Abstract
Heparin, known for its anticoagulant activity, is commonly used in catheter locks. Staphylococcus aureus, a versatile human and animal pathogen, is commonly associated with catheter-related bloodstream infections and has evolved a number of mechanisms through which it adheres to biotic and abiotic surfaces. We demonstrate that heparin increased biofilm formation by several S. aureus strains. Surface coverage and the kinetics of biofilm formation were stimulated, but primary attachment to the surface was not affected. Heparin increased S. aureus cell-cell interactions in a protein synthesis-dependent manner. The addition of heparin rescued biofilm formation of hla, ica, and sarA mutants. Our data further suggest that heparin stimulation of biofilm formation occurs neither through an increase in sigB activity nor through an increase in polysaccharide intracellular adhesin levels. These finding suggests that heparin stimulates S. aureus biofilm formation via a novel pathway.
Heparin, a heterogeneous glycosaminoglycan, is commonly used as an anticoagulant in catheter lock solutions. Heparin and heparan sulfate are closely related compounds, the latter found in the extracellular matrix on the surface of most mammalian cells (30). Several pathogens utilize heparin and other glycosaminoglycans to adhere to and invade mammalian cells (13-15, 19, 26, 50, 61).
Staphylococcus aureus is an important pathogen, particularly in hospital settings, where it is a major source of life-threatening bloodstream infections. S. aureus is well adapted to the human host and has a large number of factors that enable it to adhere to specific host substrates, evade host defenses, and resist antibiotic therapy (17, 23, 28, 31, 47, 49). One way in which bacteria become resistant to antibiotics and host defenses is through biofilm formation (62).
S. aureus adheres proficiently to abiotic surfaces as well as biotic ones and is a problem in medical situations where implants, such as indwelling catheters and prosthetic joints, are employed (51). Catheter infections are surprisingly ubiquitous. One group reported the observation that bacterial biofilms were present on all used catheters they inspected by electron microscopy (21). S. aureus is the infectious agent in the most severe and costly episodes of catheter-related sepsis (1). It is also associated with biofilm-related diseases such as infectious arthritis, endocarditis, and cystic fibrosis (11, 51, 54, 60).
Because of the high rates of catheter colonization and the common use of heparin in catheters, we questioned whether heparin might influence S. aureus biofilm potential. Here we characterized the phenotypic effects of heparin on S. aureus biofilm formation. A number of genes have been linked to both biofilm formation and disease, including hla, ica, sarA, and sigB (2, 4, 8, 63, 66). We explore whether these genes are important for biofilm stimulation by heparin.
MATERIALS AND METHODS
Strains and media.
Staphylococcus aureus strains were grown in tryptic soy broth (TSB) or on tryptic soy agar (TSA). For all phenotypic assays we use 66% TSB plus 0.2% glucose, as this medium promotes robust biofilm formation (data not shown) (20 g/liter of Difco Bacto tryptic soy broth, Becton Dickinson, Sparks, MD). In this report 66% TSB is written as TSB. Strain MZ100 was used as a wild-type strain unless otherwise noted. MZ100 was a gift from Microbia Inc. (Cambridge, MA) and was made as RN6390, but we found that it made more robust biofilms (36). Escherichia coli strains were grown in lysogeny broth (LB) (5). Ampicillin was used at a concentration of 100 μg/ml, chloramphenicol at 10 μg/ml, kanamycin at 20 μg/ml, tetracycline at 5 to 10 μg/ml, and erythromycin at 5 μg/ml. S. aureus strain RN4220 was used as a plasmid acceptor strain. Bacteriophages 80-α and φ11 were used to transduce mutations between strains.
Genetic manipulations.
The ica operon (icaADBC) was replaced in MZ100 by a tetracycline resistance cassette using the protocol of Crampton et al. (12). Candidates were screened with PCR and tested for polysaccharide intercellular adhesin (PIA) expression. Transduction of hla::ermC, sigB::ermC, sarA::aph3A, and agr::tetM into MZ100 by bacteriophages 80-α and φ11 were performed using published protocols and were verified by PCR and or Southern blot analysis (58).
Plasmid construction.
Plasmid pALC2201 was created by ligating the promoterless gfpuvr gene into the HincII and PstI sites of pSK236 and the asp23 promoter upstream of the gfpuvr in the EcoRI site. gfpuvr was made by the introduction of an S65T mutation into gfpuv (Clontech, Palo Alto, CA), shifting the excitation maximum from 395 to 488 nm.
pALC2111 features dsred under the transcriptional control of the strong RNAIII promoter. gfpuvr from plasmid pALC1743 was removed by digestion with XbaI and PstI, and replaced with dsred from pDsRed (Clontech, San Jose, CA) amplified with the sarA ribosome binding site engineered into the primers (2).
