Abstract
The C1 inhibitor (C1INH), a plasma complement regulatory protein, prevents endotoxin shock, at least partially via the direct interaction of its amino-terminal heavily glycosylated nonserpin region with gram-negative bacterial lipopolysaccharide (LPS). To further characterize the potential LPS-binding site(s) within the amino-terminal domain, mutations were introduced into C1INH at the three N-linked glycosylation sites and at the four positively charged amino acid residues. A mutant in which Asn3 was replaced with Ala was markedly less effective in its binding to LPS, while substitution of Asn47 or Asn59 had little effect on binding. The mutation of C1INH at all four positively charged amino acid residues (Arg18, Lys22, Lys30, and Lys55) resulted in near-complete failure to interact with LPS. The C1INH mutants that did not bind to LPS also did not suppress LPS binding or LPS-induced up-regulation of tumor necrosis factor alpha mRNA expression in RAW 264.7 macrophages. In addition, the binding of C1INH mutants to diphosphoryl lipid A was decreased in comparison with that of recombinant wild-type C1INH. Therefore, the interaction of C1INH with gram-negative bacterial LPS is dependent both on the N-linked carbohydrate at Asn3 and on the positively charged residues within the amino-terminal domain.
Lipopolysaccharide (LPS) is a component of the outer membrane of gram-negative bacteria. It activates mononuclear phagocytes to produce and release inflammatory mediators, among which tumor necrosis factor alpha (TNF-α) appears to be most important for the development of endotoxin shock (17). LPS binds to LPS-binding protein (LBP) and the complex binds to CD14 (14, 24, 38, 46). The formation of LPS-CD14 complexes initiates intracellular signaling by binding to Toll-like receptors (TLRs) expressed on mononuclear phagocytes and other cells (1). LPS interacts with a number of glycoproteins, including the three components of the cell membrane LPS receptor, CD14, MD-2, and TLR4 (9, 23, 50, 51, 58, 59). MD-2 has two N-glycosylation sites at Asn26 and Asn114, while TLR4 has nine N-linked sites within its amino-terminal extracellular domain (9). Binding of LPS to the LPS receptor requires N-linked glycosylation of both MD-2 and TLR4 (9) but not of CD14 (50). However, the binding sites for LPS (or lipid A) on many LPS-binding proteins, including bactericidal/permeability increasing protein (19), LBP (25), and MD-2 (31, 40, 60), contain positively charged amino acids, frequently in a similar structural motif (4, 15).
The C1 inhibitor (C1INH) is the only inhibitor of the classical complement pathway proteases C1r and C1s (48) and is the major inhibitor of factor XII and prekallikrein of the contact system (11, 44). The complement system has been implicated in both the pathogenesis of, and protection from, endotoxin shock (6). The contact system also appears to play a role in the mediation of septic shock (7). Levels of proteolytically inactivated C1INH are increased in fatal septic shock, which suggests an increased turnover and a relative secondary deficiency of biologically active C1INH (35). C1INH can be inactivated by limited proteolytic cleavage by elastase released from activated neutrophils (5, 6). The inactivation of C1INH may occur locally in inflamed tissue and thereby contribute to increased local complement activation (5). Both the complement system and the contact system are implicated in the pathophysiology of sepsis. C1INH is beneficial in animal models of endotoxemia and sepsis. Both active C1INH and reactive-center-cleaved, inactive C1INH protected mice from lethal gram-negative endotoxin shock. C1INH suppressed the binding of Salmonella enterica serovar Typhimurium LPS to the murine macrophage cell line RAW 264.7 and to human blood cells. It interacted directly with LPS and blocked LPS-induced TNF-α mRNA expression. Deletion of the amino-terminal 97 amino acid residues abrogated the ability of C1INH to bind to LPS (28). Further evidence demonstrated that N deglycosylation also significantly reduced C1INH-mediated protection of mice from lethal LPS-induced shock and failed to inhibit the binding of LPS to RAW 264.7 cells or to human blood cells (29). Therefore, N-linked glycosylation within the amino-terminal domain of C1INH is essential to mediate its interaction with gram-negative LPS. There are three N-linked glycosylation sites (Asn3, Asn47, and Asn59) within the amino-terminal domain of C1INH (41). The functional roles of glycosylation of proteins include modulation of enzyme and hormone activity, regulation of intracellular trafficking, control of protein folding, ligand recognition, and cell-cell interaction (26). However, the structural and functional implications of glycosylation of C1INH have not been elucidated.
Based on the lipid-binding motifs of other LPS-interacting proteins (15, 25, 31, 45, 60), we hypothesized that the four positively charged amino acids within the amino-terminal domain of C1INH (Arg18, Lys22, Lys30, and Lys55) might interact with the negatively charged phosphate groups of the diglucosamine backbone of lipid A. Using site-directed mutagenesis, we prepared single amino acid substitutions at each site, in addition to combinations of substitutions. Because previous data had shown a dependence on N-linked glycosylation, we also prepared substitution mutants of Asn3, Asn47, and Asn59, together with combinations of these three substitutions. The results show that replacement of Asn3 with Ala leads to nearly complete loss of the ability to interact with LPS, while little effect was observed with substitution at the other N-linked sites. On the other hand, the effect of replacement of the positively charged residues was additive. Nearly complete loss of the LPS interaction was observed only after replacement of all four amino acids.
MATERIALS AND METHODS
Reagents.
C1INH and C1s were from Advanced Research Technologies (San Diego, CA). A rabbit anti-human C1INH antibody was purchased from DAKO (Glostrup, Denmark). Goat anti-rabbit immunoglobulin G (heavy plus light chains) [IgG (H+L)] conjugated with horseradish peroxidase was purchased from Pierce Biotechnology (Rockford, IL). LPS and fluorescein isothiocyanate-conjugated LPS (FITC-LPS) from Salmonella serovar Typhimurium, Salmonella serovar Typhimurium LPS-derived diphosphoryl lipid A (dLPA), and ο-phenylenediamine dihydrochloride substrates were obtained from Sigma Chemical Co. (St. Louis, MO).
Cell culture.
The murine macrophage cell line RAW 264.7 and COS-7 cell lines from the American Type Culture Collection (Manassas, VA) were cultured in Dulbecco's minimum essential medium (DMEM; GIBCO, Invitrogen, CA) supplemented with 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA) at 37°C under 5% CO2. Confluent cells were detached by washing with phosphate-buffered saline (PBS) (pH 7.4).
Site-directed mutagenesis.
