Abstract
The O26 serogroup of enteropathogenic Escherichia coli (EPEC) is one of the serogroups most frequently implicated in infant diarrhea and is also common among enterohemorrhagic E. coli (EHEC) strains. The most common O26 strains belong to EPEC/EHEC serotype O26:H11 and are generally Shiga toxin (Stx) positive. Stx-negative E. coli strains that are negative for the EPEC EAF plasmid and bundle-forming pilus (Bfp) are classified as atypical EPEC. Here, we report a novel adhesin present in an stx-negative bfpA-negative atypical EPEC O26:H11 strain isolated from an infant with diarrhea. A cloned 15-kb genomic region from this strain, designated the locus for diffuse adherence (lda), confers diffuse adherence on HEp-2 cells when expressed in E. coli K-12. Sequence analysis of lda revealed a G+C content of 46.8% and 15 open reading frames sharing homology with the E. coli K88 fae and CS31A clp fimbrial operons. The lda region is part of a putative 26-kb genomic island inserted into the proP gene of the E. coli chromosome. Hybridization studies have demonstrated the prevalence of the minor structural subunit gene, ldaH, across E. coli serogroups O5, O26, O111, and O145. A second plasmid-encoded factor that contributed to the Hep-2 adherence of this strain was also identified but was not characterized. Null mutations that abolish adherence to HEp-2 cells can be restored by plasmid complementation. Antiserum raised against the major structural subunit, LdaG, recognizes a 25-kDa protein from crude heat-extracted protein preparations and inhibits the adherence of the E. coli DH5α lda+ clone to HEp-2 cells. Electron microscopy revealed a nonfimbrial structure surrounding the bacterial cell.
Diarrheagenic strains of Escherichia coli can be subdivided into at least six categories depending on the virulence properties they possess (23, 38). Two of these E. coli categories are enteropathogenic E. coli (EPEC), a major cause of infant diarrhea, and enterohemorrhagic E. coli (EHEC), which produces Shiga toxin (Stx) and is associated with bloody diarrhea and hemolytic uremic syndrome. Multiple serogroups and serotypes of E. coli comprise these two categories. One serogroup shared between EPEC and EHEC is O26. The virulence characteristics of serotype O26:H11 strains vary in accordance with their geographical distribution. Many O26:H11 strains isolated in North America, Europe, and Japan produce Stx and are classified as EHEC (28). In Brazil, with the exception of one reported case isolate (17), O26:H11 and O26:H− isolates from infants with diarrhea have consistently been stx negative and have not been associated with hemolytic uremic syndrome (42, 48).
A key aspect of enteric infection caused by E. coli is the colonization of the gastrointestinal tract, which is mediated by specific adherence factors. EPEC and EHEC strains produce an adherence factor, called intimin, which is chromosomally encoded by the eae (EPEC attaching and effacing) gene located within the locus for enterocyte effacement (LEE) pathogenicity island (21, 31). The more restrictive designation of EHEC refers to those Stx-producing E. coli strains that possess the LEE. In EPEC, a plasmid-encoded type IV bundle-forming pilus (Bfp) has also been shown to be required for intestinal colonization (16). Bfp mediates bacterium-to-bacterium adherence, resulting in the formation of compact microcolonies on HEp-2 epithelial cell monolayers after 3 hours, which is referred to as localized adherence (LA) (11, 46). E. coli O26 strains that are LEE+ stx negative and EAF− bfp negative are classified as atypical EPEC and differ from typical EPEC in several characteristics. Typical EPEC, found primarily in developing countries, affects infants and children under the age of 2 years, whereas atypical EPEC strains have caused outbreaks of diarrheal disease in both children and adults in industrialized countries (22, 54). In the absence of Bfp, atypical EPEC strains can still adhere to HEp-2 cells in either a diffuse-adherence or aggregative-adherence pattern (37). Some clinical isolates of serogroups O26, O55, and O111 show a Bfp-independent localized-adherence pattern similar to the LA, called localized-adherent-like (43). The majority of these strains form microcolonies on HEp-2 epithelial monolayers, but only after 6 hours of infection, whereas LA is apparent after 3 hours. While the presence of Bfp correlates well with LA, not all LA+ EPEC strains are Bfp positive. Studies by other investigators (15, 43, 47) have reported EAF− Bfp− EPEC serotypes that exhibit localized adherence, but so far, no characterization of the factor(s) responsible for this phenotype has been reported. Fischer et al. (13) reported an EAF− stx-negative eae+ O26:NM strain isolated from a calf with diarrhea that was LA+ on HEp-2 cells, but the adherence factor was not identified. E. coli 22, an O26:H11 isolate originally obtained from an infant with diarrhea in Brazil, is stx negative eae+ and exhibits the LA phenotype within 3 hours, identical to the Bfp-mediated phenotype. However, E. coli 22 does not harbor the EAF plasmid or the bfpA gene. The aim of this study was to identify the genetic determinants conferring adherence in this strain and to compare it to those encoding other known E. coli adherence factors. We report the cloning, nucleotide sequence, and organization of a chromosomal locus encoding an afimbrial adhesin with homologies to the operons encoding enterotoxigenic E. coli (ETEC) K88 and CS31A fimbriae and demonstrate its role in mediating bacterial adherence to epithelial cells. In addition, we present data showing the prevalence of these gene sequences across diverse E. coli serogroups. Our data indicate that this new fimbria is a member of the K88 family of adhesins.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, and media.
The bacterial strains and plasmids used in constructions are listed in Table 1. Wild-type E. coli strains tested for the presence of ldaH were obtained from the bacterial culture collection at the Center for Vaccine Development. All bacterial strains were maintained in Luria-Bertani medium or tryptic soy broth. Antibiotics were used at the following concentrations unless otherwise indicated: ampicillin (Ap), 100 μg/ml; chloramphenicol (Cm), 20 μg/ml; and kanamycin (Km), 50 μg/ml.
TABLE 1.
Bacterial strains used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | Host strain | Invitrogen, Carlsbad, CA |
| SM10λpir | Host strain | 49 |
| MG1655 | Sequenced K-12 strain | 9 |
| XL-10 | Host strain | Stratagene, La Jolla, CA |
| 22 | O26:H11 stx negative eae+bfp negative exhA negative | 43 |
| ICS1 | ΔldaG::kan derivative of E. coli 22 | This study |
| LDA1 | ΔldaG derivative of E. coli 22 | This study |
| LDA3 | LDA1 cured of pIJ22 | This study |
| ORN103 | Δfim | 41 |
| Plasmids | ||
| pHM5 | Suicide vector; sacB oriR6K Apr | 44 |
| pHC79 | Cosmid vector; Apr | 19 |
| pACYC177 | Cloning vector; Kmr Apr | 10 |
| pACYC184 | Cloning vector; Cmr Tcr | 10 |
| pKD3 | Source of Cmr cassette | 12 |
| pKD4 | Source of Kmr cassette | 12 |
| pCP20 | flp recombinase | 12 |
| pIJ9 | pV-B-6 ΔBssHII | This study |
| pIJ13 | ΔldaG::kan Apr KmroriR6K sacB | This study |
| pIJ20 | pIJ9 ΔStuI fragment; lda minimal subclone; Apr | This study |
| pIJ22-1 | E. coli 22 plasmid encoding LA; Kmr | This study |
| pIJ22 | Spontaneous deletion product of pIJ22-1; Kmr | This study |
| pIJ30 | pIJ22::cat | This study |
| pV-B-6 | pHC79::lda adherence+ Apr | This study |
| pV-B-6TN | ldaH::kan adherence− Kmr | This study |
| pBSL180 | mini-Tn10::kan donor plasmid; Kmr | 3 |
| pLDAH | pUC18::ldaH::kan subclone; Kmr | This study |
Standard methods of DNA manipulation and transformation were employed (45). Plasmid isolations were performed using alkaline lysis and CsCl2 gradient centrifugation (45). Restriction nucleases and enzymes were obtained from Invitrogen (Carlsbad, CA).
Primer sequences are listed in Table 2. The extension times for primer pairs used for determining the upstream junction of the LDA island were 2 min (K1/K3), 30 s (K2/K3), 3 min (K4/K6), 2 min (K5/K7), and 7 min (K3/K2792).
TABLE 2.
Primers used in PCR amplifications and plasmid constructions
| Primer | Sequence |
|---|---|
| K1 | 5′-ACATCCGCGAATTGATGAATAA |
| K2 | 5′-GACGCTGATCAGCTCAAACA |
| K3 | 5′-CGACGCCATAATAATCAGGG |
| K4 | 5′-ATCCGCAGCCTGGTAATACT |
| K5 | 5′-TGGCTACCCGCTTTATTTACA |
| K6 | 5′-GACGTAATTTATTGTCACGAAG |
| K7 | 5′-ATTGCTGCAAACAAAAATGTCAT |
| K2792 | 5′-GCATGCCTGTACTCAGTGCG |
| K2937 | 5′-ATATATCTAGACATATGAATATCCTCCTTAGTTCCT |
| K2938 | 5′-ATATATCTAGATGTGTAGGCTGGAGCTGCTTC |
| K3094 | 5′-AAAGATCTGTGATGAGGTTCAGGTGAAG |
| K3095 | 5′-AAATCTAGATGCAGACGCAACTACAGCCA |
| K3096 | 5′-AAATCTAGAGCTAACAGTACCTTGGTTATG |
| K3097 | 5′-AAAGAGCTCACGATGCATAAGCTGCAGCA |
Cell culture adhesion and adhesion inhibition assays.