Growth curves.
MZ100 cells were removed from a −80°C frozen stock and streaked to single colonies on a TSA plate incubated overnight at 37°C. Cells from three single colonies were inoculated into 5 ml of TSB broth and incubated overnight at 37°C with aeration. After a 14 to 16 h incubation, each culture was subcultured (1:1,000) into four test tubes, two with 5 ml of TSB plus 0.2% glucose plus saline (10% vol/vol) and two with 5 ml of TSB plus 0.2% glucose plus sodium heparin (10% vol/vol; a final concentration of 1,000 U/ml). Optical density readings were obtained at 600 nm with a Spectronic 20Dplus (Spectronic Instruments Inc., Rochester, NY), after cultures had been sonicated to disassociate cell-cell interactions that could potentially underestimate optical density readings; 1:10 and 1:100 dilutions were measured at later time points in order to remain within the linear range of the spectrophotometer.
Microtiter plate biofilm assay.
Overnight cultures of cells were normalized by optical density, then diluted 1:50 into TSB plus 0.2% glucose with either sodium heparin 10% volume/volume (1,000 U/ml; heparin sodium, NDC 63323-542-01, American Pharmaceutical Partners, Inc., Schaumburg, IL) or saline (Abbott Labs, Abbott Park, IL), 10% vol/vol, or other concentrations when specified. This heparin solution contains two preservatives, methylparaben (0.15% vol/vol) and propylparaben (0.015% vol/vol). These compounds were tested individually for an effect on biofilm formation and we noted that at the concentration found in heparin preparations (NDC 63323-542-01) propylparaben has a slight inhibitory effect, while methylparaben has no apparent effect on biofilm formation (data not shown). Additional heparin compounds were tested at 5.9 mg/ml (1,000 U/ml for the heparins) including sodium heparin (Sigma H-1027 and H-9399; Sigma-Aldrich, St. Louis, MO) ammonium heparin (H-6279), heparan sulfate (H-7640), dextran sulfate (D-4911), chondroitin B (C-3788), and chondritin C (C-4384). When indicated, chloramphenicol was added to 10 μg/ml.
To assess biofilm formation, cultures were added to tissue culture-treated, 96-well polystyrene microtiter dishes (Costar 3595, Corning Inc., Corning, NY) incubated in a closed, humidified plastic container for the indicated period of time, then assayed for biofilm formation using a modified version of the Christenson plate assay using changes noted by Caiazza (8, 10). Photographs of the inverted plate were taken with a digital camera (Nikon 990, Nikon, Mellvile, NY) and then crystal violet was solubilized using 30% glacial acetic acid for 15 min. Relative biofilm formation was assayed by reading optical density at 550 nm using a Vmax kinetic microplate reader (Molecular Devices, Sunnyvale, CA).
Cell surface/primary attachment assays.
Attachment of cells to polystyrene was assessed as follows. Optical density-normalized overnight cultures of cells grown in TSB were sonicated (model VC 505 equipped with a 3-mm stepped microtip, Sonics and Materials Inc., Newton, CT) to minimize preexisting cell-cell interactions. These cultures were diluted 1:50 into TSB plus 0.2% glucose with either sodium heparin (1,000 U/ml final) or saline added to 10% vol/vol, and then 0.5 ml of each sample was added to polystyrene 24-well plates (Costar 3624, Corning Inc., Corning, NY). Plates were placed at 37°C without shaking. At designated times nonadherent cells and medium were removed by aspiration followed by 10 washes of the well with 1 ml of phosphate-buffered saline (PBS); 0.5 ml of PBS was added to the wells and attached cells were observed microscopically. The same area of these 24-well plates was assessed in all treatments, as only the centers of each well are sufficiently flat and optically clear for microscopic analysis. Digital images of several fields were taken and later counted for the number and type of adherent foci. Surface coverage was determined using Kodak 1D image analysis software (Kodak Inc., Rochester, NY).
Adherence to silicone elastomer.