A full-length human C1INH cDNA construct in the pcDNA 3.1 vector (Invitrogen, San Diego, CA) was used. Site-directed mutagenesis was performed using a QuikChange site-directed mutagenesis kit (Stratagene Inc., La Jolla, CA) according to the manufacturer's instructions. Briefly, several pairs of complementary primers (sense and antisense oligonucleotides) were obtained from Invitrogen (Carlsbad, CA). In order to create the mutation of Asn to Ala at residues 3, 47, and 59, and to change Arg to Ala at position 18 or Lys to Ala at positions 22, 30, and 55, the nucleotide substitutions were placed in the middle of the primers (Table 1). Parental C1INH cDNA inserted into vector pcDNA 3.1 was amplified using 1 μl of Pfu DNA polymerase (2.5 U/μl) with these primers for 16 cycles (95°C for 30 s, 55°C for 1 min, 65°C for 5 min) in a GeneAmp PCR system 9600 (Perkin Elmer, Norwalk, CT). After digestion of the parental DNA with 1 μl of DpnI (10 U/μl) at 37°C for 1 h, the amplified DNA containing the nucleotide substitution was transformed into supercompetent cells. In the multisite mutagenesis strategy, single-site mutations were performed first, and then double-, triple-, and quadruple-site mutations were performed progressively. The mutated sites within the cDNA encoding the amino-terminal domain were confirmed by automated DNA sequencing using a Taq DyeDeoxy Terminator Cycle Sequencing kit and an ABI Prism 377 DNA sequencer (Applied Biosystems, Foster City, CA).
TABLE 1.
Primers used for mutagenesis
| Amino acid | Mutation | Sense primer sequencea |
|---|---|---|
| 3 | Asn→Ala | 5′-GATAGAGCCTCCTCAAATCCAGCTGCTACCAGCTCCAGCTCCCAG-3′ |
| 47 | Asn→Ala | 5′-GTTTCCAGCTTGCCGACAACCGCGTCAACAACCAATTCAGCCACC-3′ |
| 59 | Asn→Ala | 5′-TCAGCCACCAAAATAACAGCTGCGACCACTGATGAACCCACCACA-3′ |
| 18 | Arg→Ala | 5′-GATCCAGAGAGTTTGCAAGACGCTGGCGAAGGGAAGGTCGCAACA-3′ |
| 22 | Lys→Ala | 5′-TTGCAAGACAGAGGCGAAGGGGCTGTCGCAACAACAGTTATCTCC-3′ |
| 30 | Lys→Ala | 5′-GTCGCAACAACAGTTATCTCCGCTATGCTATTCGTTGAACCCATC-3′ |
| 55 | Lys→Ala | 5′-ACAACCAATTCAGCCACCGCTATAACAGCTAATACCACT-3′ |
Mutated nucleotides are boldfaced.
Cell transfection and recombinant proteins.
The purified expression plasmids containing the various mutant C1INH cDNAs or wild-type C1INH cDNA were transfected into COS-7 cells using the GenePORTER transfection reagent (GTS Inc., San Diego, CA) as described by the manufacturer. Briefly, C1INH cDNAs in pcDNA 3.1 were prepared using a QIAGEN maxiplasmid DNA preparation kit (QIAGEN Inc., Chatsworth, CA) and were introduced into COS-7 cells using the GenePORTER transfection method. The quality and quantity of plasmid DNA was electrophoretically assessed on 1.0% agarose gels after measurement of the optical density at 260 and 280 nm. For transfection, plasmid DNA (2.5 μg) was diluted in DMEM (1 ml) and mixed with GenePORTER transfection reagent, and the mixture was incubated at room temperature for 1 h. The DNA-GenePORTER transfection reagent complex was added to 80 to 90% confluent COS-7 cells (2 × 105 cells/per well) plated in six-well tissue culture plates. After a 7-h incubation, DMEM (1 ml) containing 20% FBS was added; the medium was replaced 72 h after transfection with serum-free DMEM. Media containing the recombinant protein mutants and a control medium from COS-7 cells that had been transfected with pcDNA3.1 alone were collected after an additional 48-h incubation and were concentrated ∼20-fold using a Vivaspin (molecular weight cutoff, 100,000) (Millipore) on the day of harvest. Samples then were analyzed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting.
SDS-PAGE and Western blotting.
Recombinant full-length C1INH or recombinant C1INH mutants were subjected to electrophoresis using a 10% SDS-Tris-glycine polyacrylamide gel (Invitrogen, San Diego, CA). In some experiments, C1INH mutants were incubated with C1s (molar ratio, 1:1) for 20 min at 37°C prior to SDS-PAGE. Proteins were electrophoretically transferred to nitrocellulose membranes (Invitrogen), blocked with 5% fat-free milk (Bio-Rad, Hercules, CA) in PBS (pH 7.4) containing 0.05% Tween at 4°C overnight, incubated with a rabbit anti-human C1INH antibody (1:1,000) in 5% fat-free milk for 2 h at room temperature, washed with 1× PBS for 20 min, and incubated with a 1:10,000 dilution of Immunopure goat anti-rabbit IgG (H+L) conjugated with horseradish peroxidase (Pierce Biotechnology) for 2 h at room temperature. Development was performed using a SuperSignal chemiluminescent substrate kit (Pierce Biotechnology).
LPS binding assay.
Plates (polyvinyl chloride, 96-well U-bottom plates) (Becton Dickinson, Franklin Lakes, NJ) were coated with dLPA (1.0 μg/ml) or LPS (175 ng/ml) at room temperature overnight (28, 29). Medium from COS-7 cells transfected with pcDNA3.1 alone or medium containing recombinant full-length C1INH (40 μg/ml) or recombinant C1INH mutants (40 μg/ml) was incubated with dLPA- or LPS-coated plates for 1 h at room temperature. Rabbit anti-human C1INH antibody (1:1,000) was added, and the plates were incubated for an additional 1 h at room temperature. After a wash, the plates were incubated with ImmunoPure goat anti-rabbit IgG (H+L) conjugated with horseradish peroxidase (1:100,000). Color reactions were developed for 3 min at room temperature and terminated with 3 N HCl. The absorbance within each well was measured at 490 nm using Revelation Microsoft in an MRX microplate reader (DYNEX Technologies, Chantilly, VA). Standard samples were detected using different concentrations of human plasma-derived C1INH binding to rabbit anti-human C1INH antibody. Data were analyzed using Tukey's multiple comparison test. All experimental data were presented as means ± standard deviations (SD).
FACS.
Murine RAW 264.7 macrophages were incubated with FITC-LPS (175 ng/ml) in the absence or presence of the various recombinant C1INH mutant preparations (40 μg/ml) in DMEM containing 10% FBS for 15 min at 37°C. The cells were fixed with a fluorescence-activated cell sorter (FACS) solution after three washes with PBS. The binding of FITC-LPS was analyzed on a FACSCalibur (Becton Dickinson, San Jose, CA) using CellQuest software.
