Abstract
An automated PCR with fluorescent probes (molecular beacons) detected Mycobacterium avium subsp. paratuberculosis in bovine feces. When the PCR was compared with culture in testing 41 fecal samples, kappa scores of 0.94 to 0.96, a sensitivity of 93 to 96%, and a specificity of 92% were obtained. Results were quantitated by using a standard curve derived from a plasmid containing IS900. A minimum quantity of 1.7 × 10−4 pg of DNA, correlating to 1 to 8 CFU, was detected.
Johne’s disease is a chronic, progressive enteritis of ruminants caused by Mycobacterium avium subsp. paratuberculosis (4, 16, 17, 25, 30). A possible association between M. avium subsp. paratuberculosis infection and Crohn’s disease in humans has also been described (3, 7, 13, 32). Due to these factors and economic considerations, many states within the United States and other countries have instituted Johne’s disease certification programs to eradicate the disease. This necessitates the development of high-throughput, sensitive diagnostic methods for the detection of infected animals and animal products. Traditional diagnostic methods currently include culture of feces and tissues and enzyme-linked immunosorbent assays. A disadvantage of using conventional culturing is the long incubation time (12 to 16 weeks). Enzyme-linked immunosorbent assays can be performed in a few hours, but their sensitivity is estimated at 45% since antibodies may not be detectable until late in infection (8). Several PCR tests to detect M. avium subsp. paratuberculosis DNA have been developed (2, 5, 9, 27, 36, 38). However, the real-time PCR described in the present report is unique, since it is the first quantitative PCR for the detection of M. avium subsp. paratuberculosis and it has a sensitivity equal to that of fecal culture. This is important since feces are a nonhomogeneous, complex material and have been known to contain substances inhibitory to PCR (1, 35, 42). A quantitative PCR may be useful in evaluating treatments to eliminate M. avium subsp. paratuberculosis from animals or animal by-products, to identify animals shedding large and small quantities of M. avium subsp. paratuberculosis, to identify clinical specimens with a consistently high bacterial load for diagnostics, and for pathogenesis studies. Additionally, this real-time, automated PCR is considered high-throughput since it is performed as a single-step PCR, it can be used for multiple samples at one time in a 96-well plate format, and it eliminates time-consuming postdetection methods.
This real-time PCR uses fluorescent probes (molecular beacons) to detect the IS900 sequence. Molecular beacons consist of a stem-and-loop structure with the loop containing a probe sequence complementary to the target sequence. A fluorophore and a nonfluorescent “quencher,” one on each arm, form a nonfluorescent hairpin structure. When a molecular beacon encounters a target, the loop sequence hybridizes with the target sequence, the stem hybrid disassociates, and the fluorophore and quencher separate, allowing for fluorescence to be detected with an ABI PRISM 7700 sequence detection system (Perkin-Elmer Applied Biosystems, Foster City, Calif.) (33, 34). Molecular beacons have been used extensively in genetic analysis studies and have some advantages over linear fluorescent probes (11, 12, 14, 15, 18, 21, 22, 23, 24, 26, 31, 37, 39).
Forty-one bovine fecal samples of known M. avium subsp. paratuberculosis status were obtained (1999 and 2000 Proficiency Tests, National Veterinary Services Laboratory, Ames, Iowa). Aliquots of each fecal sample were used for fecal culture, nested PCR (nPCR), and real-time PCR. Culture 1, for the detection of M. avium subsp. paratuberculosis, was performed using a centrifugation method (41), except for samples 21, 29, and 30, which were cultured using a sedimentation method (40). Culture 2 was performed using a previously described centrifugation method (National Animal Disease Center method) (29) with commercial media (Becton Dickinson, Sparks, Md.). A designation of “too numerous to count” (TNTC) was used if >75 colonies were observed in the culture tube. The positive control culture was prepared by isolating M. avium subsp. paratuberculosis (ATCC 19698) and adjusting a colony suspension to a turbidity equivalent to the no. 5 McFarland standard (approximately 108 CFU/ml). Aliquots were stored at −70°C and used for DNA extractions.
DNA was extracted from 1 g of fecal material and a positive control culture as described previously (28). To estimate quantitative variation due to the extraction procedures, positive control DNA was obtained from six separate extractions and quantitated by real-time PCR.
nPCR was performed as described previously (5). The presence of a 167-bp band was indicative of the presence of M. avium subsp. paratuberculosis DNA. Primer sequences were specific for M. avium subsp. paratuberculosis (6).