A 1-mm-thick sheet of silicone elastomer was acquired from Goodfellow Cambridge Limited (LS269875, Huntingdon, England). Coupons (1/8-in. diameter) were acquired using a hole punch (FSK-2351, Fiskars Inc., Madison, WI) and three were glued to the bottom of a well in a 24-well Costar dish with silicone sealant (Super Siliconee sealant 08661, 3M Inc, St. Paul, MN). TSB plus 0.2% glucose and cells were added as with the microtiter dish assay, except that 0.5 ml was added per well. Cells were incubated with the coupons for 24 h and then were washed five times with sterile dH20, stained with crystal violet, washed 3 times with sterile distilled H2O and allowed to dry overnight. Individual crystal violet-stained coupons were then moved to individual wells of a 96-well plate containing 0.125 ml of 30% glacial acetic acid for 15 min; 0.1-ml aliquots of solubilized crystal violet were then moved to 96-well dishes and A550 readings were determined as noted above.
Cell-cell interactions.
The adherence of cells to one another was determined using conditions identical to those for the cell surface experiments noted above, except that samples of stationary planktonic cultures were removed and observed directly with phase microscopy at various time points. Digital images of several fields were tallied for the number and type of foci (clustered or unclustered). For the protein synthesis inhibition experiment, a logarithmic culture of MZ100 grown in TSB was treated as above except that chloramphenicol (30 μg/ml) was added to inhibit protein synthesis. A double inoculum of cells was added at the onset of the experiment to cultures exposed to chloramphenicol to control for the difference in cell number that could affect the frequency of cell-interactions. At 180 min, planktonic samples from four wells per condition were assessed microscopically for cell-cell interactions.
Microscopy.
Phase-contrast and epifluorescent microscopy was performed with a model DM IRBE inverted microscope (Leica Microsystems, Wetzlar, Germany) with an attached charge-coupled device camera (model Orca C4742-5, Hamamatsu, Hamamatsu City, Japan) and analyzed with Open Lab 4.0.2 software (Improvision, Coventry, England). Cells in biofilms were stained with Syto-9, a fluorescent nucleic acid stain (Molecular Probes, Eugene, Oregon) by adding 1 μl of Syto-9 per 1 ml PBS to biofilms, incubating for 30 min, followed by washing several times with PBS. Biofilms were stained with calcofluor (fluorescent brightener 28, F-3543 Sigma-Aldrich, St. Louis, MO) using the protocol of Hamon et al. (24).
Scanning electron microscopy was performed as reported (33), except that biofilms were allowed to form on a polyvinylchloride plastic coverslip (12 by 22 mm; Fisher Scientific, Pittsburgh, PA) that was placed in the bottom of a six-well polystyrene cell culture dish (Falcon 35-3046, Becton Dickinson and Company, Franklin Lakes, NJ). One corner of the coverslip was bent for orientation purposes and to facilitate the use of forceps. The biofilm-coated coverslips were washed, dipped 15 times into PBS to remove nonadherent cells, and then fixed and prepared for scanning electron microscopy analysis as reported (33).
Polysaccharide adhesin levels.
Polysaccharide adhesin levels were assessed as previously described (12). Antiserum was a kind gift from G. B. Pier.
Sigma B activity.
The levels of the sigma B-dependent promoter asp23 were determined indirectly using a green fluorescent protein (GFP) reporter construct as has been previously described (56).
Statistical analysis.
P values were determined using Student's t test with Excel software. Error bars are shown as one standard deviation.
RESULTS
Heparin stimulates S. aureus biofilm formation in vitro.
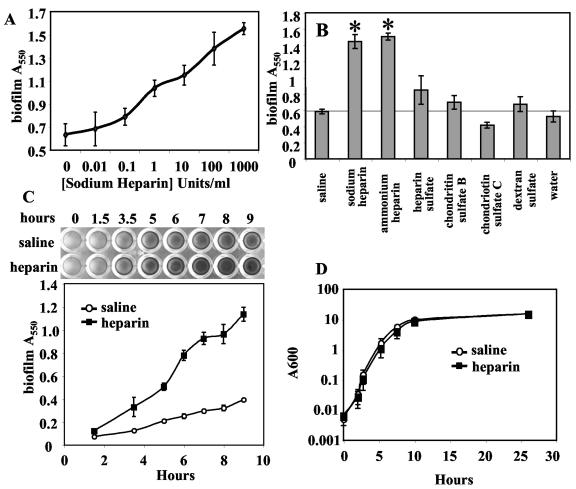
The widespread use of sodium heparin as a catheter-lock solution led us to test whether this compound has an impact upon the adherence of S. aureus to abiotic surfaces. S. aureus strain MZ100, a laboratory wild-type strain, was grown overnight in TSB, then diluted into TSB plus 0.2% glucose with the addition of serial dilutions of sodium heparin, and allowed to form a biofilm for 16 h in a 96-well polystyrene microtiter plate. Nonadherent cells were removed, and biofilms were stained with crystal violet. The relative amount of biofilm formation was determined by solubilizing crystal violet in acetic acid and determining optical density with a spectrophotometer.