The HEp-2 cell adhesion assay was performed as described previously (46). Monolayers of ca. 105 HEp-2 cells were grown in 24-well tissue culture plates in Dulbecco modified Eagle medium containing 10% fetal bovine serum. Bacterial strains were grown statically in 3 ml of tryptic soy broth or Luria broth for 16 to 18 h at 37°C. The monolayers were infected with 40 μl of bacterial culture and incubated for 3 hours at 37°C with 5% CO2 in the presence of 0.5% d-mannose. The cells were then washed six times with phosphate-buffered saline (PBS; pH 7.2), fixed with methanol, stained with May-Grünwald-Giemsa stain, and examined under a light microscope. For the inhibition assay, 50 μl of an overnight bacterial culture was mixed with a 1:50 dilution of LdaG rabbit antiserum and incubated for 2 h at 37°C prior to the adherence assay (7). The effect of the antiserum upon bacterial adherence to HEp-2 cells was determined by comparing bacteria preincubated with preimmune antiserum to bacteria preincubated with LdaG antisera under the same conditions. All experiments were performed in triplicate.
DNA hybridization.
Colony blot and Southern blot hybridizations (45) were performed at 65°C. Gene probes, bfpA (15), stx (39), ehxA (pO157 plasmid sequences) (28), ldaG, ldaH, and kanamycin were generated either by PCR or by restriction digestion, gel purified, and labeled with [32P]dCTP using a Rediprime kit (Amersham Biosciences, Piscataway, NJ) according to the manufacturer's instructions.
Transposon mutagenesis.
Mini-Tn10::kan was introduced into E. coli 22 or E. coli DH5α(pV-B-6) by electroporation with pBSL180 (3). Isolates were selected on L agar with kanamycin or kanamycin-ampicillin and tested for adherence in the HEp-2 assay.
Cloning of LDA locus.
Chromosomal DNA was isolated from E. coli 22 using the EZ-DNA kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions and then partially digested with Sau3AI and ligated into the BamHI-digested cosmid vector pHC79. The ligation was packaged into bacteriophage lambda particles using the GigaPak III Gold packaging kit (Stratagene), transfected into E. coli DH5α, and plated onto L agar with ampicillin. Colonies were tested for localized adherence in the HEp-2 adherence assay. One Apr LA+ colony harboring a cosmid, designated pV-B-6, was selected for further analysis.
A minimal-size subclone that still possessed the ability to adhere to HEp-2 cells was obtained by digesting pV-B-6 with BssHII, followed by self-ligation. The resulting clone, pIJ9, was digested with StuI and self-ligated, yielding pIJ20, which is ca. 21.5 kb in size. pIJ20 was transformed into E. coli DH5α and tested for HEp-2 adherence.
Construction of bacterial deletion mutants and plasmids.
A kanamycin-resistant subclone of plasmid pV-B-6-Tn, designated pLDAH, was generated by ligating an EcoRI-HindIII restriction fragment that contained the kan gene (present in the mini Tn10::kan cassette) and flanking lda sequences into pUC18.
A 699-bp nonpolar chromosomal deletion in ldaG was constructed in E. coli 22 by inserting sequences flanking the ldaG gene into suicide plasmid pHM5 (44). Primer sets K3094/K3095 and K3096/K3097 were used in conjunction with Pfx polymerase to generate fragments containing 950 bp upstream and 920 bp downstream of the ldaG gene, respectively. The fragments were ligated into suicide vector pHM5 to create pIJ12. The deletion was marked by the insertion of a kanamycin gene cassette that contained flanking inverted-repeat FLP sites amplified from pKD4 (12). A 1.6-kb kanamycin cassette was amplified from pKD4 using primer set K2938/K2937 and blunt-end ligated into the XbaI-digested/Klenow-treated site of pIJ12, yielding pIJ13. Plasmid pIJ13 was introduced into E. coli 22 by conjugal mating with E. coli SM10λpir and directly plated onto modified L agar containing 5% sucrose and 25 μg/ml kanamycin. Individual colonies were then patched onto McConkey media to distinguish E. coli 22 (lac+) from E. coli SM10λpir (lac negative). One lactose-positive isolate exhibiting Kmr and Aps was called ICS1. The kanamycin marker was then eliminated from the chromosome with plasmid pCP20, which harbors the flp recombinase gene on a temperature-sensitive replicon (12). E. coli ICS1 was electroporated with pCP20 and plated on L agar with ampicillin. After overnight incubation at 30°C, colonies were picked and grown under nonselective pressure on L agar at 42°C. Colonies were separately patched onto media containing kanamycin or ampicillin to check for loss of the kanamycin cassette and pCP20, respectively. One Kms Aps isolate that tested negative for ldaG by Southern blotting was designated LDA1.
A pIJ22-1 plasmid-cured derivative of LDA1 was created using plasmid incompatibility. pIJ22 was digested with EcoRI and ligated to a chloramphenicol gene cassette (cat), creating pIJ30. Plasmid pIJ30 was electroporated into the ldaG mutant LDA1 and plated onto L agar with chloramphenicol. A Cmr Kms mutant that was negative for Hep-2 adherence was called LDA3.
Sequencing and analysis.
The nucleotide sequence of the cosmid pV-B-6 was determined by first subcloning small overlapping PstI, BamHI, or EcoRI restriction fragments into plasmid vector pAYCY177 or pACYC184. Plasmid DNA was prepared by cesium chloride density gradient centrifugation, and cycle sequencing was performed using the ABI 3100 Gene Analyzer. Nucleotide sequences were analyzed for open reading frames (ORFs) using Redasoft Visual Cloning 2000 DNA software (Whitehead Institute for Biomedical Research). BLAST searches and comparisons, as well as DNA-protein analysis, were conducted using databases at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/), The Institute for Genomic Research (http://www.tigr.org), and the CLUSTAL W (http://clustalw.genome.jp) websites.
Preparation of whole-cell lysates and heat-extracted proteins and identification of LdaG by N-terminal sequencing.
E. coli strains grown on MacConkey agar at 37°C were harvested from the plates with 1 ml PBS (pH 7.4). The samples were aliquoted into portions of approximately 4.0 × 108 bacteria for whole-cell lysates and 4.0 × 109 bacteria for heat-extracted proteins. The whole-cell lysate samples were harvested by centrifugation at 12,000 × g for 5 min at room temperature, washed in 1 ml PBS (pH 7.4), centrifuged again, resuspended in 200 μl of sodium dodecyl sulfate (SDS) sample buffer, and lysed at 100°C for 10 min. The cell debris was removed by centrifugation, and the supernatant was transferred to a fresh tube and stored at −20°C. The heat-extracted protein samples were harvested by centrifugation at 3,000 × g for 10 min, resuspended in 160 μl of PBS (pH 7.4), and incubated at 60°C for 30 min. The samples were then pelleted by centrifugation at 3,000 × g for 10 min, and the supernatant was transferred to a fresh tube; 40 μl of 5× SDS sample buffer was added, and the samples were boiled at 100°C for 5 min. The samples were separated by SDS-12% PAGE minigels, and proteins were stained with Coomassie blue. The 25-kDa protein that was highly expressed in strain 22 was excised from the gel (whole-cell lysate and heat-extracted proteins) and sent for N-terminal sequencing at the Stanford University sequencing facility (Palo Alto, CA).
Production of LdaG antiserum.
A heat extraction of E. coli DH5α(pV-B-6) was prepared and separated on a 12% SDS-PAGE gel and stained with Coomassie blue. The 25-kDa LdaG protein was excised from the gel, eluted using a Mini Whole Gel Eluter (Bio-Rad), and extensively dialyzed in water. New Zealand White rabbits were immunized subcutaneously with 100 μg of purified LdaG protein conjugated with complete Freund's adjuvant. The animals were inoculated 3 weeks later with the same dose of protein in complete Freund's adjuvant. Ten days later, blood was drawn and the serum was collected.
Western blot assay.
Heat-extracted proteins were separated on a 12% SDS-PAGE gel and transferred to Immobilon-P membranes (53). The membranes were blocked overnight with 5% bovine serum albumin in Tris-buffered saline (pH 7.4) plus 0.1% Tween 20 (TBS-Tween 20). Rabbit anti-LdaG antiserum (1:10,000) was incubated for 1 h and washed with TBS-Tween 20. Goat anti-rabbit immunoglobulin G-horseradish peroxidase (1:50,000) (Sigma-Aldrich, St. Louis, MO) was incubated for 1 h and washed with TBS-Tween 20. The protein was detected using the ECL Chemiluminescent Western Blotting Detection kit (Amersham Biosciences, Piscataway, NJ) according to the manufacturer's instructions.
