0.92 myosin molecules.
As shown earlier, the effect of caldesmon on the binding of various myosin subfragments to actin is nucleotide dependent. This dependence is certainly due, at least in part, to the nucleotide dependence of the myosin binding to actin. However, the interaction between caldesmon and myosin may also be nucleotide dependent. Therefore, the binding of caldesmon to myosin filaments was measured in the absence of nucleotide. These data, shown in Fig. 4, are from 10 different experiments using different preparations of myosin and caldesmon. The interaction of myosin with caldesmon is weak, in the absence of ATP, and determination of the stoichiometry is difficult. The data may be described by a theoretical curve for a single class of binding sites with a stoichiometry of binding of either 2 or 3 molecules of caldesmon/myosin molecule. The binding constants are 5.6 × 104 m−1 and 2.8 × 104 m−1 for the theoretical curves of stoichiometries of 2 and 3, respectively.
Fig. 4. Binding of caldesmon to smooth muscle myosin filaments in the absence of nucleotide.
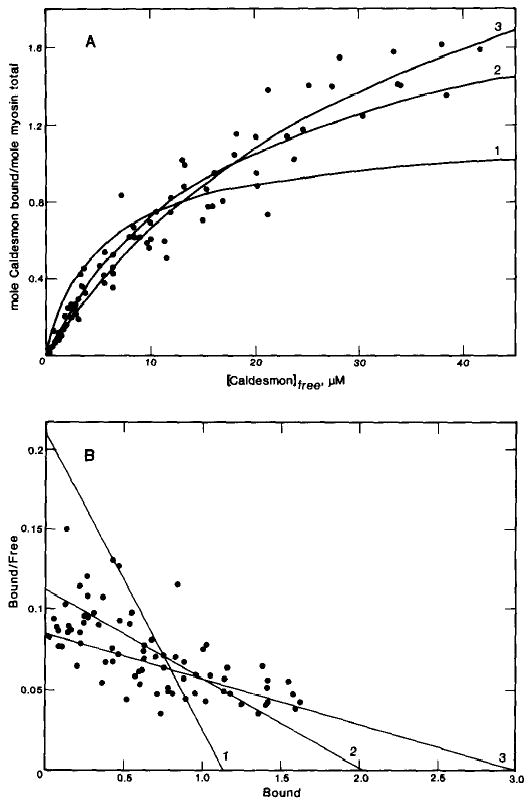
A, binding was measured in the presence of 10 μm smooth muscle myosin and varied concentrations of [14C]iodoacetamide-labeled caldesmon. The conditions were the same as described in the legend of Fig. 3, except the ATP was omitted. The theoretical curves drawn through the points are for a single class of binding sites with a stoichiometry of 1 caldesmon molecule/myosin molecule and association constant of 1.9 × 105 m−1, or a stoichiometry of 2 caldesmon molecules/myosin molecule and association constant of 5.6 × 104 m−1, or a stoichiometry of 3 caldesmon molecules/myosin molecule and association constant of 2.8 × 104 m−1. B, Scatchard plot of the data.
The ionic strength dependence of the binding of caldesmon to smooth muscle myosin is shown in Fig. 5. These experiments were done in the absence of ATP to avoid loss of filament assembly at higher ionic strength (33). Because saturation of binding could not be reached at 75 or 100 mm ionic strengths, the stoichiometry was fixed at 2 mol of caldesmon bound per myosin molecule and the binding constants were varied until a reasonable fit of the data was obtained. The inset to Fig. 5 shows the changes in the binding constant, estimated in this way, as a function of the square root of the ionic strength. For comparison, the inset also shows the ionic strength dependence of the binding of smooth muscle S-1·ATP to actin taken from Greene et al. (21). While the strength of binding of caldesmon to smooth muscle myosin decreases with increasing ionic strength, it remains stronger than the binding of smooth muscle S-1·ATP to actin. In actuality, the binding of caldesmon to smooth muscle myosin at high ionic strength is probably greater than that shown since the affinity of caldesmon for myosin increases in the presence of ATP.
Fig. 5. Binding of caldesmon to smooth muscle myosin filaments in the absence of nucleotide and at variable ionic strengths.