Stimulation of bacterial adhesion to plastic by heparin was observed over a range from 0.1 unit/ml to the maximum tested dose of 1,000 units/ml (Fig. 1A). These are relevant levels as the concentration of heparin used in catheter lock solution is commonly up to 10,000 units/ml. Heparin, in the absence of cells did not stimulate crystal violet staining of wells (data not shown). We tested other heparin-like glycosaminoglycans and anionic polysaccharides to determine if the effect is specific to heparin and observed statistically significant biofilm stimulation with the addition of sodium heparin, ammonium heparin (P < 0.01), and to a lesser degree heparan sulfate, chondroitin sulfate B, and dextran sulfate, but not chondroitin sulfate C (Fig. 1B). We also found that heparin-stimulated biofilms displayed antibiotic resistance levels indistinguishable from nonstimulated biofilms, including highly elevated levels of vancomycin resistance compared to planktonic cells (data not shown) (42).
FIG. 1.
Sodium heparin increases the adherence of S. aureus to polystyrene. A. Dose response. Serial dilutions of sodium heparin were added to cultures and biofilms were allowed to form for 16 h, at which time nonadherent cells were removed by vigorous washing. Adherent cells were stained with crystal violet. Spectrophotometric analysis of solubilized crystal violet is shown as a function of increasing sodium heparin concentration in U/ml. B. Biofilm formation in response to various charged molecules and glycosaminoglycans. Microtiter biofilms were analyzed as above, and significant differences from the saline-alone control are shown with an asterisk (P < 0.01). C. Adherence kinetics. The effect of sodium heparin (1,000 U/ml, 10% vol/vol) or saline (10% vol/vol) upon biofilm formation was assessed over time. A photograph of crystal violet-stained biofilms is shown at the top of the panel and spectrophotometric results are charted with respect to time at the bottom of the panel. D. Growth curve. The effect of heparin (1,000 U/ml) on planktonic growth in TSB plus glucose was analyzed and plotted versus time.
Biofilm formation in the presence and absence of heparin was assessed over time using the microtiter plate assay. A twofold increase in crystal violet staining was seen as early as 3.5 h; at 6 to 9 h approximately three times as much staining was observed (see Fig. 1C).
The adherence of S. aureus to silicone elastomer coupons was also determined. Silicone elastomer is a common component of catheters. At 24 h, a twofold increase in adherence to silicone elastomer coupons was recorded (0.11 ± 0.04 A550 units for saline versus 0.25 ± 0.09 A550 units for heparin, P < 0.05).
Microscopic observation of heparin-stimulated biofilms.
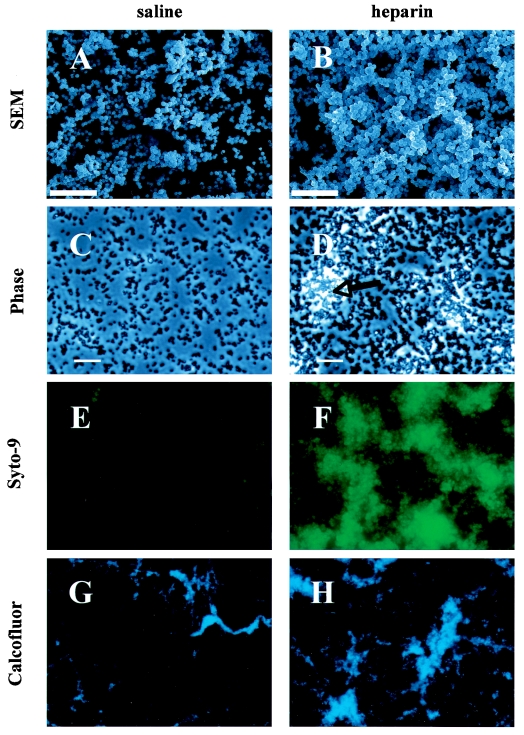
S. aureus biofilms were allowed to form overnight on polyvinylchloride under static conditions and then washed repeatedly and prepared for scanning electron microscopy. Scanning electron microscopy images suggest that surface coverage and the three-dimensional structure of biofilms were stimulated by heparin at 1,000 U/ml (Fig. 2, panels A versus B).
FIG. 2.