Reverse transcription-PCR (RT-PCR).
Total RNA was isolated from RAW 264.7 macrophages induced with LPS (175 ng/ml) in the presence or absence of either plasma C1INH (40 μg/ml), medium containing recombinant C1INH (40 μg/ml) or C1INH mutants (40 μg/ml), or medium from COS cells transfected with vector alone and was reverse transcribed using Moloney murine leukemia virus reverse transcriptase with Oligo(dT)20 primers (Invitrogen, Carlsbad, CA) (1 h at 37°C). PCR primers were designed to generate mouse TNF-α and β-actin fragments, each of which is approximately 200 bp. Mouse TNF-α primers were 5′-ATGAGCACAGAAAGCATGATCC-3′ (sense) and 5′-GAGGCCATTTGGGAACTTCTC-3′ (antisense). Mouse β-actin primers were 5′-TGGATGACGATATCGCTGC-3′ (sense) and 5′-AGGGTCAGGATACCTCTCTT-3′ (antisense). PCR products were analyzed on 1.2% (wt/vol) agarose gels containing 0.5 μg/ml ethidium bromide and were visualized under UV light. Band density was analyzed and quantified using ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
RESULTS
Characterization of the mutated C1INH proteins.
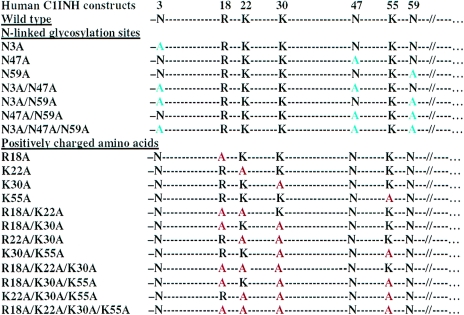
The binding of LPS or LPA to LPS-binding proteins is dependent on the interaction of the phosphate groups on LPA with specific positively charged residues within the protein (13, 15, 22, 25, 33, 47, 49, 55). We previously demonstrated that in the case of C1INH both N-linked glycosylation and its amino-terminal domain are required for its interaction with LPS (28, 29). Therefore, we predict that the LPS-binding site is located within the amino-terminal heavily glycosylated nonserpin domain of C1INH. On the basis of this prediction, the N-linked glycosylation sites (Asn3, Asn47, and Asn59) and the positively charged amino acid residues (Arg18, Lys22, Lys30, and Lys55) were replaced, both individually and in combination, with Ala residues (Fig. 1). The wild-type and mutated cDNA constructs listed were each transfected into COS-7 cells. Each recombinant mutant protein was analyzed by SDS-PAGE followed by Western blotting (data not shown). The C1INH mutants with substitutions at Arg18, Lys22, Lys30, or Lys55 were each of the same apparent size as wild-type C1INH. On the other hand, the absence of expression of one, two or three N-linked carbohydrates due to replacement of the relevant Asn residues resulted in a progressive decrease in the apparent molecular weight of the recombinant proteins. To examine whether the C1INH mutants retained protease inhibitory activity, the ability of the recombinant proteins to complex with C1s was tested. The C1INH mutants each retained the ability to complex with C1s, which indicates that the mutated molecules retained protease inhibitory activity (data not shown).
FIG. 1.
Site-directed mutagenesis of N-linked glycosylation sites and positively charged amino acids within the amino-terminal domain of C1INH. Each of the mutant recombinant proteins used in the studies described is shown.
Analysis of the LPS-binding site within the amino-terminal domain of C1INH.
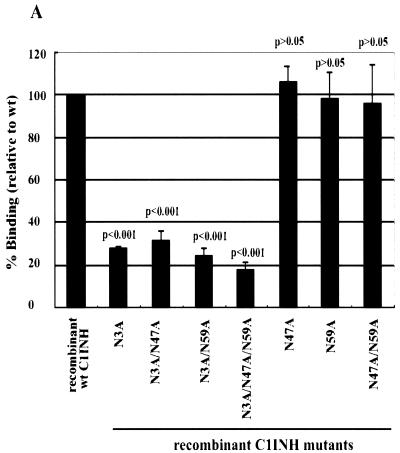
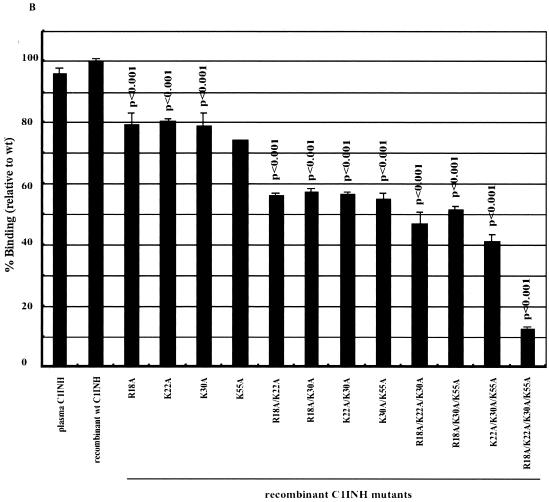
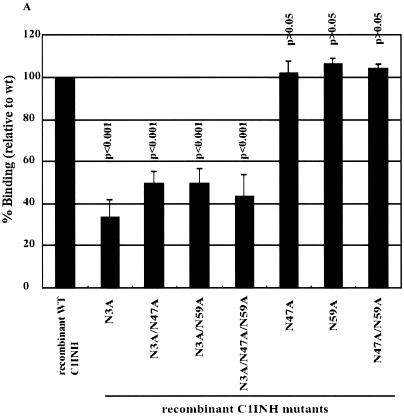
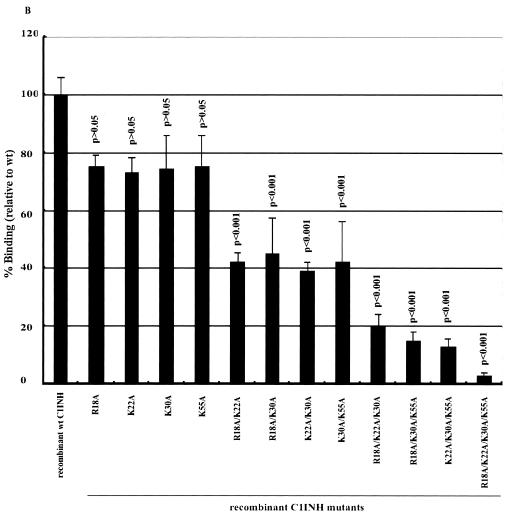
We immobilized LPS in microtiter wells and measured the binding of the recombinant C1INH mutants by enzyme-linked immunosorbent assay (ELISA). The mutants with substitutions at Asn47 and/or Asn59 were virtually unchanged in their ability to bind to LPS (∼100%) (Fig. 2A). However, all mutants with substitutions at Asn3 showed weak binding: 28.2% ± 1% that of the wild-type control for the Asn3→Ala mutant, 32.1% ± 4.4% for the Asn3→Ala Asn47→Ala mutant, 24.2% ± 4% for the Asn3→Ala Asn59→Ala mutant, and 17.2% ± 4.8% for the Asn3→Ala Asn47→Ala Asn59→Ala mutant (Fig. 2A). Negative controls consisting of medium from cells transfected with vector DNA that contained no C1INH cDNA showed no binding (data not shown). This demonstrates that N-linked glycosylation at Asn3 is critical for the binding of C1INH to LPS. To further elucidate the molecular requirements for C1INH binding, the interaction of the recombinant C1INH mutants with substitutions at Arg18, Lys22, Lys30, and/or Lys55 with LPS was analyzed. Substitution of each individual positively charged amino acid decreased binding by ∼20 to 25%; the double and triple substitutions diminished binding by 40 to 50% and 50 to 60%, respectively, while substitution of all four positively charged residues reduced binding by ∼90% (Fig. 2B). Taken together, these results demonstrate that the binding of C1INH to gram-negative bacterial LPS is dependent on both the N-linked carbohydrate at Asn3 and the positively charged amino acid residues Arg18, Lys22, Lys30, and Lys55, all of which are within the amino-terminal domain.