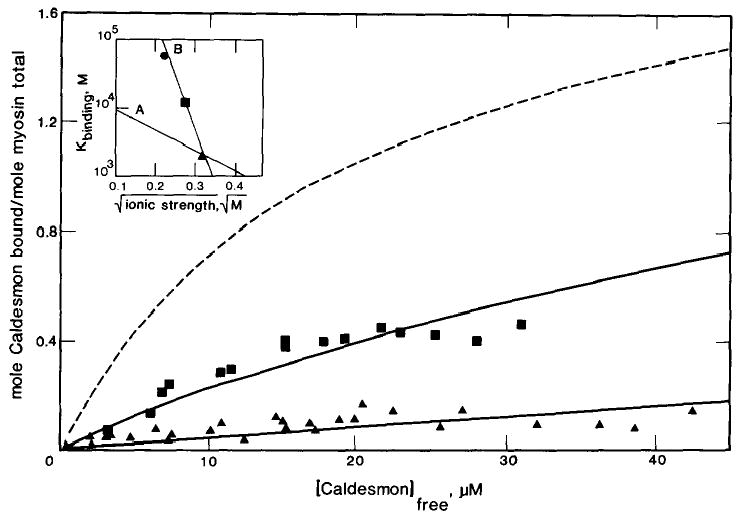
Binding was measured in the presence of 10 μm smooth muscle myosin and varied concentrations of [14C]iodoacetamide-labeled caldesmon under the conditions described in the legend to Fig. 4 except the concentration of NaCl was increased to 55 mm and 80 mm for the 75 mm (▪) and 100 mm (▴) ionic strength measurements, respectively. The theoretical curves drawn through the points are for a single class of binding sties with a stoichiometry of 2 caldesmon molecules/myosin molecule and association constants of 5.6 × 104 m−1, 50 mm (dashed line), 1.3 × 104 m−1, 75 mm, or 2.1 × 103 m−1, 100 mm. The inset shows the binding constant as a function of the square root of the ionic strength (B). For comparison, the inset contains a plot of the ionic strength dependence of smooth muscle S-1·ATP binding to actin (A) (19).
To demonstrate reversibility, the binding of [14C]iodoacetamide-labeled caldesmon to smooth muscle myosin filaments was measured in the presence of proteins which were expected to compete with this binding. Fig. 6 shows the binding of caldesmon to smooth muscle myosin as a function of the competitor protein concentration. As expected, smooth muscle HMM was very effective in displacing caldesmon from the smooth muscle myosin. Smooth muscle S-1 and, to a lesser extent, skeletal muscle S-1 displaced bound caldesmon from the smooth muscle myosin filaments. The order of effectiveness of competition (smooth muscle HMM > smooth muscle S-1 > skeletal muscle S-1) was the same as the relative strength of binding of these subfragments to caldesmon. These data show that caldesmon is not trapped by the myosin but is bound reversibly. The inset to Fig. 6 is a plot of moles of caldesmon bound per mol of total myosin as a function of the free caldesmon concentration. Increasing concentrations of any of the subfragments resulted in a linear decrease in the bound caldesmon. Extrapolation to infinite subfragment concentrations suggested that caldesmon can be displaced completely by any of these myosin subfragments. Bovine serum albumin had no effect on the binding of caldesmon to smooth muscle myosin; this demonstrates the specificity of the competition. As a further test of nonspecific trapping, the effect of the order of addition of components on the binding of caldesmon to myosin was studied. Smooth muscle HMM was equally effective in decreasing the amount of caldesmon bound to smooth muscle myosin if the smooth muscle HMM was added before or after caldesmon was mixed with smooth muscle myosin (data not shown).
Fig. 6. Effect of myosin subfragments on the binding of caldesmon to smooth muscle myosin filaments.