For real-time PCR, the molecular beacon (5′ TET-CGGACCGTAACTACCCGCGGCGTGATGGGTCCG-dabcyl3′) was designed to detect the IS900 gene of M. avium subsp. paratuberculosis (underlined bases are arm sequences). Tetrachloro-6-carboxyfluorescein-5′ (TET) is the reporter fluorochrome, and 4-dimethylaminophenylazobenzoic acid-3′ (dab cyl) is the quencher. The 50-μl master mixture contained 3 μl of template DNA, 5 μl of 10× PCR buffer (200 mM Tris-HCl [pH 8.4], 500 mM KCl), 1.5 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate, a 1.32 μM concentration of each of the nested sense and antisense primers, 2.5 U of Platinum Taq DNA polymerase (Gibco Life Technologies, Gaithersburg, Md.), and a 0.3 μM concentration of the molecular beacon. Thermal cycler conditions were as follows: 95°C for 2 min and 45 cycles of PCR amplification at 95°C for 30 s, 58°C for 1 min, and 72°C for 30 s. Exposure time was 45 ms, and data were collected at the annealing phase of each cycle. The threshold value (CT) was defined as the cycle at which the fluorescence exceeded 10 times the standard deviation of the mean baseline emission for cycles 3 to 15.
For quantitation, a standard curve was created by using a plasmid containing the IS900 gene. This gene was amplified by PCR, ligated into pGEM-T Easy Vector (Promega, Madison, Wis.), and quantitated by spectrophotometry. Plasmid DNA amounts were 100, 10, 1, and 0.1 pg. The amount of sample DNA was calculated by interpolation of the CT with the standard curve. Calculations of percent sensitivity, percent specificity, and kappa scores to determine a measure of the agreement between diagnostic tests used for this study have been described previously (20).
Kappa scores of 0.96, 0.94, and 0.85 were obtained when the results of culture 1, culture 2, and nPCR, respectively, were compared to those of real-time PCR (Table 1). The sensitivities of the real-time PCR compared to those of culture 1 and culture 2 were 96 and 93%, respectively. The specificity was 92% between either culture 1 or culture 2 and real-time PCR.
TABLE 1.
Results of culture, nPCR, and real-time PCR (with molecular beacons) assays of bovine fecal samples
| Sample no. | No. of coloniesa
|
nPCR resultb | Amt of DNA (pg) detected by real-time PCRc | |
|---|---|---|---|---|
| Culture 1 | Culture 2 | |||
| 1 | 0 | 0 | − | 0 |
| 2 | 0 | 0 | − | 0 |
| 3 | 0 | 0 | − | 0 |
| 4 | 0 | 0 | − | 0 |
| 5 | 0 | 0 | − | 0 |
| 6 | 0 | 0 | − | 0 |
| 7 | 0 | 0 | − | 0 |
| 8 | 0 | 0 | − | 0 |
| 9 | 0 | 0 | − | 0 |
| 10 | 0 | 0 | − | 0 |
| 11 | 0 | 1 | − | 0 |
| 12 | 0 | 0 | + | 0 |
| 13 | 0 | 0 | − | 6.1 × 10−4 |