Immunogold labeling and electron microscopy.
Strains were grown statically overnight at 37°C in MacConkey broth (Difco), pelleted at 5,000 × g, resuspended in PBS (pH 7.4), and allowed to adhere to Formvar-carbon-coated copper grids (200 mesh; Electron Microscopy Sciences). The grids were floated on drops of rabbit anti-LdaG antiserum (1:100 dilution) for 30 min. After being washed, the grids were placed onto 10-nm-gold-labeled goat anti-rabbit sera (1:20 dilution; Electron Microscopy Sciences) for 30 min. After being further washed in PBS and distilled water, the grids were negatively stained with 1% phosphotungstic acid, pH 6.8 (Kodak), and visualized in a Joel JEM 1200 EX II transmission electron microscope at 80 kV.
Nucleotide sequence accession number.
The GenBank accession number for the lda gene sequence is AY858803.
RESULTS
Identification and cloning of a locus for diffuse adherence.
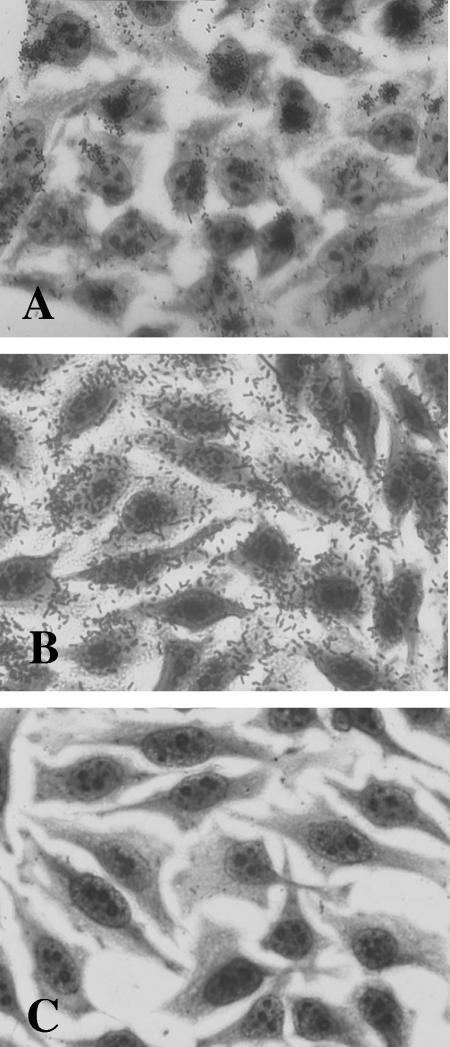
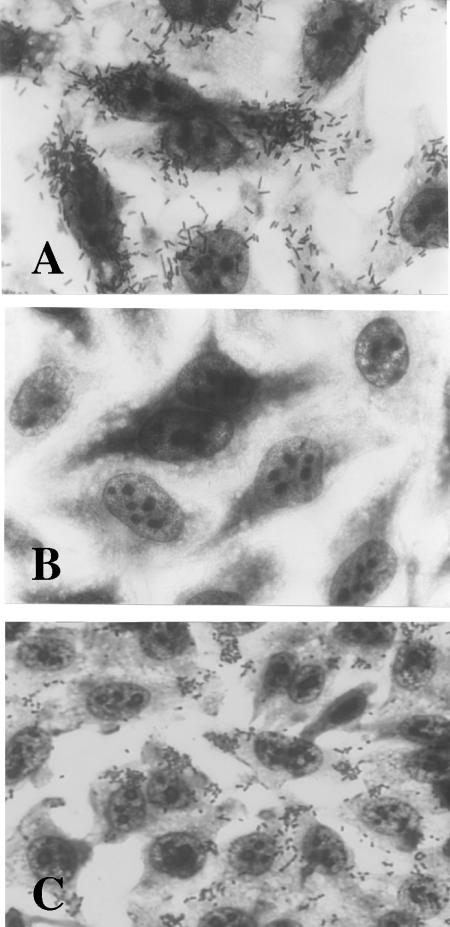
In a previous report (43), Escherichia coli strains isolated from Brazilian children with watery diarrhea were tested for adherence to HEp-2 cell monolayers. Isolates that exhibited the LA phenotype in 3 h were then analyzed by colony hybridization for the presence/absence of sequences encoding EPEC Bfp (bfpA), EHEC Stx (stx), and hemolysin (exhA-hlyA), the last serving as a marker for the pO157 plasmid. One LA+ bfpA-negative stx-negative exhA-negative isolate, E. coli 22, was selected for use in this study (Fig. 1A).
FIG. 1.
HEp-2 adherence assay. (A) E. coli 22 wild type. (B) E. coli DH5α(pV-B-6). (C) E. coli DH5α(pV-B-6-Tn).
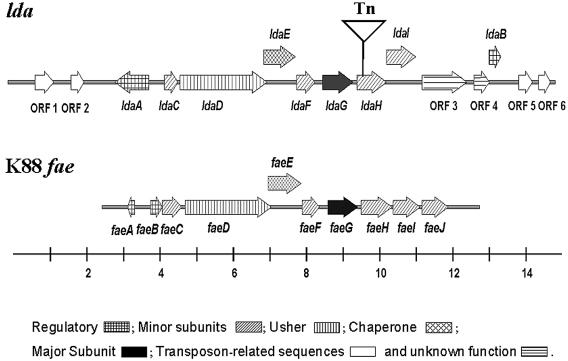
A genomic cosmid library of E. coli 22 was generated in E. coli K-12, and the resulting clones were screened for localized adherence to HEp-2 cells. One cosmid clone, pV-B-6, exhibited adherence in a diffuse pattern over the entire epithelial cell rather than the LA microcolony formation seen with E. coli 22 (Fig. 1B). Transposon mutagenesis was utilized to identify the region of cosmid pV-B-6 responsible for the adhesive phenotype. Plasmid pBSL180 carrying mini-Tn10::kan was electroporated into E. coli DH5α(pV-B-6), and isolates were selected on L agar with kanamycin and ampicillin. One isolate resistant to ampicillin and kanamycin that tested negative for adherence (pV-B-6-Tn) was chosen for further analysis (Fig. 1C). DNA sequencing of a subclone of pV-B-6 (pLDAH) revealed an open reading frame, ldaH, whose predicted protein product shares 73% amino acid identity with that of the E. coli K88 fimbrial gene faeH (6), suggesting that this region might include genes involved in a novel adherence factor (Fig. 2). This region was named the locus for diffuse adherence (lda).
FIG. 2.
Comparison of predicted amino acid sequence of LdaH with E. coli K88 fimbrial subunit FaeH. Identical residues in both proteins are indicated by asterisks, and conserved residues are indicated by dots (:, strongly similar; ., weakly similar). The alignment was created by CLUSTAL W.
Sequence analysis of lda and homology with other fimbrial operons.
Subsequent mapping and subcloning of pV-B-6 using restriction enzymes HindIII, PstI, EcoRI, and BamHI generated overlapping clones, which were purified and whose DNA sequences were determined. Sequence analysis revealed a 15-kb region having a G+C content of 46.8% comprised of 15 ORFs whose predicted protein products are similar to those of the K88 fae (33) and CS31A (14, 32) operons. Because of its well-characterized nature, we have chosen to use the K88 operon as the family prototype of chaperone-usher fimbria in which to compare the LDA operon (Fig. 3). Putative functions of the predicted proteins have been assigned on the basis of their homology to other proteins (Table 3). To maintain parallel nomenclature, we have named the lda genes to correspond to their counterparts in the K88 fae operon. These proteins include a structural subunit (LdaG), an outer membrane usher protein (LdaD), a periplasmic chaperone (LdaE), and four accessory proteins (LdaC, LdaF, LdaH, and LdaI). LdaC shows 85% amino acid identity to FaeC and is predicted to be a minor-subunit protein. Although slightly shorter than FaeD (812 amino acids), LdaD (790 amino acids) shares 82% amino acid identity and is predicted to be an outer membrane usher protein. Like FaeD, LdaD contains four conserved cysteine residues, two at the N terminus and two at the C terminus, which are necessary for proper folding of the protein (32). Compared to the periplasmic chaperone protein FaeE, LdaE is slightly larger and shares ca. 75% identity. Like other chaperone proteins within the gram-negative immunoglobulin superfamily, LdaE contains the highly conserved Arg-8, Lys-112, and Met-172 residues (numbered according to the PapD sequence) necessary for pilin production, as demonstrated by site-directed mutagenesis in PapD (20). The predicted minor subunit LdaF shows 72% identity to FaeF. The predicted major structural subunit, LdaG, shows 56% identity to the K99 adhesin, F41 (4), and shows only 23% identity with the K88ac major subunit. On the nucleotide level, ldaG shares no striking homology with any bacterial genes in the National Center for Biotechnology Information GenBank database. The predicted minor pilin subunits LdaH and LdaI show 73% and 60% identity, respectively, to FaeH/ClpH and FaeI/ClpL.