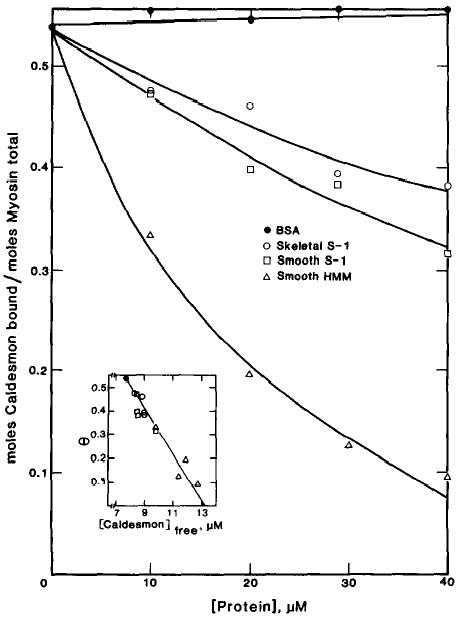
Binding was measured in the presence of 10 μm smooth muscle myosin, 18 μm [14C]iodoacetamide-labeled caldesmon, and variable concentrations of bovine serum albumin (BSA) (•), skeletal muscle S-1 (○), smooth muscle (S-1) (□), and smooth muscle HMM (▵). The conditions were the same as described in the legend of Fig. 4. The moles of caldesmon bound per mol of myosin total in the absence of myosin subfragment was 0.54. The inset contains a plot of the moles of caldesmon bound per mol of myosin total as a function of free caldesmon.
To confirm the interaction of the subfragment-2 region of myosin with caldesmon, the binding of caldesmon directly to smooth muscle light meromyosin and to smooth muscle rod was measured. As shown in Fig. 7, [14C]iodoacetamide-labeled caldesmon does not bind to smooth muscle light meromyosin. However, caldesmon does bind to smooth muscle rod. Since the rod contains the S-2 region and LMM does not, the S-2 region is necessary for the binding of caldesmon to myosin. The binding of caldesmon to smooth muscle rod, in the absence of ATP, resembles the binding of caldesmon to smooth muscle myosin in the presence of ATP. Caldesmon binds to smooth muscle rod with a stoichiometry of 0.9 caldesmon molecules/myosin molecule and a binding constant of 2.2 × 105 m−1.
Fig. 7. Binding of caldesmon to smooth muscle rods and light meromyosin.
Binding was measured in the presence of either 10 μm rods (•) or 10 μm light meromyosin (○) and varied concentrations of [14C]iodoacetamide-labeled caldesmon. The conditions were the same as described in the legend of Fig. 4. The theoretical curve for rods was generated assuming a single class of binding sites with an association constant of 2.2 × 105 m−1 and a stoichiometry of 1 caldesmon molecule/0.9 rod molecules.
A chymotryptic peptide derived from the NH2 terminus of caldesmon binds to myosin but not to actin (15). The binding of this caldesmon fragment to smooth muscle myosin was measured to demonstrate its ability to bind independently of the remaining portions of caldesmon and to compare its stoichiometry and strength of the binding to that of the whole molecule. Fig. 8 shows the binding of the [14C]iodoacetamide-labeled myosin-binding fragment of caldesmon to smooth muscle myosin at 50 mm ionic strength and in the absence of ATP. The myosin-binding fragment binds to myosin filaments with a stoichiometry of 1:1 and a binding constant of 9.3 × 104 m−1. This binding resembles the binding of caldesmon to smooth muscle myosin-ATP and the binding of caldesmon to smooth muscle rod.
Fig. 8. Binding of purified myosin-binding fragment of caldesmon to smooth muscle myosin filaments.