| 14 | 0–4 | TNTC | + | 1.4 × 10−2 |
| 15 | 0–4 | TNTC | − | 2.9 × 10−3 |
| 16 | 0–6 | 7–18 | + | 3.2 × 10−3 |
| 17 | 0–6 | 3–12 | − | 1.4 × 10−3 |
| 18 | 1–8 | 1–3 | + | 1.7 × 10−4 |
| 19 | 1–8 | 1–4 | + | 8.1 × 10−4 |
| 20 | 1–8 | 9–15 | + | 0 |
| 21 | 4–14 | TNTC | − | 1.6 × 10−3 |
| 22 | 9–12 | 3–12 | + | 5.1 × 10−4 |
| 23 | 2–21 | 3–5 | + | 1.4 × 10−2 |
| 24 | 2–21 | 20–75 | + | 1.3 × 10−2 |
| 25 | 3–27 | 20–25 | + | 3.2 × 10−3 |
| 26 | 3–27 | 15–40 | + | 3.0 × 10−3 |
| 27 | 3–29 | 5–12 | + | 5.4 × 10−4 |
| 28 | 6–27 | 23–30 | + | 2.2 × 10−4 |
| 29 | 12–22 | 50–75 | + | 2.6 × 10−1 |
| 30 | 16–38 | 40–50 | + | 8.3 × 10−2 |
| 31 | 3-TNTC | TNTC | + | 1.3 × 100 |
| 32 | 3-TNTC | TNTC | + | 9.5 × 10−1 |
| 33 | 3-TNTC | TNTC | + | 1.6 × 10−1 |
| 34 | 27-TNTC | TNTC | + | 4.7 × 10−2 |
| 35 | 27-TNTC | TNTC | + | 3.8 × 10−2 |
| 36 | 27-TNTC | TNTC | + | 2.0 × 10−2 |
| 37 | TNTC | 20–40 | + | 5.1 × 10−2 |
| 38 | TNTC | 20–40 | + | 2.3 × 10−1 |
| 39 | TNTC | TNTC | + | 1.3 × 10−2 |
| 40 | TNTC | TNTC | + | 3.7 × 10−1 |
| 41 | TNTC | TNTC | + | 1.2 × 100 |
Results for cultures 1 and 2 represent the number or a range of the number of bacterial colonies for three replicate culture tubes per sample.
+, presence of a 167-bp band on an ethidium bromide-stained agarose gel representative of the IS900 gene for M. avium subsp. paratuberculosis; −, absence of the 167-bp product.
Values indicate mean amounts of M. avium subsp. paratuberculosis DNA from three replicate samples.
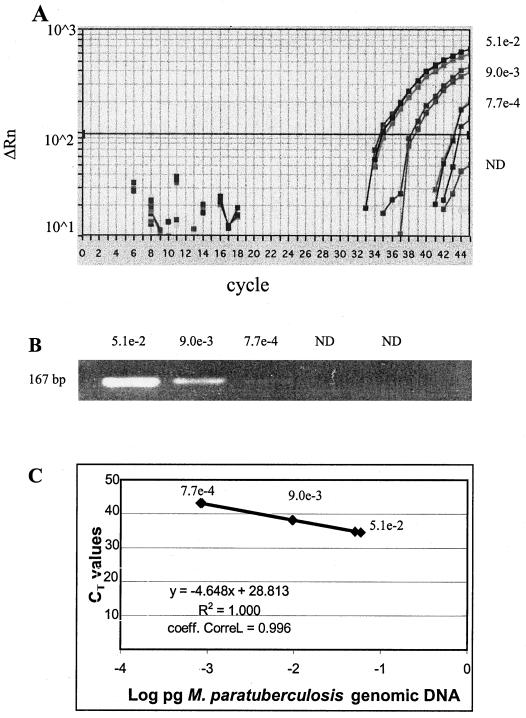
The analytical sensitivities derived from a 10-fold serial dilution of M. avium subsp. paratuberculosis DNA from a fecal sample were similar for nPCR and real-time PCR, with a threshold level of 7.7 × 10−4 pg of DNA, although the fluorescence intensity of the nPCR product was very faint at this level (Fig. 1A and B). An absolute threshold was not determined, since smaller amounts of M. avium subsp. paratuberculosis DNA were found in other fecal specimens (samples 13, 18, 22, 27, and 28), with a minimum detectable level of 1.7 × 10−4 pg (Table 1).
FIG. 1.
Comparison of assay sensitivities for detection of M. avium subsp. paratuberculosis DNA by real-time PCR and nPCR. (A) Fluorescence intensities of serial 10-fold dilutions of fecal sample 37 determined in triplicate (Table 1) by real-time PCR. Quantities on the right of the y axis are DNA amounts in picograms. ND, nondetectable amount of DNA. (B) Quantities of DNA corresponding to those shown in panel A but detected by nPCR on an ethidium bromide-stained 4% agarose gel. (C) The plot of the CT values and DNA concentrations fits a linear function (r2 = 1.00).