FIG. 3.
Maps of lda and K88 fae loci (drawn to scale). The locations and directions of genes are indicated by arrows, and the point of Tn5 insertion in pV-B-6-Tn is shown by an inverted triangle. A line scale drawn below indicates the size in kb. The shading patterns within the arrows indicate the predicted functions of the proteins as indicated.
TABLE 3.
Description of lda open reading frames
| Protein or ORF | No. of amino acids | Protein mass (kDa) | % Identity | Similar protein(s) and putative function | Protein accession no. |
|---|---|---|---|---|---|
| ORF1 | 513 | 20 | 38 | Putative transposase; Yersinia pestis | NP_669742.1 |
| 37 | Hypothetical protein encoded within IS1240 of Pseudomonas syringae | AAB81643.1 | |||
| ORF2 | 118 | 13.5 | 63 | Putative transposase; Pseudomonas syringae | AAB81642.1 |
| 54 | Putative transposase; Yersinia pestis | CAC90690.1 | |||
| LdaA | 309 | 36.4 | 53 | CS12 transcriptional regulator (YdeR) | AAK09057.1 |
| 49 | Transcriptional activator (AfrR) in E. coli RDEC-1a | AAC28314.1 | |||
| 39 | Caf1 of Yersinia pestis | S19097 | |||
| LdaC | 119 | 12.8 | 85 | E. coli K88 fimbrial protein (FaeC) | AAL47165.1 |
| 54 | E. coli CS31A fimbrial protein (ClpC) | AAB39316.1 | |||
| LdaD | 790 | 83.5 | 82 | E. coli K88 outer membrane usher (FaeD) | CAA27310.1 |
| 60 | RalD of rabbit E. coli (REPEC) | AAB97611 | |||
| LdaE | 287 | 31.9 | 82 | E. coli CS31A periplasmic chaperone (ClpE) | I41172 |
| 75 | E. coli K88 periplasmic chaperone (FaeE) | S15227 | |||
| LdaF | 163 | 17.8 | 72 | E. coli CS31A minor subunit (ClpF) | AAA23584.1 |
| 63 | E. coli K88 minor fimbrial subunit (FaeF) | B45725 | |||
| LdaG | 277 | 29 | 56 | E. coli K99 adhesin F41 | I41116 |
| 27 | CS31A fimbrial subunit of E. coli K88 (ClpG) | A41663 | |||
| 23 | E. coli K88 fimbrial subunit K88ac (FaeG) | I41318 | |||
| LdaH | 268 | 28.9 | 73 | E. coli K88 minor fimbrial subunit (FaeH) | S24810 |
| 73 | CS31A minor subunit (ClpH) | AAA23780.1 | |||
| 71 | Minor fimbrial subunit; Salmonella enterica | AAC09003.1 | |||
| LdaI | 265 | 28.9 | 61 | E. coli minor subunit CS31A (ClpL) | AAA23586.1 |
| 59 | E. coli K88 minor fimbrial subunit (FaeI) | F45725 | |||
| 59 | Minor fimbrial subunit; Salmonella enterica | AAC09004.1 | |||
| ORF3 | 407 | 48.2 | 87 | Conserved hypothetical protein (YdbN); E. coli | NP754377 |
| ORF4 | 143 | 16.4 | 85 | Hypothetical protein; Shigella flexneri | AAK18415.1 |
| 82 | Hypothetical protein; Salmonella enterica | NP_458668.1 | |||
| 79 | Hypothetical protein; E. coli O157:H7 | NP_286739.1 | |||
| LdaB | 108 | 12.7 | 73 | Putative transcriptional regulator; E. coli O157:H7 | NP_309415.1 |
| 38 | PerC transcriptional regulator; E. coli | AAG16672.1 | |||
| ORF5 | 126 | 14.3 | 55 | ISEc8-related protein within prophage CP-933X of E. coli O157:H7 | NP_286015.1 |
| ORF6 | 107 | 12.3 | 55 | ISEc8-related protein within prophage CP-933X of E. coli O157:H7 | NP_286015.1 |
RDEC-1, rabbit diarrheagenic E. coli type 1.
There are two ORFs with similarity to transcriptional regulators. The first ORF, LdaA, has 53% and 49% identity to YdeR of ETEC O159:H4 and AfrR of rabbit diarrheagenic E. coli type 1, respectively. The second ORF, LdaB, has 73% and 38% amino acid identity to a putative transcriptional regulator in E. coli O157:H7 and to EPEC plasmid regulator PerC, respectively. Interestingly, LdaA and LdaB have little identity (<11%) to either of their regulatory counterparts, FaeA and FaeB, and unlike the fae operon, in which the faeA and faeB regulator genes are next to each other, ldaA and ldaB are separated by the structural genes of the lda locus.
There are four ORFs (ORF1, -2, -5, and -6) that share 37 to 39% nucleotide homology with genes encoding transposases and insertion sequence-like sequences, suggesting that this region could have been horizontally acquired from another organism. ORF3 shows 87% identity to the hypothetical E. coli YbdN protein (9), which is a member of the phosphoadenosine phosphosulfate reductase family and may play a role in amino acid transport and metabolism. Lastly, ORF4 is similar to conserved hypothetical proteins of unknown function but shows some limited amino acid identity to YdbM (9), so it may function similarly to ORF3.
The lda locus is part of a putative pathogenicity island.
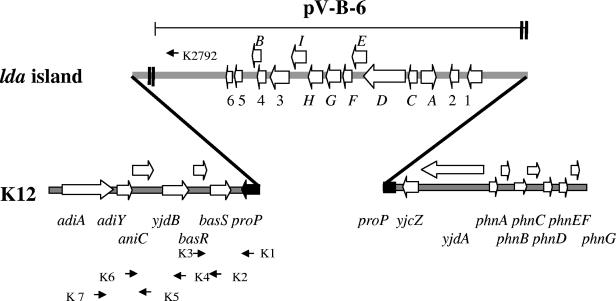
The flanking transposase-like sequences and lower G+C content suggested that this region could be part of a mobile element introduced as a block into the E. coli chromosome. To determine the chromosomal location of the lda region in E. coli 22, we examined the sequences flanking the 5′ and 3′ ends of the lda locus in cosmid clone pV-B-6 and compared the sequence to the published E. coli K-12 MG1655 genome (9). DNA sequence analysis revealed that the upstream LDA junction was within the proP gene (Fig. 4). Unfortunately, sequences within pV-B-6 did not extend far enough downstream, and so we relied on PCR analysis to determine the downstream junction. Based on E. coli K-12 and LDA sequence data, we designed primers downstream of the proP gene and one primer (K2792) within the region of lda in cosmid pV-B-6. Primer sets K1/K3, K2/K3, K4/K6, and K5/K7 all yielded the same size products from E. coli MG1655 and E. coli 22, indicating that the region downstream of proP was the same in both strains. PCR amplification using primer set K3/K2792 yielded a 6.5-kb product with E. coli 22. No PCR product was obtained with E. coli MG1655, indicating that this region was not present in K-12. These results confirmed that the lda island (ca. 26 kb) is inserted within the proP gene.
FIG. 4.
Chromosomal location of LDA island. Wide arrows indicate the locations and directions of ORFs and genes. The interrupted proP gene is represented as a discontinuous filled arrow. The small arrows indicate the locations of PCR primers. The region encompassing the cosmid clone is represented by a thin horizontal line. Double bold vertical lines indicate gaps in the sequence.
Presence of ldaH and ldaG in other E. coli serotypes.
To determine whether the lda locus was specific to our O26:H11 strain or if it is present in other E. coli strains, we performed colony blots and HEp-2 adherence assays on a collection of eae+ bfp-negative E. coli isolates representing 22 O serogroups. Nineteen strains in four groups, O5, O26, O111, and O145, were positive for the ldaH gene probe, indicating that this adhesin is not limited to O26:H11 (Table 4). All ldaH probe-positive isolates were positive for HEp-2 localized-adherence assays, but not all LA+ strains were ldaH probe positive, suggesting the presence of another adherence factor(s) in these isolates. ldaH was found in both stx-positive and -negative isolates. Because the CS31A and K88 operons are reported to share high homology between coding regions of the accessory proteins but to have distinct structural genes (30, 36), we tested for the presence of the major structural subunit gene. Six ldaH+ isolates, including those representing O26:H strains and all non-O26 lda+ strains, were tested with an ldaG gene probe. E. coli strains 22 (O26:H11), 86-280 (O26:H11), and 84-8198 (O26:H−) were positive and 88-0643 (O5:H−), 85-0839 (O111:H−), and 87-1713 (O145:H16) were negative with the ldaG gene probe, indicating that while there are homologies within a minor subunit (ldaH) among serotypes, the major structural subunit (ldaG) may be specific for the O26 serogroup. Control strains harboring genes for K88 and F41 fimbriae were all negative (data not shown).
TABLE 4.