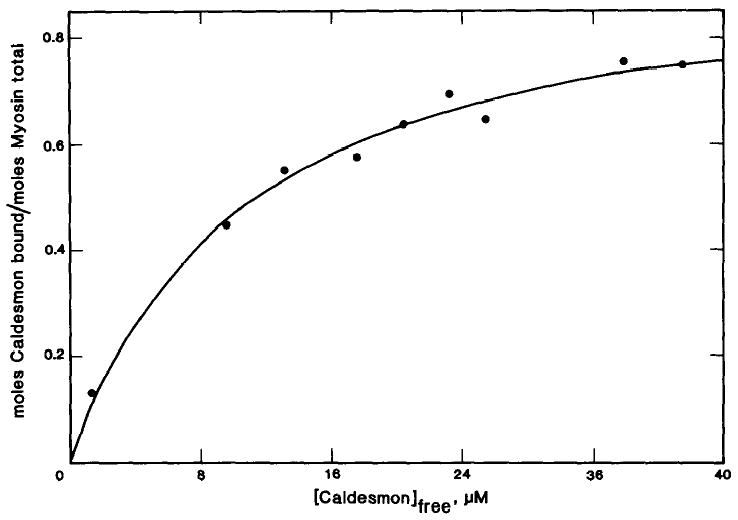
Binding was measured in the presence of 10 μm smooth muscle myosin and varied concentrations of purified [14C]iodoacetamide-labeled myosin-binding fragment. The conditions were the same as described in the legend of Fig. 4. The theoretical curve was calculated assuming a single class of binding sites with an association constant of 9.3 × 104 m−1 and a stoichiometry of 0.96 myosin binding fragment/myosin molecule.
DISCUSSION
Our earlier studies lead us to conclude that the binding of caldesmon to actin inhibited the binding of myosin-ATP complexes to actin (7). We further suggested that caldesmon also binds to myosin and can form an inactive actin-caldesmon-myosin complex (8). The competition of binding of caldesmon and myosin to actin has been confirmed (15, 16), as has the binding of caldesmon to myosin (11–13). Although we and others have shown that caldesmon binds to myosin, several aspects of this interaction required clarification. First, our earlier results indicated that caldesmon bound with a different affinity to different types of myosin subfragment. We wanted to determine which regions of caldesmon and myosin were involved in the association since this information is important in understanding the function of this interaction. Second, all earlier studies merely showed that caldesmon binds to myosin; neither the association constant nor the stoichiometry of binding had been determined. Quantitation of the binding is important in determining if this association is specific and whether this interaction occurs with such an affinity that it could occur in vivo.
Our first concern was to determine which regions of myosin and caldesmon are associated with each other and thus determine the reason for differences in affinity of various myosin species. In our earlier studies, an interaction of caldesmon with smooth muscle S-1 but not skeletal muscle S-1 was observed (7, 8). Our present competition studies show that high concentrations of skeletal muscle S-1, as well as smooth muscle S-1, can displace caldesmon from smooth muscle myosin filaments. Thus, skeletal muscle S-1 binds to caldesmon but with a very low affinity. The present results show that the difference in affinity of smooth and skeletal muscle S-1 for caldesmon is not due to light chain 2 since papain skeletal muscle S-1, containing light chain 2, appears to bind to caldesmon as weakly as chymotryptic skeletal muscle S-1 which lacks this light chain. The stronger binding of smooth muscle S-1 must be due to differences in the primary structure of either the light chains or of the heavy chains. The areas of contact between caldesmon and smooth muscle S-1 have not yet been defined.
We also noted previously that HMM from smooth or skeletal muscle myosin binds to caldesmon more tightly than S-1 prepared from the respective myosin. This increased affinity of HMM cannot be attributed to the presence of two S-1 binding sites since the effect of caldesmon on skeletal muscle single-headed HMM is indistinguishable from the effect of caldesmon on skeletal muscle two-headed HMM. Thus, the S-2 region of myosin appears to be important in the binding to caldesmon. This importance was clearly shown in Fig. 7 where caldesmon was shown to bind stoichiometrically with smooth muscle rods but not at all with smooth muscle LMM. Caldesmon binds both to the S-2 and S-1 regions of myosin although the S-2 region appears to dominate, particularly in the case of skeletal muscle myosin. The S-1 and S-2 regions of myosin may constitute a single binding site since high concentrations of skeletal or smooth muscle S-1 can displace bound caldesmon from myosin.