The intra-assay variability, determined by the coefficient of variation (CV), was consistently <2% (Table 2). Mean CT values and DNA concentrations of clinical samples also demonstrated reproducible linearity (r2 = 1.00) (Fig. 1C). To determine interassay variability, real-time PCR was performed on the standard concentrations of plasmid DNA, positive control DNA, and DNA from an M. avium subsp. paratuberculosis-positive fecal sample over a 3-day period. The CV was <4% (Table 2). Quantitative variations due to extraction procedures were estimated. The CV of CT values for six separate DNA extractions from different aliquots of positive control DNA was 6.4% (data not shown).
TABLE 2.
Intra- and interassay variabilities of real-time PCR (with molecular beacons) for plasmid, positive control, and fecal sample DNAsa
| DNA type | Results for:
|
||||||
|---|---|---|---|---|---|---|---|
| Day 1
|
Day 2
|
Day 3
|
Days 1 to 3
|
||||
| CT | % CVb | CT | % CVb | CT | % CVb | % CVc | |
| Plasmid | |||||||
| 100 pg | 18.73, 18.68, 18.79 | 0.29 | 18.96, 18.91, 19.06 | 0.40 | 18.80, 18.76, 18.84 | 0.21 | 0.63, 0.62, 0.76 |
| 10 pg | 23.08, 23.19, 23.36 | 0.61 | 24.64, 24.69, 24.74 | 0.29 | 23.18, 22.95, 23.09 | 0.50 | 3.69, 3.99, 3.73 |
| 1 pg | 28.41, 28.55, 28.84 | 0.77 | 28.53, 29.08, 29.65 | 1.93 | 27.62, 27.71, 27.80 | 0.32 | 1.75, 2.43, 3.22 |
| 0.1 pg | 32.74, 32.68, 32.92 | 0.38 | 32.83, 32.73, 32.91 | 0.27 | 32.99, 33.64, 33.75 | 1.23 | 0.39, 1.64, 1.45 |
| Positive control | 27.60, 27.53, 27.98 | 0.87 | 28.08, 28.43, 28.79 | 1.25 | 27.87, 27.95, 28.13 | 0.48 | 0.86, 1.61, 1.52 |
| Feces | 40.90, 40.74, 41.25 | 0.64 | 42.77, 41.78, 41.63 | 1.47 | 41.35, 41.50, 40.13 | 1.83 | 2.34, 1.30, 1.90 |
CT results are for three replicate samples.
Percent CV intra-assay variability.
Percent CV interassay variability.
The sensitivities of the real-time PCR of 96 and 93% relative to cultures 1 and 2, respectively, indicate a high probability of detecting animals that are shedding the bacteria. Only 1 of 41 samples was negative by real-time PCR but positive by nPCR and culture (sample 20). When a second aliquot of this fecal sample was reextracted and real-time PCR was performed, this sample remained negative. At lower bacteriologic colony counts, neither PCR was 100% sensitive, and the nonhomogeneous distribution and/or small quantity of M. avium subsp. paratuberculosis bacteria may be responsible for this effect. There is also the possibility of a polymorphism within the target sequence of the molecular beacon that prevented hybridization.
The specificity between real-time PCR and culture was affected in 1 of 41 samples (sample 13). By using another aliquot of this fecal sample, M. avium subsp. paratuberculosis DNA was again detected, which suggested that a low level of DNA, but not viable bacteria, was present in this sample. Since the nPCR result was also negative, this suggested that the real-time PCR was more sensitive than the nPCR. A higher sensitivity of the real-time PCR was also observed in results with samples 15, 17, and 21. DNA contamination may have occurred when the real-time PCR was performed. However, with a closed-tube format and with fewer manipulations required to perform real-time PCR, and since extraction and PCR were repeated with the same results, contamination seems unlikely.
The results with real-time PCR agreed well with those of culture. Kappa scores were lower for the comparison of nPCR and real-time PCR results than for the comparison of culture and real-time PCR results due to the higher sensitivity of the real-time PCR. Since the extraction techniques are the same for both PCR methods, it appears very possible that loss of DNA is limited by the fewer manipulations that are associated with real-time PCR.
Intra- and interassay variabilities representing the repeatability and reproducibility of the real-time PCR, respectively, appeared similar to those for other real-time PCR assays and were fairly low (10, 18, 19). This allows comparisons of bacterial loads to be obtained over time for pathogenesis studies and for accurate diagnostic testing.