Prevalence of ldaH sequences in E. coli eae+ bfp-negative strainsa
| Serotype | stx probed | ldaH probed | Localized adherenceb,d | No. of strains |
|---|---|---|---|---|
| O26:H− | − | − | − | 2 |
| O26:H− | − | + | + | 7 |
| O26:H8 | − | − | − | 2 |
| O26:H11 | − | − | − | 6c |
| O26:H11 | − | + | + | 7 |
| O26:H11 | + | − | − | 14 |
| O26:H11 | + | − | + | 5 |
| O26:H11 | + | − | +/− | 8 |
| O26:H11 | + | + | + | 2 |
| O26:H25 | − | − | − | 1 |
| O26:H32 | − | − | − | 2 |
| O26:H33 | − | − | − | 2 |
| O1:H6 | − | − | − | 1 |
| O1:H7 | − | − | − | 1 |
| O5:H− | + | + | + | 1 |
| O16:H16 | + | − | − | 1 |
| O22:H16 | + | − | − | 1c |
| O38:H21 | + | − | − | 1 |
| O45:H2 | + | − | − | 1 |
| O70:H11 | + | − | − | 1 |
| O82:H8 | + | − | − | 1 |
| O84:H2 | + | − | − | 1 |
| O91:H14 | + | − | + | 1 |
| O103:H2 | + | − | + | 1 |
| O111:H− | + | − | + | 1 |
| O111:H− | + | + | + | 1 |
| O111:H8 | + | − | − | 2c |
| O111:H34 | + | − | − | 1 |
| O113:H21 | − | − | − | 1 |
| O118:H30 | + | − | + | 1 |
| O121:H− | + | − | − | 1 |
| O21:H19 | − | − | − | 1 |
| O126:H | + | − | − | 1 |
| O126:H8 | + | − | − | 1 |
| O128:H12 | − | − | + | 1 |
| O145:H− | + | − | + | 1 |
| O145:H− | + | − | +/− | 1 |
| O145:H16 | + | + | + | 1c |
| O153:H25 | + | − | − | 1 |
| O165:H− | + | − | − | 1 |
| O157:H7 | + | − | − | 3 |
All strains are of human fecal origin except for the bovine fecal isolates indicated below. All strains are bfpA negative.
Localized adherence assays were recorded at 3 hours with a +/− indicating an equivocal phenotype.
One strain was isolated from bovine feces.
+, positive; −, negative.
Construction and complementation of an ldaG mutant and identification of a second adherence factor.
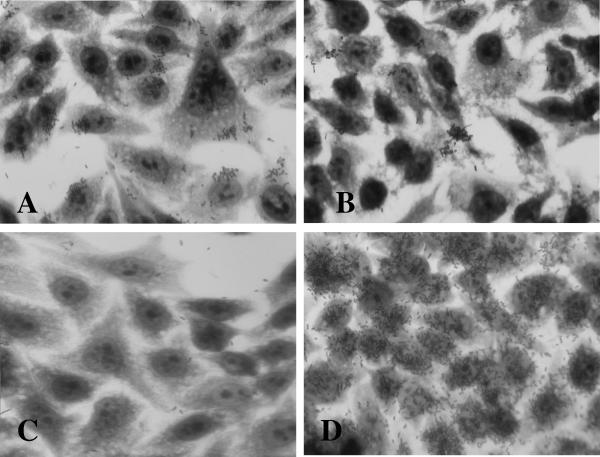
To demonstrate the requirement for lda for adherence in E. coli 22, we made a null mutation in the putative structural gene ldaG (Table 3) to yield strain LDA1 (see Materials and Methods). The construct was confirmed by Southern blotting and PCR analysis (data not shown). Surprisingly, E. coli LDA1 exhibited the same adherence pattern as wild-type E. coli 22, albeit to a lesser extent (compare Fig. 5A to Fig. 1A).
FIG. 5.
HEp-2 adherence assay. (A) E. coli LDA1. (B) E. coli XL-10(pIJ22). (C) E. coli LDA3 mutant. (D) E. coli LDA3(pIJ20) complement.
Because the cloned LDA region conferred adherence on E. coli DH5α and an insertion mutation within the cosmid abolished this phenotype, we hypothesized that the failure to abolish adherence following mutation of ldaG in E. coli 22 indicated the existence of a second adherence factor in the strain. Plasmid analysis revealed that this strain contains at least five plasmids (Fig. 6), and the possibility of a plasmid-encoded adhesin was investigated. Plasmids were extracted from a collection of transposon-mutagenized E. coli 22 isolates (see Materials and Methods), purified by CsCl2 gradient centrifugation, transformed into E. coli XL-10, and selected on L agar with kanamycin. The resulting transformants were tested for adherence on HEp-2 cells, and one Kmr transformant, designated E. coli(pIJ22), exhibited a localized adherence phenotype (Fig. 5B). The size of pIJ22 did not correspond to that of any plasmid present in the wild-type strain (taking into account the addition of the mini-Tn10::kan), and we therefore presumed that the parent plasmid had suffered a deletion. To identify which of the five wild-type plasmids encoded adherence in E. coli 22 ΔldaG (LDA1), an EcoRI restriction fragment of pIJ22 was used to probe a Southern blot of the unrestricted plasmids isolated from wild-type E. coli 22. A large plasmid (pIJ22-1) was identified as containing homologous sequences (Fig. 6) and was targeted for curing by plasmid incompatibility using a chloramphenicol derivative of pIJ22 (see Materials and Methods). The resulting plasmid-cured strain, called LDA3, was negative for adhesion on HEp-2 cells (Fig. 5C) and was confirmed by agarose gel electrophoresis to have lost pIJ22-1 (Fig. 6, lane 2).
FIG. 6.
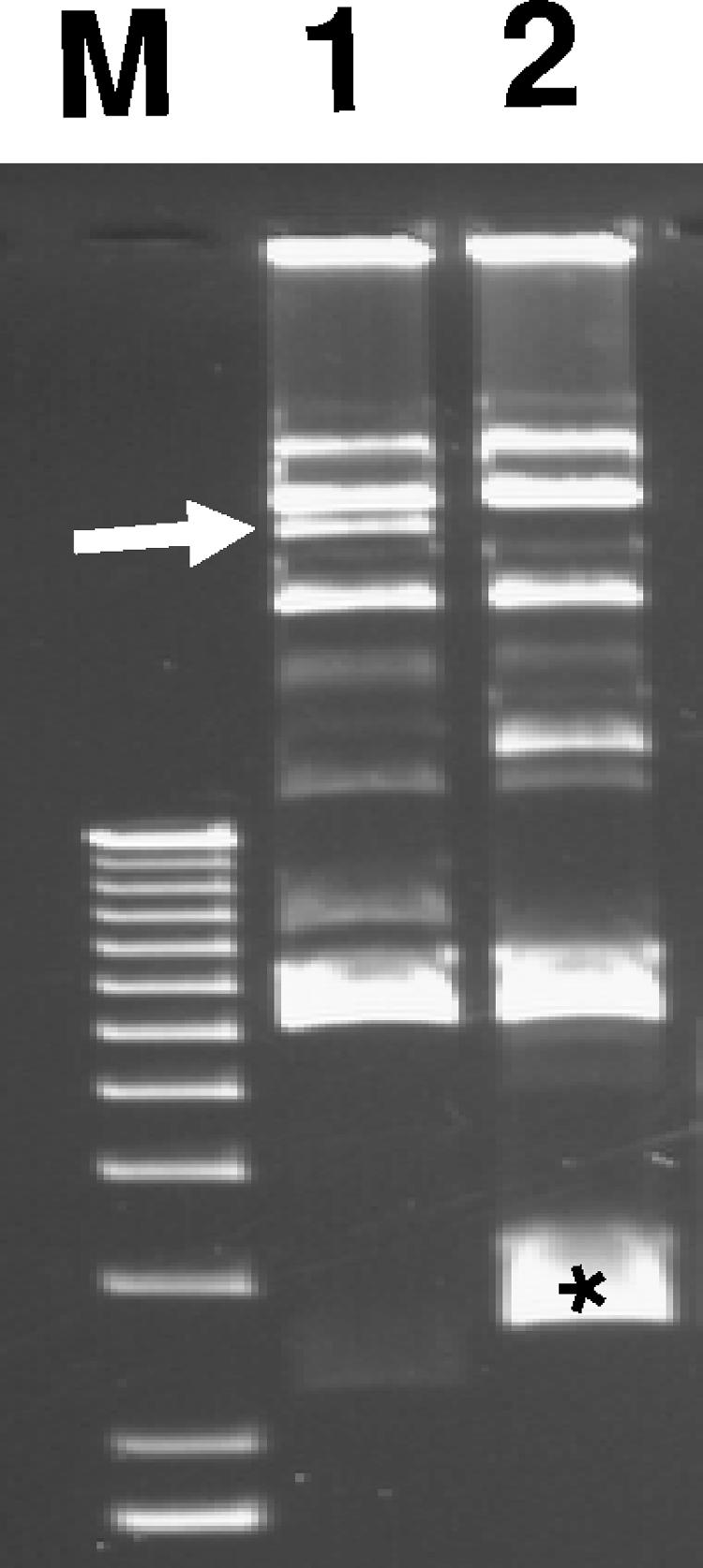
Plasmid profiles. M, 1-kb DNA ladder markers (Invitrogen); lane 1, E. coli 22; lane 2, E. coli LDA3. The arrow indicates the location of pIJ22-1. The asterisk in lane 2 identifies pIJ30, used to cure pIJ22-1.