In addition to directly defining the regions of myosin responsible for the interaction with caldesmon, we have also confirmed earlier reports that the NH2 terminal region of caldesmon binds to myosin (11, 15). The chymotryptic peptide of caldesmon containing the NH2 terminus binds to smooth muscle myosin stoichiometrically and with an affinity even greater than that of the intact caldesmon under the same conditions. The addition of other caldesmon peptides did not enhance the binding of NH2-terminal peptide to smooth muscle myosin (data not shown). Thus, this chymotryptic peptide, which has an apparent molecular weight of 25,000 on sodium dodecyl sulfate-gel electrophoresis, contains most, if not all, of the myosin-binding site. In another report a purified presumptive myosin-binding fragment of caldesmon was unable to bind to myosin (13). The reason for this difference is unknown.
The large effect of ATP on the binding of caldesmon to smooth muscle myosin is interesting and is further evidence for the specificity of the interaction between caldesmon and myosin. ATP effects the stability and structure of smooth muscle myosin filaments (33). Under our conditions, smooth muscle myosin remains filamentous, even in the presence of ATP. The binding of caldesmon to smooth muscle rods is similar to the binding to filamentous smooth muscle myosin in the presence of ATP. Therefore, the structural changes in the smooth muscle myosin filament, which occur upon the addition of ATP and are responsible for tight binding to caldesmon, can be partially duplicated by cleaving the S-1 heads from the myosin. The stoichiometry of binding, in the absence of ATP, is 1:1 in the case of the myosin-binding fragment of caldesmon, whereas it is greater than 2 caldesmons/myosin with intact caldesmon. In the absence of ATP, the structure of smooth muscle myosin may change so as to expose additional binding sites which may interact with regions of caldesmon other than that of the NH2-terminal chymotryptic fragment.
The binding of caldesmon to smooth muscle myosin was shown to be reversible and saturable by direct binding studies with filamentous smooth muscle myosin. The binding of caldesmon to smooth muscle myosin, in the absence of ATP, is at least as strong as the interaction of smooth muscle S-1·ATP with actin over a wide range of ionic strengths. Since smooth muscle myosin binds to caldesmon in the presence of ATP with an affinity even greater than in the absence of ATP, we have underestimated the binding of caldesmon to smooth muscle myosin at high ionic strength by a factor of about 20. The strength of the binding of caldesmon to myosin and the specificity of the interaction, shown by the stoichiometry and the reversibility, indicate that this interaction could occur in vivo. An interesting problem of future research is to demonstrate the interaction of caldesmon with myosin in a cell and define the function of this interaction.
Acknowledgments
We wish to thank Michael V. Freedman and Caryl E. Benson for their excellent technical assistance, Dr. Yi-der Chen for his helpful conversations, Perdue Inc. for supplying chicken gizzards, and Swift-Eckrich Inc. for supplying turkey gizzards.
Footnotes
The abbreviations used are: S-1, subfragment-1; HMM, heavy meromyosin; LMM, light meromyosin.
This work was supported by Grant AR35216 from the National Institutes of Health and Grant 1988-89-A16 (to M. E. H.) from the American Heart Association, North Carolina Affiliate. A preliminary report of this work was presented at the Biophysical Society Meeting, Baltimore, MD, February 19, 1990 (1).