An examination of scatterplots revealed no consistent correlation between the quantity of DNA obtained by real-time PCR and colony counts. DNA concentrations would not necessarily correlate with viable bacteria since DNA may be obtained from nonviable bacteria. Even within replicate tubes of a given culture method, colony counts varied significantly. These differences may be due to a lack of homogenization of the sample, factors affecting the growth of different colonies, or possibly the presence of bacterial, fungal, or unknown contaminants within fecal material that might affect growth.
In conclusion, real-time PCR with molecular beacons compares very favorably with culture and nPCR for the direct detection of M. avium subsp. paratuberculosis, in bovine feces. The real-time PCR with molecular beacons has the advantages over the other methods of a rapid testing time, a single-tube format, and the ability to obtain reproducible, quantitative results.
Acknowledgments
Funding for this study was provided by the South Dakota Agricultural Experiment Station Cooperative Grants Program, the Animal Disease Research and Diagnostic Laboratory at South Dakota State University, and USDA CSREES grant 9803756 (equipment grant).
We thank Fred Kramer and Sanjay Tyagi of the Department of Molecular Genetics, Public Health Research Insititute, New York, N.Y., for their assistance in the design and use of molecular beacons. The M. avium subsp. paratuberculosis culture facility support given by Connie Gates at the Animal Disease Research and Diagnostic Laboratory at South Dakota State University was very much appreciated. We give special thanks to Janet Payeur of the National Veterinary Services Laboratory, Ames, Iowa, for providing proficiency test samples.
REFERENCES
- 1.Al-Soud, W. A., and P. Rådström. 2000. Effects of amplification facilitators on diagnostic PCR in the presence of blood, feces, and meat. J. Clin. Microbiol. 38:4463–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauerfeind, R., S. Benazzi, R. Weiss, T. Schliesser, H. Willems, and G. Baljer. 1996. Molecular characterization of Mycobacterium paratuberculosis isolates from sheep, goats, and cattle by hybridization with a DNA probe to insertion element IS900. J. Clin. Microbiol. 34:1617–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiodini, R. J. 1989. Crohn’s disease and the mycobacterioses: a review and comparison of two disease entities. Clin. Microbiol. Rev. 2:90–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiodini, R. J., H. J. Van Kruiningen, and R. S. Merkal. 1984. Ruminant paratuberculosis (Johne’s disease): the current status and future prospects. Cornell Vet. 74:218. [PubMed] [Google Scholar]
- 5.Collins, D. M., D. M. Stephens, and G. W. De Lisle. 1993. Comparison of polymerase chain reaction tests and fecal culture for detecting Mycobacterium paratuberculosis in bovine feces. Vet. Microbiol. 36:289–299. [DOI] [PubMed] [Google Scholar]
- 6.Collins, D. M., D. M. Gabric, and G. W. de Lisle. 1989. Identification of a repetitive sequence specific to Mycobacterium paratuberculosis. FEMS Microbiol. Lett. 60:165–178. [DOI] [PubMed] [Google Scholar]
- 7.Collins, M. T., G. Lisby, C. Moser, D. Chicks, S. Christensen, M. Reichelderfer, N. Høiby, B. A. Harms, O. Ø. Thomsen, U. Skibsted, and V. Binder. 2000. Results of multiple diagnostic tests for Mycobacterium avium subsp. paratuberculosis in patients with inflammatory bowel disease and in controls. J. Clin. Microbiol. 38:4373–4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins, M. T. 1996. Diagnosis of paratuberculosis, p.357–371. In R. W. Sweeney (ed.), The Veterinary Clinics of North America Food animal practice. Paratuberculosis (Johne’s Disease). W. B. Saunders Co., Philadelphia, Pa. [DOI] [PubMed]
- 9.Dell’Isola, B., C. Poyart, O. Goulet, J. F. Mougenot, E. Sadoun-Journo, N. Brousse, J. Schmitz, C. Ricour, and P. Berche. 1994. Detection of Mycobacterium paratuberculosis by polymerase chain reaction in children with Crohn’s disease. J. Infect Dis. 169:449–451. [DOI] [PubMed] [Google Scholar]