FIG. 2.
Binding of recombinant C1INH mutants to immobilized LPS. LPS was immobilized on the surfaces of polyvinyl chloride plates as described in Materials and Methods. Binding was detected using an anti-human C1INH antibody followed by ImmunoPure goat anti-rabbit IgG (H+L) conjugated with horseradish peroxidase. After the color reaction, the absorbance of each well was measured at 490 nm. Results (level of binding as a percentage of the binding of recombinant wild-type [wt] C1INH) were expressed as the means ± SD from at least three independent experiments. (A) Binding of the recombinant Asn substitution mutants. (B) Binding of the recombinant Lys/Arg substitution mutants. The P value for the binding by each mutant versus that of the recombinant wild-type control is shown.
Effects of C1INH substitution mutations on the binding of LPS to RAW 264.7 cells.
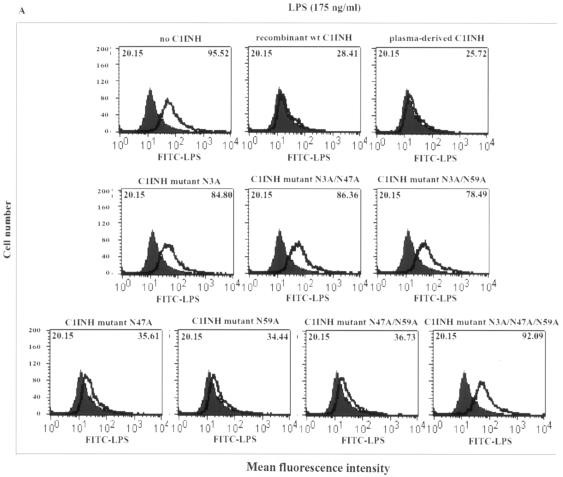
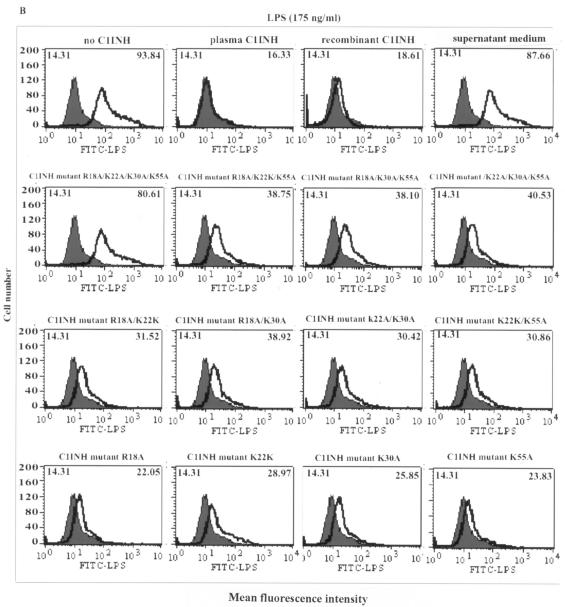
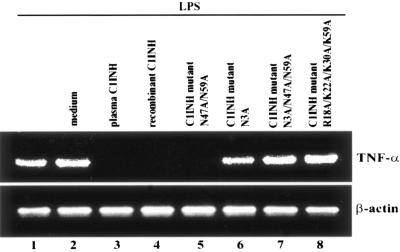
We tested whether the C1INH mutants suppressed the binding of FITC-LPS to RAW 264.7 macrophages. The recombinant mutant in which Asn3 was replaced with Ala lost the ability to block the binding of FITC-LPS to RAW 264.7 cells, while the mutants with substitutions at Asn47 and/or Asn59 retained the ability to suppress LPS binding (Fig. 3A). With replacement of the positively charged amino acids, each individual substitution progressively decreased the suppression of binding of FITC-LPS to macrophages (Fig. 3B). In addition, the induction of TNF-α mRNA by LPS in RAW 264.7 macrophages was tested by RT-PCR. Both wild-type and plasma-derived C1INH suppressed the LPS-induced expression of TNF-α mRNA, as did the mutant in which only Asn47 and Asn59 were replaced. However, mutants in which Asn3 was replaced lost this ability (Fig. 4). Substitution of all four positively charged residues eliminated the ability of C1INH to suppress TNF-α mRNA production by RAW 264.7 cells (Fig. 4). Medium from cells transfected with pcDNA3.1 that contained no C1INH cDNA had no effect on FITC-LPS binding, nor did it suppress LPS-induced TNF-α mRNA expression (Fig. 4). Therefore, the N-linked carbohydrate site at Asn3 and the positively charged residues (Arg18, Lys22, Lys30, and Lys55) within the amino-terminal domain are critical for inhibition of LPS binding to, and of LPS-induced production of TNF-α by, RAW 264.7 macrophages.
FIG. 3.
FACS analysis of C1INH-mediated inhibition of binding of LPS to the murine macrophage cell line RAW 264.7. Thick line, LPS binding; shaded field, control. Mean fluorescence intensities for control and treated cells are given in the upper left and upper right corners, respectively, of each panel. RAW 264.7 cells were incubated with FITC-LPS in the presence of plasma-derived C1INH, recombinant wild-type (wt) C1INH, and either the recombinant Asn substitution mutants (A) or the recombinant Lys/Arg substitution mutants (B).
FIG. 4.