References
- 1.Hemric M. Biophys J. 1990;57:152a. [Google Scholar]
- 2.Sobue K, Muramoto Y, Fujita M, Kakuichi S. Proc Natl Acad Sci U S A. 1981;78:5652–5655. doi: 10.1073/pnas.78.9.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ngai P, Walsh M. Biochem J. 1985;230:695–707. doi: 10.1042/bj2300695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sobue K, Takahashi K, Wakabayashi I. Biochem Biophys Res Commun. 1985;132:645–651. doi: 10.1016/0006-291x(85)91181-7. [DOI] [PubMed] [Google Scholar]
- 5.Smith CWJ, Marston S. FEBS Lett. 1985;184:115–118. doi: 10.1016/0014-5793(85)80665-7. [DOI] [PubMed] [Google Scholar]
- 6.Lim M, Walsh M. Biochem J. 1986;238:523–530. doi: 10.1042/bj2380523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chalovich J, Cornelius P, Benson C. J Biol Chem. 1987;262:5711–5716. [PubMed] [Google Scholar]
- 8.Hemric ME, Chalovich JM. J Biol Chem. 1988;263:1878–1885. [PubMed] [Google Scholar]
- 9.Lash JA, Sellers JR, Hathaway DR. J Biol Chem. 1986;261:16155–16160. [PubMed] [Google Scholar]
- 10.Marston SB. FEBS Lett. 1988;238:147–151. doi: 10.1016/0014-5793(88)80245-x. [DOI] [PubMed] [Google Scholar]
- 11.Sutherland C, Walsh MP. J Biol Chem. 1989;264:578–583. [PubMed] [Google Scholar]
- 12.Ikebe M, Reardon S. J Biol Chem. 1988;263:3055–3058. [PubMed] [Google Scholar]
- 13.Katajama E, Horuichi KY, Chacko S. Biochem Biophys Res Commun. 1989;160:1316–1322. doi: 10.1016/s0006-291x(89)80147-0. [DOI] [PubMed] [Google Scholar]
- 14.Marston SB, Pritchard K, Redwood C, Taggard M. Biochem Soc Trans. 1988;16:494–497. doi: 10.1042/bst0160494. [DOI] [PubMed] [Google Scholar]
- 15.Velaz L, Ingraham RH, Chalovich JM. J Biol Chem. 1990;265:2929–2934. [PubMed] [Google Scholar]
- 16.Horiuchi KY, Chacko S. Biochemistry. 1989;28:9111–9116. doi: 10.1021/bi00449a023. [DOI] [PubMed] [Google Scholar]
- 17.Brenner B, Yu LC, Chalovich JM. Biophys J. 1990;57:397a. [Google Scholar]
- 18.Persechini A, Hartshorne D. Biochemistry. 1983;22:470–476. doi: 10.1021/bi00271a033. [DOI] [PubMed] [Google Scholar]
- 19.Kielley W, Harrington W. Biochim Biophys Acta. 1960;41:401–421. doi: 10.1016/0006-3002(60)90037-8. [DOI] [PubMed] [Google Scholar]
- 20.Seidel J. J Biol Chem. 1980;255:4355–4361. [PubMed] [Google Scholar]
- 21.Greene LE, Sellers JR, Eisenberg E, Adelstein RS. Biochemistry. 1983;22:530–535. doi: 10.1021/bi00272a002. [DOI] [PubMed] [Google Scholar]
- 22.Weeds A, Taylor R. Nature. 1975;257:54–56. doi: 10.1038/257054a0. [DOI] [PubMed] [Google Scholar]
- 23.Margossian SS, Lowey S. Methods Enzymol. 1982;85:66–67. doi: 10.1016/0076-6879(82)85009-x. [DOI] [PubMed] [Google Scholar]
- 24.Spudich J, Watt S. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 25.Eisenberg E, Kielley W. Cold Spring Harbor Symp Quant Biol. 1972;37:145–152. [Google Scholar]
- 26.Bretscher A. J Biol Chem. 1984;259:12873–12880. [PubMed] [Google Scholar]
- 27.Velaz L, Hemric ME, Benson CE, Chalovich JM. J Biol Chem. 1989;264:9602–9610. [PubMed] [Google Scholar]
- 28.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 29.Peterson G. Methods Enzymol. 1983;91:95–119. doi: 10.1016/s0076-6879(83)91014-5. [DOI] [PubMed] [Google Scholar]
- 30.Yanagisawa M, Hamada Y, Katsuragawa Y, Imamura M, Mikawa T, Masaki T. J Biol Chem. 1987;198:143–157. doi: 10.1016/0022-2836(87)90302-0. [DOI] [PubMed] [Google Scholar]
- 31.Bryan J, Imai M, Lee R, Moore P, Cook RG, Lin WG. J Biol Chem. 1989;264:13873–13879. [PubMed] [Google Scholar]
- 32.Chalovich JM, Eisenberg E. J Biol Chem. 1982;257:2432–2437. [PMC free article] [PubMed] [Google Scholar]
- 33.Onishi H, Suzuki H, Nakamura K, Takahashi K, Watanabe S. J Biochem (Tokyo) 1978;83:835–847. doi: 10.1093/oxfordjournals.jbchem.a131980. [DOI] [PubMed] [Google Scholar]