- 10.Desjardin, L. E., Y. Chen, M. D. Perkins, L. Teixeira, M. D. Cave, and K. D. Eisenach. 1998. Comparison of the ABI 7700 system (TaqMan) and competitive PCR for quantification of IS6110 DNA in sputum during treatment of tuberculosis. J. Clin. Microbiol. 36:1964–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eun, A. J. C., and S. M. Wong. 2000. Molecular beacons: a new approach to plant virus detection. Phytopathology 90:269–275. [DOI] [PubMed] [Google Scholar]
- 12.Giesendorf, B. A. J., J. A. M. Vet, S. Tyagi, E. J. M. G. Mensink, F. J. M. Trijbels, and H. J. Blom. 1998. Molecular beacons: a new approach for semiautomated mutation analysis. Clin. Chem. 44:482–486. [PubMed] [Google Scholar]
- 13.Green, E. P., M. L. Tizard, M. T. Moss, J. Thompson, D. J. Winterborne, J. J. McFadden, and J. Hermon-Taylor. 1989. Sequence and characteristics of IS900, an insertion sequence element identified in a human Crohn’s isolate of Mycobacterium paratuberculosis. Nucleic Acids Res. 17:9063–9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kostrikis, L. G., S. Tyagi, M. M. Mhlanga, D. D. Ho, and F. R. Kramer. 1998. Spectral genotyping of human alleles. Science 279:1228–1229. [DOI] [PubMed] [Google Scholar]
- 15.Kota, R., T. A. Holton, and R. J. Henry. 1999. Detection of transgenes in crop plants using molecular beacon assays. Plant Mol. Biol. Rep. 17:363–370. [Google Scholar]
- 16.Larsen, A. B., R. S. Merkal, and R. C. Cutlip. 1975. Age of cattle as related to resistance to infection with Mycobacterium paratuberculosis. Am. J. Vet. Res. 36:255–257. [PubMed] [Google Scholar]
- 17.Lawrence, W. E. 1956. Congenital infection with Mycobacterium johnei in cattle. Vet. Rec. 68:312–314. [Google Scholar]
- 18.Lewin, S. R., M. Vesanen, L. Kostrikis, A. Hurley, M. Duran, L. Zhang, D. D. Ho, and M. Markowitz. 1999. Use of real-time PCR and molecular beacons to detect virus replication in human immunodeficiency virus type 1-infected individuals on prolonged effective antiretroviral therapy. J. Virol. 73:6099–6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Locatelli, G., F. Santoro, F. Veglia, A. Gobbi, P. Lusso, and M. S. Malnati. 2000. Real-time quantitative PCR for human herpesvirus 6 DNA. J. Clin. Microbiol. 38:4042–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin, W. W., and B. Bonnett. 1987. Clinical epidemiology. Can. Vet. J. 28:318–325. [PMC free article] [PubMed] [Google Scholar]
- 21.Martinson, J. J., L. Hong, R. Karanicolas, J. P. Moore, and L. G. Kostrikis. 2000. Global distribution of the CCR2-64I/CCR5-59653T HIV-1 disease-protective haplotype. AIDS 14:483–489. [DOI] [PubMed] [Google Scholar]
- 22.McKillip, J. L. and M. Drake. 2000. Molecular beacon polymerase chain reaction detection of Escherichia coli 0157:H7 in milk. J. Food Prot. 63:855–859. [DOI] [PubMed] [Google Scholar]
- 23.Park, S., M. Wong, S. A. E. Marras, E. W. Cross, T. E. Kiehn, V. Chaturvedi, S. Tyagi, and D. S. Perlin. 2000. Rapid identification of Candida dubliniensis using a species-specific molecular beacon. J. Clin. Microbiol. 38:2829–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piatek, A. S., S. Tyagi, A. C. Pol, A. Telenti, L. P. Miller, F. R. Kramer, and D. Alland. 1998. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat. Biotechnol. 16:359–486. [DOI] [PubMed] [Google Scholar]
- 25.Rankin, J. D. 1959. Johne’s disease in cattle. IV. experimental infection. Vet. Rec. 71:1157–1160. [Google Scholar]
- 26.Schofield, P., A. N. Pell, and D. O. Krause. 1997. Molecular beacons: trial of a fluorescence-based solution hybridization technique for ecological studies with ruminal bacteria. Appl. Environ. Microbiol. 63:1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Secott, T. E., A. M. Ohme, S. Barton, C. C. Wu, and F. A. Rommel. 1999. Mycobacterium paratuberculosis detection in bovine feces is improved by coupling agar culture enrichment to an IS900-specific polymerase chain reaction. J. Vet. Diagn. Invest. 11:441–447. [DOI] [PubMed] [Google Scholar]
- 28.Singh, S. N. 1998. Mycobacterium paratuberculosis PCR assay of feces, p.27. In L. H. Lauerman (ed.), Nucleic acid amplification assays for diagnosis of animal diseases. American Association of Veterinary Laboratory Diagnosticians, Davis, Calif.