Effects of C1INH mutants on LPS-induced TNF-α mRNA expression. Total RNA from RAW 264.7 macrophages was isolated after treatment with LPS (175 ng/ml) in the presence of recombinant mutant or wild-type C1INH for 30 min at 37°C. The control for this experiment consisted of LPS (175 ng/ml) treatment in the presence of medium from COS cells that had been transfected with vector without C1INH cDNA (LPS plus medium). RT-PCR was performed using mouse TNF-α cDNA and β-actin cDNA primers.
The LPS-binding site within the amino-terminal domain of C1INH is recognized by the lipid A moiety of LPS.
Gram-negative bacterial LPS is composed of two structural regions: the hydrophilic repeating polysaccharide of the core and O-antigen structures and the hydrophobic LPA domain (56). Virtually all the biological responses induced by LPS are dependent on LPA (18, 43, 53). As with most other proteins that bind to LPS, LPA likely represents the primary structure in the LPS molecule that is recognized by C1INH (29). To further characterize the molecular requirements for the interaction of C1INH with endotoxin, the recombinant C1INH mutants were analyzed for binding to immobilized dLPA by using ELISA. Replacement of Asn47 and/or Asn59 with Ala had no effect on the binding of C1INH to dLPA (Fig. 5A). As observed with binding to LPS, substitution of Asn3, either alone or in combination with Asn47 and/or Asn59, led to a marked reduction in binding (Fig. 5A). However, as observed previously with deglycosylated C1INH, the reduction in binding to LPA was not as great as the reduction in binding to LPS (29). Similarly, mutation of the basic residues led to a progressive decrease in the binding to dLPA. Single substitutions reduced binding 25 to 35%, double substitutions by 50 to 60%, triple substitutions by 80 to 85%, and substitution of all four basic residues reduced binding by ∼95% (Fig. 5B). These reductions are even greater than those observed for the binding of the mutants to LPS (Fig. 2B).
FIG. 5.
Binding of C1INH mutants to immobilized dLPA. dLPA was immobilized on the surfaces of polyvinyl chloride plates as described in Materials and Methods. Binding was detected and results expressed as in Fig. 2. (A) Binding of the recombinant Asn substitution mutants; (B) binding of the recombinant Lys/Arg substitution mutants. Results (level of binding as a percentage of the binding of recombinant wild-type [wt] C1INH) were expressed as the means ± SD from at least three independent experiments. The P value for the binding by each mutant versus that of the recombinant wild-type control is shown.
DISCUSSION
The studies described here demonstrate that the binding of LPS by C1INH is dependent on the glycosylation of C1INH at Asn3 and on the presence of the positively charged residues Arg18, Lys22, Lys30, and Lys55. Each of these basic residues contributes approximately equally to binding. The binding of LPS to a variety of LPS-binding proteins takes place via the interaction of the phosphate groups on the LPA component of LPS with specific basic residues within the protein (2, 3, 15, 21, 25, 27, 33, 34, 36, 42, 47, 54, 57). Antimicrobial peptides such as bactericidal/permeability increasing protein, cathelicidins, and the Limulus anti-lipopolysaccharide factor all appear to function in this manner, as has been suggested by structural studies and by studies using synthetic peptides corresponding to the positively charged endotoxin-binding regions (3, 21, 27, 33, 34). The LPS binding domains of these and other proteins share sequence homology with LBP (3, 10, 12, 19, 25). Based on both synthetic peptide and site-directed mutagenesis experiments, Arg 94 and Lys 95 of LBP are required for LPS binding activity (25, 42, 47, 54). A similar positively charged region of MD-2, a component of the macrophage LPS receptor, is responsible for binding to LPS (20, 31, 40, 60). Analysis of the structure of protein-LPS complexes indicates that at least two groups of positively charged residues on the protein contact the LPA phosphate groups (15, 16, 39).
Glycosylation has been shown to influence the activity of some endotoxin binding proteins, particularly MD-2 and TLR4 (9, 37). Although CD14 is glycosylated, carbohydrate does not play a role in its function (52). Glycosylation of TLR4 at Asn526 and Asn575 is required for proper expression of the protein at the cell surface and therefore for appropriate function of the LPS receptor (9). MD-2 is glycosylated at Asn26 and Asn114. Glycosylation at these sites does not affect cell surface expression and does not appear to have a major effect on folding (9, 37). The role of these N-linked carbohydrates in the binding of LPS by MD-2 remains incompletely defined. One study suggested that glycosylation might play a direct role in LPS recognition and binding or that it might function to stabilize the LPS receptor complex (9), while another study suggested that carbohydrate is not directly involved in LPS binding to MD-2 (37).
Previous data indicated that the LPS binding site of C1INH was within its amino-terminal 97 amino acid residues (28) and that binding was dependent on the presence of N-linked carbohydrate (29). Recombinant C1INH that consisted of amino acids 98 through 478 (the carboxyl terminus) retained full protease inhibitory activity (8). However, it lost the ability to bind to LPS, as demonstrated both by ELISA and by gel shift assays using nondenaturing PAGE (28). Furthermore, C1INH in which N-linked carbohydrate had been removed by use of N-glycosidase F lost the ability to bind to LPS and to protect mice from lethal endotoxin shock (29). These findings, together with studies characterizing the LPS binding sites in other proteins, led us to investigate the amino-terminal domain more extensively. Analysis of the role of the positively charged residues might be approached through the use of synthetic peptides. However, studies with other LPS-binding proteins have shown that the findings of experiments with substituted peptides frequently do not correspond to the results of experiments using mutated intact proteins (25, 42, 47). Furthermore, the only practical approach to analysis of the three individual N-linked sites was through the use of recombinant mutated proteins.
The C1INH amino-terminal domain contains no sequences that are highly homologous with the binding regions of other proteins that interact with LPS. It, in fact, contains only four positively charged amino acids: Arg 18, Lys 22, Lys 30, and Lys 55. Substitution of each of these amino acids individually led to a decrease in binding to LPS of approximately 20% in comparison with wild-type C1INH. Sequential replacements resulted in progressive decreases in binding to a maximum of a nearly 90% reduction with substitution of all four (Fig. 2B). Therefore, these positively charged residues are required for, and may be directly involved in, the binding of LPS to C1INH. These findings are strengthened by the observation that the experiments that analyzed the inhibition of binding of LPS to RAW 264.7 cells (Fig. 3B) and the inhibition of TNF-α production in LPS-stimulated cells (Fig. 4) paralleled the results of the binding experiments. The observation that LPS binding was nearly completely dependent on the single N-linked site at Asn3, however, was a surprise (Fig. 2A). All of the mutants in which this residue was replaced, either alone or in combination with the other mutations, showed a 70 to 80% reduction in binding, while replacement of either or both of the other two Asn residues had no effect on binding.