To confirm the expression pattern of the LDA phenotype and restore the diffuse-adherence phenotype of LDA3, a minimal subclone of the original cosmid clone pV-B-6 that still possessed the ability to adhere to HEp-2 cells, termed pIJ20 (21.5 kb), was introduced into E. coli LDA3 to complement the ΔldaG mutation. As seen in Fig. 5D, LDA3(pIJ20) adheres to the HEp-2 cells in a diffuse pattern identical to that of E. coli DH5α(pV-B-6) (compare Fig. 1B and 5D). These results confirm that E. coli 22 contains two genetic loci that encode HEp-2 adherence factors. One adhesin, encoded by the chromosomal lda locus, expresses diffuse adherence, and the other adhesin (or an essential regulator of adhesion expression), expressing localized adherence, is encoded by a plasmid.
Expression and detection of LdaG.
Crude fimbrial preparations of E. coli 22 and LDA3 were obtained through whole-cell lysates and heat extracts. A broad band with an apparent molecular mass of 25 kDa was present in the heat extracts of E. coli 22 and to a lesser degree in the whole-cell lysates and was absent in the LDA3 extracts (Fig. 7A). The abundance of this protein is consistent with its being a major fimbrial structural subunit. The 25-kDa band was excised from the gel, and its N-terminal amino acid sequence was determined. The sequence, SDWTDNQPAGDI, corresponds to the mature form of LdaG after a 22-amino-acid putative signal sequence, MKKTLLALAVVASAVVSGSALA, has been removed.
FIG. 7.
Detection of LdaG. (A) SDS-PAGE of whole-cell lysates (lanes 1 and 2) and heat-extracted proteins (lanes 3 and 4) from E. coli 22 (lanes 1 and 3) and E. coli LDA1 (lanes 2 and 4). The arrow indicates the location of LdaG (lanes 1 and 3). (B) Western blot analysis of LdaG. E. coli 22 (lane 1), E. coli LDA3 (lane 2), and E. coli DH5α(pIJ20) (lane 3) are shown.
Antibody inhibition of LDA adhesion.
To provide further evidence that the LDA adhesin was responsible for the diffuse adherence seen on HEp-2 cells, we raised polyclonal rabbit antiserum against the major structural subunit LdaG. Western blot analysis (Fig. 7B) showed that the antiserum recognized the 25-kDa band present in wild-type E. coli 22 and E. coli ORN103(pIJ20), but not the LDA3 mutant. To determine whether the antiserum could inhibit adhesion of the bacteria, overnight bacterial cultures were first incubated with the LdaG antiserum for 2 h at 37°C and then added to the HEp-2 cell monolayer. Addition of the LdaG antiserum at a 1:50 dilution completely inhibited diffuse adherence of E. coli DH5α(pV-B-6), containing the cloned lda region, to the HEp-2 cells, whereas the preimmune serum from the same rabbit did not inhibit adhesion (Fig. 8A and B). Treatment of wild-type E. coli 22 with the antiserum did not inhibit LA adhesion even at a 1:5 dilution, providing further evidence that there are two factors that mediate Hep-2 adherence by this strain (Fig. 8C).
FIG. 8.
Antibody inhibition of HEp-2 adherence. (A) E. coli DH5α(pV-B-6) incubated with preimmune serum (1:50 dilution). (B) E. coli DH5α(pV-B-6) incubated with α-LdaG antiserum (1:50 dilution). (C) E. coli 22 incubated with α-LdaG antiserum (1:5 dilution).
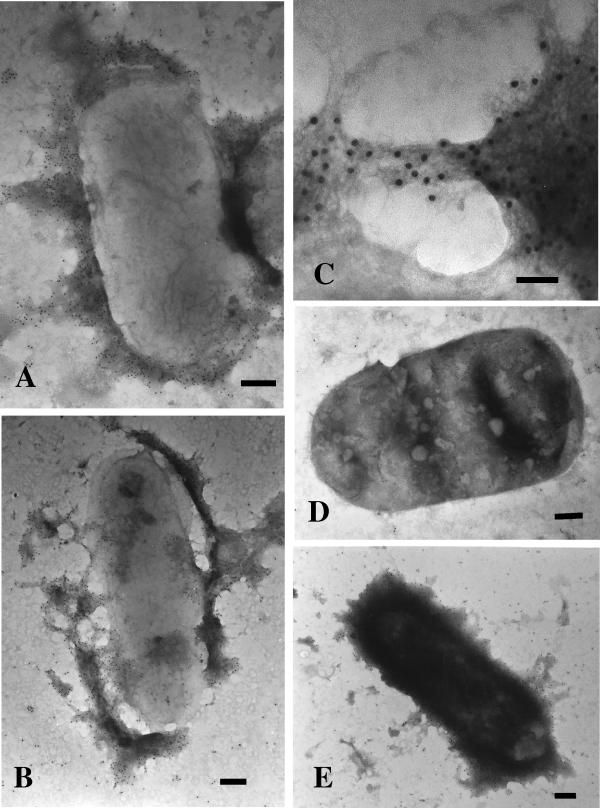
Detection of LDA by electron microscopy.
E. coli 22 and the LDA isogenic mutant, LDA1, were examined by electron microscopy using immunogold labeling and negative-staining techniques. As seen in Fig. 9A and B, immunogold labeling with LdaG antiserum identified an amorphous surface matrix with no obvious fimbrial structure. At higher magnification (×120,000), very fine wiry fibrils forming mesh-like structures can be visualized, but the details are beyond the resolution of the electron microscope (Fig. 9C). This surface matrix is specific for the presence of LdaG, as 10-nm gold particles can be seen decorating the entire surface of E. coli 22 while no gold particles decorate the ldaG mutant LDA1 (Fig. 9D). The gold label also recognizes LdaG expressed in the E. coli XL-10(pIJ20) subclone (Fig. 9E), which exhibits the same decorative pattern as the wild-type strain (Fig. 9A and B). Controls for nonspecific gold particle binding were performed in which the LdaG primary antibody was omitted prior to incubation with the goat anti-rabbit-labeled 10-nm gold particles. The results were identical to those seen with the null mutant, LDA1 (data not shown). The appearance of LDA is very similar to that seen with other afimbrial adhesins, such as Ral from rabbit EPEC (1), CS6 from ETEC (56), and Afa, originally identified in a uropathogenic E. coli serotype O2 strain (26) and later in EPEC strain 132/12 (O55:H−) (24). Thus, despite the amino acid homologies to the K88 and CS3A1 fimbriae, LDA appears to be afimbrial in structure.
FIG. 9.
Immunogold labeling and negative staining of LDA. (A and B) E. coli 22. The bars represent 200 nm. (C) E. coli 22. The bar represents 50 nm. (D) E. coli LDA1. The bar represents 200 nm. (E) E. coli XL10(pIJ20). The bar represents 200 nm.
DISCUSSION
It has long been known that EHEC serogroup O26 is an important pathogen in both human and animal infections (27, 35). The EPEC O26 serogroup has been implicated in human diarrheal infections in several countries worldwide (2, 42, 43), but its incidence in animal infections has only recently been reported (29, 55). Although EPEC differs from EHEC in its inability to produce Stx, both share the same mechanism for intimate attachment and effacement (38). Multiple factors, including Bfp, flagella, and the LEE-encoded proteins, have been implicated in adherence in typical EPEC strains (reviewed in reference 40), but to date, no distinct adhesins have been characterized in atypical EPEC. The work of Szalo et al. (51) identified several bfpA lpfA-negative O26 bovine isolates that have a gene sequence whose predicted protein sequence shows 89% identity to that of ClpE, the chaperone protein of CS31A pili. These same isolates were negative for the gene encoding the major structural subunit (clpG), suggesting the presence of a novel adhesin, but it has yet to be elucidated. Here, we report the identification and characterization of a novel gene cluster, locus for diffuse adherence (lda), present in an LA+ stx-negative eae+ EAF− bfpA-negative atypical O26:H11 E. coli strain, that is responsible for mediating diffuse adherence to HEp-2 cells.