Sodium heparin enhances S. aureus biofilm formation. The effect of heparin on the formation of S. aureus MZ100 biofilms on abiotic surfaces was assessed microscopically. Scanning electron micrographs of 12-hour-old S. aureus biofilms on polyvinylchloride are shown with (B) and without (A) heparin (12,500× magnification, bar = 10 μm). S. aureus biofilms (4 h) formed on polystyrene were viewed with phase-contrast microscopy (C and D, bar = 20 μm, the arrow indicates a phase-bright microcolony), or were stained with a fluorescent bacterial stain (Styo-9) with a 250-ms exposure and at 400× magnification with epifluorescent microscopy (E, without heparin, and F, with heparin at 1,000 U/ml). The 5-hour-old biofilms were stained with calcofluor and viewed with epifluorescent microscopy at 100× magnification, with (H) and without (G) heparin.
Phase microscopy of heparin-treated biofilms formed on polystyrene similarly reveal that heparin promotes biofilm formation and that heparin-grown biofilms subject to shear forces through repeated washes with PBS appeared to be more tenacious than those formed with control medium (Fig. 2, panels C and D).
Epifluorescent staining of biofilms was also utilized to assess the effect of heparin on biofilms. Syto-9, a quantitative DNA-binding fluorescent dye, was used to determine relative amounts of biofilm. Figure 2, panels C and D, depicts digital micrographs of Syto-9-stained 4-hour biofilms taken with the same exposure time. Heparin-treated biofilms were more brightly stained and exhibited larger but not more numerous microcolonies. When relative fluorescence was measured, we reproducibly found a 2- to 10-fold increase in the heparin-treated sample. In a sample experiment taken 5 hours after the inoculation, we found a 3.5-fold increase in biofilm fluorescence in the heparin-treated sample [7.5 × 1010± 1.1 × 1010 relative fluorescent units (RFU) for the heparin-treated strain and 2.1 × 109± 1.4 × 109 RFU for the control, n ≥ 5 fields, P < 0.01].
Biofilms are often characterized by the presence of an extracellular matrix. We used calcofluor, a fluorescent dye that has been used to stain the extracellular matrix of biofilms in other species, to determine whether heparin-treated biofilms exhibit this hallmark of biofilm formation (Fig. 2 G and H) (24). At 5 hours, biofilms formed with and without heparin both consisted of monolayers with little to no calcofluor staining interspersed with large phase-bright microcolonies that stained brightly with calcofluor (Fig. 3). The relative fluorescence of several fields was quantified using Open Lab software. A significant difference in brightness was observed from heparin-treated biofilms at both 5 and at 21 hours; at 5 h the average RFU/visual field was found to be 1.9 × 1010 ± 3.3 × 109 for heparin-treated biofilms versus 7.4 × 109 ± 5.5 × 109 for control biofilms (P < 0.001, n ≥ 8 fields); data are not shown for 21 h. Similar results were observed for Styo-9 at 4 h.
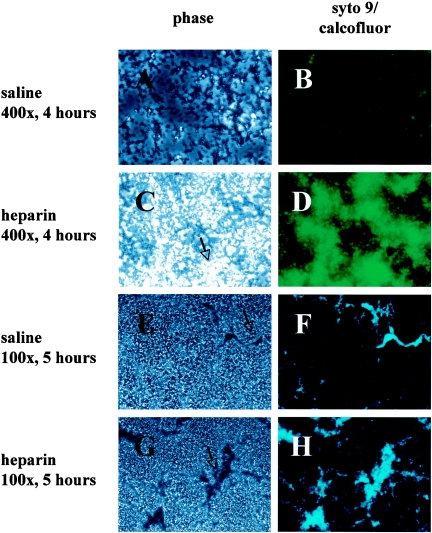
FIG. 3.
Sodium heparin enhances S. aureus biofilm formation. The effect of heparin formation of S. aureus (MZ100) biofilms on abiotic surfaces was assessed microscopically. S. aureus biofilms (4 hours) formed on polystyrene were viewed with phase 2 microscopy (A, without heparin, and C, with heparin, the arrow indicates one of several phase-bright microcolonies), or were stained with a fluorescent bacterial stain (Styo-9) with a 250-ms exposure and at 400× magnification with epifluorescent microscopy (B, without heparin, and D, with heparin at 1,000 U/ml). The 5-h-old biofilms were stained with calcofluor and viewed with phase 2 microscopy (E, without heparin, and G, with heparin, the arrow indicates a macrocolony), or epifluorescent microscopy at 100× magnification, without (F) or with (H) heparin.