Taken together, the data are most compatible with the following hypothesis: the binding site for LPS on C1INH is discontinuous and contains Arg18, Lys22, Lys30, and Lys55. This hypothesis also is consistent with the previous observations that the binding of LPS to most, if not all, other LPS-binding proteins is dependent on positively charged sites (2, 3, 15, 21, 25, 27, 33, 34, 36, 42, 47, 54, 57). Removal of the carbohydrate at Asn3 very likely results in a conformational rearrangement that obscures the positively charged binding site. The possibility of structural rearrangement of the Asn3→Ala mutant, in theory, could be tested by analysis of the three-dimensional structure of the recombinant mutant in comparison with the wild type. Unfortunately, the small amounts of recombinant protein available do not lend themselves to direct structural studies. Another approach would be to use a panel of monoclonal antibodies to test for loss of epitopes in the mutant as a probe for three-dimensional alterations. Unfortunately, no appropriate antibodies are available. However, it is well established that glycosylation frequently is important for maintenance of protein conformation (26, 30, 32). In addition, the LPA binding data support our interpretation. Although the substitution of Asn3 led to reduced binding to LPA, the reduction was not as great as the reduction in binding to LPS (Fig. 2A and 5A). This is similar to previous data using deglycosylated plasma-derived C1INH and suggests that the smaller LPA molecule is able to gain access to the binding site while the larger LPS cannot (29). Replacement of the positively charged residues with the uncharged Ala led to an even greater reduction in binding with LPA than with LPS (Fig. 2B and 5B). This may suggest that only the positively charged residues are required for LPA binding but that other residues may be involved in the binding, or the stabilization of binding, to LPS.
In conclusion, the ability of C1INH to prevent endotoxin shock via a direct interaction with LPS is mediated via binding to the amino-terminal heavily glycosylated domain of C1INH. Although this domain contains at least seven O-linked carbohydrates, these play no role in binding (29). On the other hand, a single N-linked site at Asn3 is absolutely required for binding. The four positively charged residues are also required, and each appears to contribute equally to binding.
Acknowledgments
This work was supported by Public Health Service grants HD22082 and HD33727 from the National Institute of Child Health and Human Development
Editor: A. D. O'Brien
REFERENCES
- 1.Aderem, A., and R. J. Ulevitch. 2000. Toll-like receptors in the induction of the innate immune response. Nature 406:782-787. [DOI] [PubMed] [Google Scholar]
- 2.Appelmelk, B. J., Y.-Q. An, M. Geerts, B. G. Thijs, H. A. De Boer, D. M. MacLaren, J. de Graaff, and J. H. Juijens. 1994. Lactoferrin is a lipid A-binding protein. Infect. Immun. 62:2628-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battafarano, R. J., P. S. Dahlberg, C. A. Ratz, J. W. Johnston, B. H. Gray, J. R. Haseman, K. H. Mayo, and D. L. Dunn. 1995. Peptide derivatives of three distinct lipopolysaccharide binding proteins inhibit lipopolysaccharide-induced tumor necrosis factor-alpha secretion in vitro. Surgery 118:318-324. [DOI] [PubMed] [Google Scholar]
- 4.Beamer, L. J., S. F. Carroll, and D. Eisenberg. 1998. The BPI/LBP family of proteins: a structural analysis of conserved regions. Protein Sci. 7:906-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brower, M. S., and P. C. Harpel. 1982. Proteolytic cleavage and inactivation of α2 plasmin inhibitor and C1 inactivator by human polymorphonuclear leukocyte elastase. J. Biol. Chem. 257:9849-9854. [PubMed] [Google Scholar]
- 6.Caliezi, C., W. A. Wuillemin, S. Zeerleder, M. Redondo, B. Eisele, and C. E. Hack. 2001. C1-esterase inhibitor: an anti-inflammatory agent and its potential use in the treatment of diseases other than hereditary angioedema. Pharmacol. Rev. 52:91-112. [PubMed] [Google Scholar]
- 7.Colman, R. W. 1999. Biologic activities of the contact factors in vivo. Potentiation of hypotension, inflammation, and fibrinolysis, and inhibition of cell adhesion, angiogenesis and thrombosis. Thromb. Haemost. 82:1568-1577. [PubMed] [Google Scholar]
- 8.Coutinho, M., K. S. Aulak, and A. E. Davis III. 1994. Functional analysis of the serpin domain of C1 inhibitor. J. Immunol. 153:3648-3654. [PubMed] [Google Scholar]
- 9.da Silva Correia, J., and R. J. Ulevitch. 2002. MD-2 and TLR4 N-linked glycoproteins are important for a functional lipopolysaccharide receptor. J. Biol. Chem. 277:1845-1854. [DOI] [PubMed] [Google Scholar]
- 10.Day, J. R., J. J. Albers, C. E. Lofton-Day, T. L. Gilbert, A. F. Ching, F. J. Grant, P. J. O'Hara, S. M. Marcovina, and J. L. Adolphson. 1994. Complete cDNA encoding human phospholipid transfer protein from human endothelial cells. J. Biol. Chem. 269:9388-9391. [PubMed] [Google Scholar]
- 11.de Agostini, A., H. R. Lijnen, R. A. Pixley, R. W. Colman, and M. Schapira. 1984. Inactivation of factor-XII active fragment in normal plasma: predominant role of C1-inhibitor. J. Clin. Investig. 93:1542-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drayna, D., A. S. Jarnagin, J. McLean, W. Henzl, W. Kohr, C. Fielding, and R. Lawn. 1987. Cloning and sequencing of human cholesteryl ester transfer protein cDNA. Nature 327:632-634. [DOI] [PubMed] [Google Scholar]
- 13.Farnaud, S., C. Spiller, L. C. Moriarty, A. Patel, V. Gant, E. W. Odell, and R. W. Evans. 2004. Interactions of lactoferricin-derived peptides with LPS and antimicrobial activity. FEMS Microbiol. Lett. 233:193-199. [DOI] [PubMed] [Google Scholar]
- 14.Fenton, M. J., and D. T. Golenbock. 1998. LPS-binding proteins and receptors. J. Leukoc. Biol. 64:25-32. [DOI] [PubMed] [Google Scholar]