Atypical EPEC strain 22 was originally investigated because of its ability to form microcolonies on HEp-2 cells and its lack of hybridization to known fimbrial-gene probes. Our analysis revealed that there are two factors contributing to the adherence phenotype of this strain. The first adhesin is encoded by a chromosomal locus, lda, which gives a diffuse pattern of adherence on HEp-2 cells when cloned into a nonadhesive K-12 strain. A chromosomal mutation in the major structural gene, ldaG, abated but did not abolish HEp-2 adherence by wild-type E. coli 22. The second adhesin is a plasmid-encoded factor, which by itself in E. coli XL-10(pIJ22) forms isolated microcolonies on HEp-2 cells, albeit fewer and smaller than those of wild-type E. coli 22 (compare Fig. 5B to 1A). Elimination of this plasmid, along with the ldaG chromosomal mutation in E. coli 22 (LDA3), results in complete lack of adherence to HEp-2 cells. Complementation of the double mutant LDA3 (ΔldaG pIJ22−) with pIJ20 (lda+) restored the diffuse-adherence phenotype but not the localized adherence originally observed in the wild-type strain (compare Fig. 5D to 1A). Thus, it is not known if the diffuse-adherence property of LDA is masked by the expression of a second adhesin encoded on pIJ22 in wild-type E. coli 22 or if these two factors are acting concomitantly to give the full localized-adherence pattern. For example, pIJ22 could be involved in expressing another factor(s) that regulates the LA phenotype. Indeed, one previously described regulator that influences the LA phenotype in EPEC is TrcA; null mutations in trcA produced smaller than normal adherent microcolonies compared to wild-type EPEC (52). The nature of this second LA adherence phenotype associated with pIJ22 is unknown and is being investigated.
The organization and predicted protein sequence similarities of the lda locus closely resemble those of the K88 fae operon (6), although it also shows considerable identity to the ETEC CS31A clp fimbrial operon (14) (Table 4) and to a lesser extent the rabbit EPEC ral (1) fimbrial operon (data not shown). It is interesting that the amino acid sequence of the putative major structural gene, LdaG, is related more closely to the chromosomally encoded F41 adhesin structural subunit (56%) than it is to any of the variants of the plasmid-encoded K88 FaeG structural subunits (23%). The specificity of fimbrial adhesins for their host cells is well documented (32, 50). ETEC strains harboring the F41 fimbrial antigen are pathogenic for cattle, swine, and lambs, whereas ETEC K88 strains are pathogenic for swine (34). The importance of LDA-mediated adhesion in the pathogenicity of these strains in human disease is not clear, but the isolation of this O26 isolate from the stool of a patient suffering from diarrhea from whom no other pathogen could be isolated would be consistent with a role in disease. In addition, although our sample size was small (only five strains, each of a different serotype), it is also of interest to note that the two ldaG+ strains were stx negative and the three ldaG strains were stx+, further suggesting that Lda might be specific in the pathogenesis of atypical EPEC.
Three of the minor subunits, LdaC, LdaH, and LdaI, show considerable identity (85%, 73%, and 59%) to the FaeC, FaeH, and FaeI proteins of K88 fimbriae, respectively. The closely related FaeH and ClpH proteins show 88% identity to each other. Previous investigations have reported extensive DNA homologies between K88-related ancillary proteins (4, 5, 30), and in fact, some can be used interchangeably for fimbrial expression (25). Therefore, the nucleotide homologies seen within the minor subunit, ldaH, among strains of different serotypes were not so surprising (Table 3). LDA appears to belong to a family of fimbrial adhesins that may share a conserved mechanism of pilin production while possessing a unique structural subunit specific for host-fimbria recognition by O26 strains. Indeed, LdaE has striking identity (75% and 82%) to FaeE and ClpE periplasmic chaperones, respectively, suggesting that it too is a member of the chaperone-assisted pilus assembly mechanism, similar to the prototypic Pap operon (8, 50). It is not known how much homology is shared between E. coli O26:H11 and the ldaH+ ldaG-negative isolates outside the ldaH gene, but the possibility that they may be closely related is very intriguing, and we are exploring this hypothesis.
This is the first reported sequence of a novel adhesin found in an atypical EPEC strain. Like many known fimbrial loci, LDA is predicted to contain regulatory genes (ldaA and ldaB), a periplasmic chaperone (ldaE), an usher (ldaD), a major structural gene (ldaG), and a few ancillary genes (ldaC, ldaF, ldaH, and ldaI) thought to aid in the formation of the fimbriae. Unlike its plasmid-borne K88, CS31A, and Ral fimbrial counterparts, Lda is located within a genomic island that is absent from the K-12 strain MG1655 and has not been reported in any of the E. coli genomes so far described. The insertion of this region into the proP gene, along with the transposon sequences flanking the LDA locus, suggests that this region has been horizontally acquired as a pathogenicity island (18). Additionally, lda apparently does not encode a well-defined fimbrial structure but rather one with a flexible morphological appearance similar to nonfimbrial antigens. Like the Ral (1), CS6 (56), and Afa (24) fibrillae, LDA is wiry, unstructured, and rather difficult to visualize under the electron microscope. A comparative exchange of the structural proteins of these distinct fimbriae would provide a useful understanding of the pili and adhesins associated with these E. coli strains. Additional work is needed to characterize the regulation and role in virulence of the LDA adhesin.
Acknowledgments
We thank Lisa Sadzewicz for her skill in nucleotide sequencing, Rogeria Keller for her help and guidance with the immunogold labeling, Rebecca Wade and Robert Nauman for electron microscope facilities, and MaryJane Lombardo for critically reviewing the manuscript.
This work was supported by NIH grants AI21657 and DK58957 to J.B.K. A.G.T. was supported by institutional funds from the UTMB John Sealy Memorial Endowment Fund for Biomedical Research and the Gastrointestinal Research Interdisciplinary Program. I.C.A.S. was supported with the help of a 2003 ASM International Fellowship Award.
Editor: A. D. O'Brien
REFERENCES
- 1.Adams, L. M., C. P. Simmons, L. Rezmann, R. A. Strugnell, and R. M. Robins-Browne. 1997. Identification and characterization of a K88- and CS31A-like operon of a rabbit enteropathogenic Escherichia coli strain which encodes fimbriae involved in the colonization of rabbit intestine. Infect. Immun. 65:5222-5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afset, J. E., K. Bergh, and L. Bevanger. 2003. High prevalence of atypical enteropathogenic Escherichia coli (EPEC) in Norwegian children with diarrhoea. J. Med. Microbiol. 52:1015-1019. [DOI] [PubMed] [Google Scholar]
- 3.Alexeyev, M. F., I. N. Shokolenko, and T. P. Croughan. 1995. New mini-Tn5 derivatives for insertion mutagenesis and genetic engineering in Gram-negative bacteria. Can. J. Microbiol. 41:1053-1055. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, D. G., and S. L. Moseley. 1988. Escherichia coli F41 adhesin: genetic organization, nucleotide sequence, and homology with the K88 determinant. J. Bacteriol. 170:4890-4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakker, D., C. E. Vader, B. Roosendaal, F. R. Mooi, B. Oudega, and F. K. de Graaf. 1991. Structure and function of periplasmic chaperone-like proteins involved in the biosynthesis of K88 and K99 fimbriae in enterotoxigenic Escherichia coli. Mol. Microbiol. 5:875-886. [DOI] [PubMed] [Google Scholar]
- 6.Bakker, D., P. T. Willemsen, R. H. Willems, T. T. Huisman, F. R. Mooi, B. Oudega, F. Stegehuis, and F. K. de Graaf. 1992. Identification of minor fimbrial subunits involved in biosynthesis of K88 fimbriae. J. Bacteriol. 174:6350-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barros, H. C., M. L. Silva, M. Z. Laporta, R. M. Silva, and L. R. Trabulsi. 1992. Inhibition of enteropathogenic Escherichia coli adherence to HeLa cells by immune rabbit sera. Braz. J. Med. Biol. Res. 25:809-812. [PubMed] [Google Scholar]
- 8.Bertin, Y., J. P. Girardeau, M. Der Vartanian, and C. Martin. 1993. The ClpE protein involved in biogenesis of the CS31A capsule-like antigen is a member of a periplasmic chaperone family in gram-negative bacteria. FEMS Microbiol. Lett. 108:59-67. [DOI] [PubMed] [Google Scholar]
- 9.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 10.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cravioto, A., R. J. Gross, S. M. Scotland, and B. Rowe. 1979. An adhesive factor found in strains of Escherichia coli belonging to the traditional infantile enteropathogenic serotypes. Curr. Microbiol. 3:95-99. [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer, J., C. Maddox, R. Moxley, D. Kinden, and M. Miller. 1994. Pathogenicity of a bovine attaching effacing Escherichia coli isolate lacking Shiga-like toxins. Am. J. Vet. Res. 55:991-999. [PubMed] [Google Scholar]
- 14.Girardeau, J. P., M. Der Vartanian, J. L. Ollier, and M. Contrepois. 1988. CS31A, a new K88-related fimbrial antigen on bovine enterotoxigenic and septicemic Escherichia coli strains. Infect. Immun. 56:2180-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Girón, J. A., M. S. Donnenberg, W. C. Martin, K. G. Jarvis, and J. B. Kaper. 1993. Distribution of the bundle-forming pilus structural gene (bfpA) among enteropathogenic Escherichia coli. J. Infect. Dis. 168:1037-1041. [DOI] [PubMed] [Google Scholar]
- 16.Girón, J. A., A. S. Ho, and G. K. Schoolnik. 1991. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science 254:710-713. [DOI] [PubMed] [Google Scholar]
- 17.Guth, B. E., R. Lopes de Souza, T. M. Vaz, and K. Irino. 2002. First Shiga toxin-producing Escherichia coli isolate from a patient with hemolytic uremic syndrome, Brazil. Emerg. Infect. Dis. 8:535-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hacker, J., and J. B. Kaper. 1999. The concept of pathogenicity islands, p. 1-11. In J. B. Kaper and J. Hacker (ed.), Pathogenicity islands and other mobile virulence elements. American Society for Microbiology, Washington, D.C.