- 29.Stabel, J. R. 1997. An improved method for cultivation of Mycobacterium paratuberculosis from bovine fecal samples and comparison to three other methods. J. Vet. Diagn. Invest. 9:375–380. [DOI] [PubMed] [Google Scholar]
- 30.Sweeney, R. W., R. H. Whitlock, A. E. Rosenberger, and S. A. Herr. 1992. Isolation of Mycobacterium paratuberculosis after oral inoculation in uninfected cattle. Am. J. Vet Res. 53:1312–1314. [PubMed] [Google Scholar]
- 31.Täpp, L., E. Malmberg, M. Rennel, M. Wik, and A.-C. Syvänen. 2000. Homogeneous scoring of single-nucleotide polymorphisms: comparison of the 5′-nuclease TaqMan assay and molecular beacon probes. BioTechniques 28:732–737. [DOI] [PubMed] [Google Scholar]
- 32.Thompson, D. E. 1994. The role of mycobacteria in Crohn’s disease. J. Med. Microbiol. 41:74–94. [DOI] [PubMed] [Google Scholar]
- 33.Tyagi, S., D. P. Bratu, and F. R. Kramer. 1998. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 16:49–53. [DOI] [PubMed] [Google Scholar]
- 34.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303–308. [DOI] [PubMed] [Google Scholar]
- 35.Uwatoko, K., M. Sunairi, A. Yamamoto, M. Nakajima, and K. Yamaura. 1996. Rapid and efficient method to eliminate substances inhibitory to the polymerase chain reaction from animal fecal samples. Vet. Microbiol. 52:73–79. [DOI] [PubMed] [Google Scholar]
- 36.van der Giessen, J. W. B., R. M. Haring, E. Vauclare, A. Eger, J. Haagsma, and B. A. M. van der Zeijst. 1992. Evaluation of the abilities of three diagnostic tests based on the polymerase chain reaction to detect Mycobacterium paratuberculosis in cattle: application in a control program. J. Clin. Microbiol. 30:1216–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Schie, R. C. A. A., S. A. E. Marras, J. M. Conroy, N. J. Nowak, J. J. Catanese, and P. J. de Jong. 2000. Semiautomated clone verification by real-time PCR using molecular beacons. BioTechniques 29:1296–1308. [DOI] [PubMed] [Google Scholar]
- 38.Vary, P. H., P. R. Andersen, E. Green, J. Hermon-Taylor, and J. J. McFadden. 1990. Use of highly specific DNA probes and the polymerase chain reaction to detect Mycobacterium paratuberculosis in Johne’s Disease. J. Clin. Microbiol. 28:933–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vet, J. A. M., A. R. Majithia, S. A. E. Marras, S. Tyagi, S. Dube, B. J. Poiesz, and F. R. Kramer. 1999. Multiplex detection of four pathogenic retroviruses using molecular beacons. Proc. Natl. Acad. Sci. USA. 96:6394–6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whipple, D. L., D. R. Calihan, and J. L. Jarnagin. 1991. Cultivation of Mycobacterium paratuberculosis from bovine fecal specimens and a suggested standardized procedure. J. Vet. Diagn. Invest. 3:368–373. [DOI] [PubMed] [Google Scholar]
- 41.Whipple, D. L., P. A. Kapke, and P. R. Andersen. 1992. Comparison of a commercial DNA probe test and three cultivation procedures for detection of Mycobacterium paratuberculosis in bovine feces. J. Vet. Diagn. Invest. 4:23–27. [DOI] [PubMed] [Google Scholar]
- 42.Wilde, J., J. Eiden, and R. Yolken. 1990. Removal of inhibitory substances from human fecal specimens for detection of group A rotaviruses by reverse transcriptase and polymerase chain reaction. J. Clin. Microbiol. 28:1300–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]