- 15.Ferguson, A. D., W. Welte, E. Hofmann, B. Lindner, O. Holst, J. W. Coulton, and K. Diederichs. 2000. A conserved structural motif for lipopolysaccharide recognition by procaryotic and eucaryotic proteins. Structure 8:585-592. [DOI] [PubMed] [Google Scholar]
- 16.Frecer, V., B. Ho, and J. L. Ding. 2000. Interpretation of biological activity data of bacterial endotoxins by simple molecular models of mechanism of action. Eur. J. Biochem. 267:837-852. [DOI] [PubMed] [Google Scholar]
- 17.Glauser, M. P., G. Zanetti, J. D. Baumgartner, and J. Cohen. 1991. Septic shock: pathogenesis. Lancet 338:732-736. [DOI] [PubMed] [Google Scholar]
- 18.Grabarek, J., G. Her, V. N. Reinhold, and J. Hawiger. 1990. Endotoxic lipid A interaction with human platelets: structure-function analysis of lipid A homologs obtained from Salmonella Minnesota Re595 lipopolysaccharide. J. Biol. Chem. 265:8117-8121. [PubMed] [Google Scholar]
- 19.Gray, P. W., G. Flaggs, S. R. Leong, R. J. Gumina, J. Weiss, C. E. Ooi, and P. Elsbach. 1989. Cloning of the cDNA of a human neutrophil bactericidal protein: structural and functional correlations. J. Biol. Chem. 264:9505-9509. [PubMed] [Google Scholar]
- 20.Gruber, A., M. Mancek, H. Wagner, C. J. Kirschning, and R. Jerala. 2004. Structural model of MD-2 and functional role of its basic amino acid clusters involved in cellular lipopolysaccharide recognition. J. Biol. Chem. 279:28475-28482. [DOI] [PubMed] [Google Scholar]
- 21.Hoess, A., S. Watson, G. R. Siber, and R. Liddington. 1993. Crystal structure of an endotoxin-neutralizing protein from the horseshoe crab, Limulus anti-LPS factor, at 1.5 Å resolution. EMBO J. 12:3351-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang, X. C., C. Bruce, T. Cocke, S. Wang, M. Boguski, and A. R. Tall. 1995. Point mutagenesis of positively charged amino acids of cholesteryl ester transfer protein: conserved residues within the lipid transfer/lipopolysaccharide binding protein gene family essential for function. Biochemistry 34:7258-7263. [DOI] [PubMed] [Google Scholar]
- 23.Juan, T. S., E. Hailman, M. J. Kelley, L. A. Busse, E. Davy, C. J. Empig, L. O. Narhi, S. D. Wright, and H. S. Lichenstein. 1995. Identification of a lipopolysaccharide binding domain in CD14 between amino acids 57 and 64. J. Biol. Chem. 270:5219-5224. [DOI] [PubMed] [Google Scholar]
- 24.Kitchens, R. L. 2000. Role of CD14 in cellular recognition of bacterial lipopolysaccharides. Chem. Immunol. 74:61-82. [DOI] [PubMed] [Google Scholar]
- 25.Lamping, N., A. Hoess, B. Yu, T. C. Park, C.-J. Kirschning, C. Pfeil, D. Reuter, S. D. Wright, F. Herrman, and R. R. Schumann. 1996. Effects of site directed mutagenesis of basic residues (Arg 94, Lys 95, Lys 99) of LPS-binding protein on binding and transfer of LPS and subsequent immune cell activation. J. Immunol. 157:4648-4656. [PubMed] [Google Scholar]
- 26.Lis, H., and N. Sharon. 1993. Protein glycosylation. Structural and functional aspects. Eur. J. Biochem. 218:1-27. [DOI] [PubMed] [Google Scholar]
- 27.Little, R. G., D. N. Kelner, E. Lim, D. J. Burke, and P. J. Conlon. 1994. Functional domains of recombinant bactericidal/permeability increasing protein (rBPI23). J. Biol. Chem. 269:1865-1872. [PubMed] [Google Scholar]
- 28.Liu, D., S. Cai, X. Gu, J. Scafidi, X. Wu, and A. E. Davis III. 2003. C1 inhibitor prevents endotoxin shock via a direct interaction with lipopolysaccharide. J. Immunol. 171:2594-2601. [DOI] [PubMed] [Google Scholar]
- 29.Liu, D., X. Gu, J. Scafidi, and A. E. Davis III. 2004. N-linked glycosylation is required for C1 inhibitor-mediated protection from endotoxin shock in mice. Infect. Immun. 72:1946-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Machamer, C. E., and J. K. Rose. 1988. Vesicular stomatitis virus G proteins with altered glycosylation sites display temperature-sensitive intracellular transport and are subject to aberrant intermolecular disulfide bonding. J. Biol. Chem. 263:5955-5960. [PubMed] [Google Scholar]
- 31.Mancek, M., P. Pristovsek, and R. Jerala. 2002. Identification of LPS-binding peptide fragment of MD-2, a Toll-receptor accessory protein. Biochem. Biophys. Res. Commun. 292:880-885. [DOI] [PubMed] [Google Scholar]
- 32.Matzuk, M. M., and I. Boime. 1988. The role of the asparagine-linked oligosaccharides of the alpha subunit in the secretion and assembly of human chorionic gonadotropin. J. Cell Biol. 106:1049-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagaoka, I., S. Hirota, F. Niyonsaba, M. Hirata, Y. Adachi, H. Tamura, and D. Heumann. 2001. Cathelicidin family of antibacterial peptides CAP18 and CAP11 inhibit the expression of TNF-α by blocking the binding of LPS to CD14+ cells. J. Immunol. 167:3329-3338. [DOI] [PubMed] [Google Scholar]
- 34.Nagaoka, I., S. Hirota, F. Niyonsaba, M. Hirata, Y. Adachi, H. Tamura, S. Tanaka, and D. Heumann. 2002. Augmentation of the lipopolysaccharide-neutralizing activities of human cathelicidin CAP18/LL-37-derived antimicrobial peptides by replacement with hydrophobic and cationic amino acid residues. Clin. Diagn. Lab. Immunol. 9:972-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nuijens, J. H., A. J. M. Eerenberg-Belmer, C. C. M. Huijbregts, W. O. Schreuder, R. J. F. Felt-Bersma, J. J. Abbink, L. G. Thijs, and C. E. Hack. 1989. Proteolytic inactivation of plasma C1 inhibitor in sepsis. J. Clin. Investig. 84:443-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Odell, E. W., R. Sarra, M. Foxworthy, D. S. Chapple, and R. W. Evans. 1996. Antibacterial activity of peptides homologous to a loop region in human lactoferrin. FEBS Lett. 382:175-178. [DOI] [PubMed] [Google Scholar]