- 19.Hohn, B., and J. Collins. 1980. A small cosmid for efficient cloning of large DNA fragments. Gene 11:291-298. [DOI] [PubMed] [Google Scholar]
- 20.Holmgren, A., M. J. Kuehn, C. I. Branden, and S. J. Hultgren. 1992. Conserved immunoglobulin-like features in a family of periplasmic pilus chaperones in bacteria. EMBO J. 11:1617-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jerse, A. E., J. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. USA 87:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaper, J. B. 1996. Defining EPEC. Rev. Microbiol. São Paulo 27:130-133. [Google Scholar]
- 23.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123-140. [DOI] [PubMed] [Google Scholar]
- 24.Keller, R., J. G. Ordonez, R. R. de Oliveira, L. R. Trabulsi, T. J. Baldwin, and S. Knutton. 2002. Afa, a diffuse adherence fibrillar adhesin associated with enteropathogenic Escherichia coli. Infect. Immun. 70:2681-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korth, M. J., J. M. Apostol, Jr., and S. L. Moseley. 1992. Functional expression of heterologous fimbrial subunits mediated by the F41, K88, and CS31A determinants of Escherichia coli. Infect. Immun. 60:2500-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Labigne-Roussel, A. F., D. Lark, G. Schoolnik, and S. Falkow. 1984. Cloning and expression of an afimbrial adhesin (AFA-I) responsible for P blood group-independent, mannose-resistant hemagglutination from a pyelonephritic Escherichia coli strain. Infect. Immun. 46:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levine, M. M. 1987. Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic, and enteroadherent. J. Infect. Dis. 155:377-389. [DOI] [PubMed] [Google Scholar]
- 28.Levine, M. M., J. G. Xu, J. B. Kaper, H. Lior, V. Prado, B. Tall, J. Nataro, H. Karch, and K. Wachsmuth. 1987. A DNA probe to identify enterohemorrhagic Escherichia coli of O157:H7 and other serotypes that cause hemorrhagic colitis and hemolytic uremic syndrome. J. Infect. Dis. 156:175-182. [DOI] [PubMed] [Google Scholar]
- 29.Mansfield, K. G., K. C. Lin, J. Newman, D. Schauer, J. MacKey, A. A. Lackner, and A. Carville. 2001. Identification of enteropathogenic Escherichia coli in simian immunodeficiency virus-infected infant and adult rhesus macaques. J. Clin. Microbiol. 39:971-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin, C., C. Boeuf, and F. Bousquet. 1991. Escherichia coli CS31A fimbriae: molecular cloning, expression and homology with the K88 determinant. Microb. Pathog. 10:429-442. [DOI] [PubMed] [Google Scholar]
- 31.McDaniel, T. K., and J. B. Kaper. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23:399-407. [DOI] [PubMed] [Google Scholar]
- 32.Mol, O., and B. Oudega. 1996. Molecular and structural aspects of fimbriae biosynthesis and assembly in Escherichia coli. FEMS Microbiol. Rev. 19:25-52. [DOI] [PubMed] [Google Scholar]
- 33.Mooi, F. R., N. Harms, D. Bakker, and F. K. de Graaf. 1981. Organization and expression of genes involved in the production of the K88ab antigen. Infect. Immun. 32:1155-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moon, H. W. 1990. Colonization factor antigens of enterotoxigenic Escherichia coli in animals. Curr. Top. Microbiol. Immunol. 151:147-165. [DOI] [PubMed] [Google Scholar]
- 35.Morris, J. A., and W. J. Sojka. 1985. Escherichia coli as a pathogen in animals, p. 44-77. In M. Sussman (ed.), The virulence of Escherichia coli: reviews and methods. Academic Press, London, United Kingdom.
- 36.Moseley, S. L., G. Dougan, R. A. Schneider, and H. W. Moon. 1986. Cloning of chromosomal DNA encoding the F41 adhesin of enterotoxigenic Escherichia coli and genetic homology between adhesins F41 and K88. J. Bacteriol. 167:799-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nataro, J. P. 2001. Diarrhoeagenic Escherichia coli, p. 1463-1504. In M. Sussman (ed.), Molecular medical microbiology, vol. 2. Academic Press, London, United Kingdom. [Google Scholar]
- 38.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newland, J. W., and R. J. Neill. 1988. DNA probes for Shiga-like toxins I and II and for toxin-converting bacteriophages. J. Clin. Microbiol. 26:1292-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nougayrede, J. P., P. J. Fernandes, and M. S. Donnenberg. 2003. Adhesion of enteropathogenic Escherichia coli to host cells. Cell. Microbiol. 5:359-372. [DOI] [PubMed] [Google Scholar]
- 41.Orndorff, P. E., P. A. Spears, D. Schauer, and S. Falkow. 1985. Two modes of control of pilA, the gene encoding type 1 pilin in Escherichia coli. J. Bacteriol. 164:321-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peixoto, J. C. C., S. Y. Bando, J. A. G. Ordoñez, B. A. Botelho, L. R. Trabulsi, and C. A. Moreira-Filho. 2001. Genetic differences between Escherichia coli O26 strains isolated in Brazil and in other countries. FEMS Microbiol. Lett. 196:239-244. [DOI] [PubMed] [Google Scholar]
- 43.Pelayo, J. S., I. C. Scaletsky, M. Z. Pedroso, V. Sperandio, J. A. Giron, G. Frankel, and L. R. Trabulsi. 1999. Virulence properties of atypical EPEC strains. J. Med. Microbiol. 48:41-49. [DOI] [PubMed] [Google Scholar]
- 44.Runyen-Janecky, L. J., M. Hong, and S. M. Payne. 1999. The virulence plasmid-encoded impCAB operon enhances survival and induced mutagenesis in Shigella flexneri after exposure to UV radiation. Infect. Immun. 67:1415-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 46.Scaletsky, I. C., M. L. Silva, and L. R. Trabulsi. 1984. Distinctive patterns of adherence of enteropathogenic Escherichia coli to HeLa cells. Infect. Immun. 45:534-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scotland, S. M., G. A. Willshaw, H. R. Smith, and B. Rowe. 1990. Properties of strains of Escherichia coli O26:H11 in relation to their enteropathogenic or enterohemorrhagic classification. J. Infect. Dis. 162:1069-1074. [DOI] [PubMed] [Google Scholar]
- 48.Silva, M. L. M., I. C. A. Scaletsky, and L. H. Viotto. 1983. Non-production of cytotoxin among enteropathogenic strains of Escherichia coli isolated in São Paulo, Brazil. Rev. Microbiol. 14:161-162. [Google Scholar]
- 49.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 50.Soto, G. E., and S. J. Hultgren. 1999. Bacterial adhesins: common themes and variations in architecture and assembly. J. Bacteriol. 181:1059-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szalo, I. M., F. Goffaux, V. Pirson, D. Pierard, H. Ball, and J. Mainil. 2002. Presence in bovine enteropathogenic (EPEC) and enterohaemorrhagic (EHEC) Escherichia coli of genes encoding for putative adhesins of human EHEC strains. Res. Microbiol. 153:653-658. [DOI] [PubMed] [Google Scholar]
- 52.Tobe, T., I. Tatsuno, E. Katayama, C. Y. Wu, G. K. Schoolnik, and C. Sasakawa. 1999. A novel chromosomal locus of enteropathogenic Escherichia coli (EPEC), which encodes a bfpT-regulated chaperone-like protein, TrcA, involved in microcolony formation by EPEC. Mol. Microbiol. 33:741-752. [DOI] [PubMed] [Google Scholar]
- 53.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trabulsi, L. R., R. Keller, and T. A. Tardelli Gomes. 2002. Typical and atypical enteropathogenic Escherichia coli. Emerg. Infect. Dis. 8:508-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wani, S. A., M. A. Bhat, I. Samanta, Y. Nishikawa, and A. S. Buchh. 2003. Isolation and characterization of Shiga toxin-producing Escherichia coli (STEC) and enteropathogenic Escherichia coli (EPEC) from calves and lambs with diarrhoea in India. Lett. Appl. Microbiol. 37:121-126. [DOI] [PubMed] [Google Scholar]
- 56.Wolf, M. K., G. P. Andrews, B. D. Tall, M. M. McConnell, M. M. Levine, and E. C. Boedeker. 1989. Characterization of CS4 and CS6 antigenic components of PCF8775, a putative colonization factor complex from enterotoxigenic Escherichia coli E8775. Infect. Immun. 57:164-173. [DOI] [PMC free article] [PubMed] [Google Scholar]