- 37.Ohnishi, T., M. Muroi, and K. Tanamoto. 2001. N-linked glycosylations at Asn26 and Asn114 of human MD-2 are required for toll-like receptor 4-mediated activation of NF-κB by lipopolysaccharide. J. Immunol. 167:3354-3359. [DOI] [PubMed] [Google Scholar]
- 38.Poltorak, A., X. He, I. Smirnova, M. Y. Liu, C. van Huffel, X. Du, D. Birdwell, E. Alejos, M. Silva, C. Galanos, M. Freudenberg, P. Ricciardi-Castagnoli, B. Layton, and B. Beutler. 1998. Defective LPS signaling in C3H/HeJ and C57Bl/10ScCr mice: mutations in the Tlr4 gene. Science 282:2085-2088. [DOI] [PubMed] [Google Scholar]
- 39.Pristovsek, P., and J. Kidric. 1999. Solution structure of polymyxins B and E and effect of binding to lipopolysaccharide: an NMR and molecular modeling study. J. Med. Chem. 42:4603-4613. [DOI] [PubMed] [Google Scholar]
- 40.Re, F., and J. L. Strominger. 2003. Separate functional domains of human MD-2 mediate Toll-like receptor 4-binding and lipopolysaccharide responsiveness. J. Immunol. 171:5272-5276. [DOI] [PubMed] [Google Scholar]
- 41.Reboul, A., M. Prandini, and M. Colomb. 1987. Proteolysis and deglycosylation of human C1 inhibitor: effect on functional properties. Biochem. J. 24:117-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reyes, O., M. G. Vallespi, H. E. Garay, L. J. Cruz, L. J. Gonzalez, G. Chinea, W. Buurman, and M. J. Arana. 2002. Identification of single amino acid residues essential for the binding of lipopolysaccharide (LPS) to LPS binding protein (LBP) residues 86-89 using an Ala-scanning library. J. Peptide Sci. 8:144-150. [DOI] [PubMed] [Google Scholar]
- 43.Rietschel, E. T., T. Kirikae, F. U. Schade, U. Mamat, G. Schmidt, H. Loppnow, A. J. Ulmer, U. Zahringer, U. Seydel, F. di Padova, M. Schreier, and H. Brade. 1994. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB J. 8:217-225. [DOI] [PubMed] [Google Scholar]
- 44.Schapira, M., C. F. Scott, and R. W. Colman. 1982. Contribution of plasma protease inhibitors to the inactivation of kallikrein in plasma. J. Clin. Investig. 69:462-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schumann, R. R., N. Lamping, and A. Hoess. 1997. Interchangeable endotoxin-binding domains in proteins with opposite lipopolysaccharide-dependent activities. J. Immunol. 159:5599-5605. [PubMed] [Google Scholar]
- 46.Schumann, R. R., and E. Latz. 2000. Lipopolysaccharide-binding protein. Chem. Immunol. 74:42-60. [DOI] [PubMed] [Google Scholar]
- 47.Scott, M. G., A. C. E. Vreugdenhil, W. A. Buurman, R. E. W. Hancock, and M. R. Gold. 2000. Cationic antimicrobial peptides block the binding of lipopolysaccharide (LPS) to LPS binding protein. J. Immunol. 164:549-553. [DOI] [PubMed] [Google Scholar]
- 48.Sim, R. B., A. Reboul, G. J. Arlaud, C. L. Villiers, and M. G. Colomb. 1979. Interaction of 125I-labelled complement components C1r and C1s with protease inhibitors in plasma. FEBS Lett. 97:111-115. [DOI] [PubMed] [Google Scholar]
- 49.Srimal, S., N. Surolia, S. Balusubramanian, and A. Surolia. 1996. Titration calorimetric studies to elucidate the specificity of the interactions of polymyxin B with lipopolysaccharides and lipid A. Biochem. J. 315:679-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stelter, F., M. Bernheiden, R. Menzel, R. S. Jack, S. Witt, X. Fan, M. Pfister, and C. Schutt. 1997. Mutation of amino acids 39-44 of human CD14 abrogates binding of lipopolysaccharide and Escherichia coli. Eur. J. Biochem. 243:100-109. [DOI] [PubMed] [Google Scholar]
- 51.Stelter, F., H. Loppnow, R. Menzel, U. Grunwald, M. Bernheiden, R. S. Jack, A. J. Ulmer, and C. Schutt. 1999. Differential impact of substitution of amino acids 9-13 and 91-101 of human CD14 on soluble CD14-dependent activation of cells by lipopolysaccharide. J. Immunol. 163:6035-6044. [PubMed] [Google Scholar]
- 52.Stelter, F., M. Pfister, M. Bernheiden, R. S. Jack, P. Bufler, H. Engelmann, and C. Schutt. 1996. The myeloid differentiation antigen CD14 is N- and O-glycosylated. Contribution of N-linked glycosylation to different soluble CD14 isoforms. Eur. J. Biochem. 236:457-464. [DOI] [PubMed] [Google Scholar]
- 53.Takada, H., and S. Kotani. 1989. Structural requirements of lipid A for endotoxicity and other biological activities. Crit. Rev. Microbiol. 16:477-523. [DOI] [PubMed] [Google Scholar]
- 54.Taylor, A. H., G. Heavner, M. Nedelman, D. Sherris, E. Brunt, D. Knight, and J. Ghrayeb. 1995. Lipopolysaccharide neutralizing peptides reveal a lipid A binding site of LPS binding protein. J. Biol. Chem. 270:17934-17938. [DOI] [PubMed] [Google Scholar]
- 55.Tsubery, H., I. Ofek, S. Cohen, and M. Fridkin. 2001. N-terminal modifications of polymyxin B nonapeptide and their effect on antibacterial activity. Peptides 22:1675-1681. [DOI] [PubMed] [Google Scholar]
- 56.Ulevitch, R. J., and P. S. Tobias. 1995. Receptor-dependent mechanisms of cell stimulation by bacterial endotoxin. Annu. Rev. Immunol. 13:437-457. [DOI] [PubMed] [Google Scholar]
- 57.van Berkel, P. H., M. E. Geerts, H. A. van Veen, M. Mericskay, H. A. de Boer, and J. H. Nuijens. 1997. N-terminal stretch Arg2, Arg3, Arg4 and Arg5 of human lactoferrin is essential for binding to heparin, bacterial lipopolysaccharide, human lysozyme and DNA. Biochem. J. 328:145-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Viriyakosol, S., and T. N. Kirkland. 1995. A region of human CD14 required for lipopolysaccharide binding. J. Biol. Chem. 270:361-368. [DOI] [PubMed] [Google Scholar]
- 59.Viriyakosol, S., J. C. Mathison, P. S. Tobias, and T. N. Kirkland. 2000. Structure-function analysis of CD14 as a soluble receptor for lipopolysaccharide. J. Biol. Chem. 275:3144-3149. [DOI] [PubMed] [Google Scholar]
- 60.Visintin, A., E. Latz, B. G. Monks, T. Espevik, and D. T. Golenbock. 2003. Lysines 128 and 132 enable lipopolysaccharide binding to MD-2, leading to toll-like receptor-4 aggregation and signal transduction. J. Biol. Chem. 278:48313-48320. [DOI] [PubMed] [Google Scholar]