Observation of biofilms formed in the presence of heparin (Fig. 2 B, D, F, and H) suggests that they have more three-dimensional architecture than do biofilms grown with control medium (Fig. 2A, C, E, and G and data not shown) and are consistent with the microtiter plate analysis results that heparin promotes biofilm formation.
Heparin does not promote S. aureus biofilm formation by accelerating growth.
Increased growth in the presence of the polysaccharide heparin could account for the more robust biofilms we observed. To determine whether sodium heparin stimulates or impedes the growth of a S. aureus population, the optical density of MZ100 cultures grown in TSB plus 0.2% glucose in the presence of either saline or sodium heparin (1,000 U/ml) was assessed over 24 h. Sonication was used to disrupt any cell-cell interactions that could alter optical density readings. Heparin at 1,000 U/ml had no discernible impact upon the growth rate of S. aureus strain MZ100 (see Fig. 1D), suggesting that it does not stimulate biofilm formation by accelerating growth. The same results were found for cultures grown without the addition of 0.2% glucose (data not shown).
Sodium heparin promotes cell-cell interactions but not primary attachment.
Staphylococcal biofilm formation has been divided into two developmental stages. Primary attachment is the first stage, wherein cells form stable interactions with a surface (22). This is followed by a growth and cell-cell interaction-dependent accumulation phase. We examined the effect of sodium heparin on each stage of S. aureus biofilm formation.
Cells adherent to plastic were observed with phase-contrast microscopy, and the foci were counted, with a single cell or a group of associated cells counted as one focus. Heparin did not affect primary attachment at 10 or 30 min (see Table 2).
TABLE 2.
Heparin does not promote primary cell surface attachment
| Strain | Treatment | Avg no. of focia ± SD
|
|
|---|---|---|---|
| 10 min | 30 min | ||
| MZ100 | Saline | 210.7 ± 9.5 | 537.3 ± 35.8 |
| MZ100 | Heparinb | 214 ± 19.6 | 549.3 ± 42.4 |
The average of three microscope fields is shown, n > 630 foci per group at 10 min and >1,600 at 30 min.
Sodium heparin was used at 1,000 U/ml in TSB with 0.2% glucose.
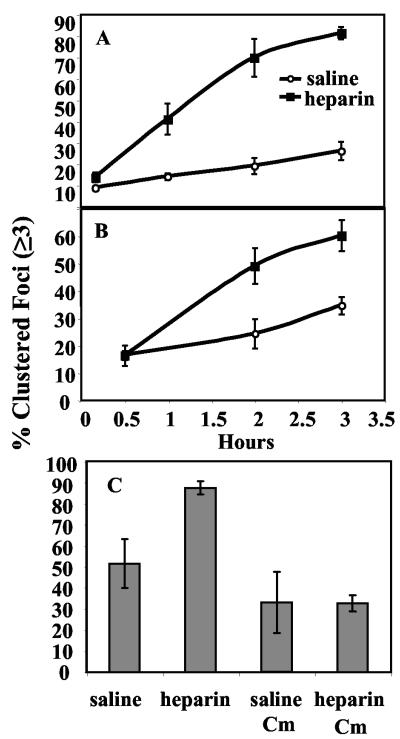
Cell-cell interactions were assessed both in the planktonic phase and on a plastic surface. Overnight planktonic cultures were sonicated to disrupt the majority of existing cell-cell interactions and then subcultured into TSB plus 0.2% glucose with either heparin or saline added to 10% (vol/vol). Samples were placed in 24-well dishes and incubated at 37°C without shaking. Aliquots of cells were removed from the planktonic phase of nonshaking cultures, observed microscopically, and assessed for cell clustering over the course of 3 h. The frequency of cell clusters of three or more bacteria was up to fourfold greater for cells in the presence of heparin (Fig. 4A). By 1 hour, 41% of microscopic foci from heparin-containing cultures consisted of clusters of three or more cells, compared to only 14% foci for cells grown without heparin. By 2 hours heparin cultures exhibited 70% clustered foci compared to 19% foci in the saline control. Planktonic cells grown overnight in heparin and incubated on a rapidly rotating wheel at 37°C also exhibit elevated levels of clustered foci compared to a no-heparin control (data not shown).
FIG. 4.
Heparin indirectly stimulates S. aureus cell-cell interactions. The effect of heparin on cell-cell interactions was assessed using phase-contrast microscopy with strain MZ100. Foci were counted and classified as either clusters (≥3 cells/focus) or nonclusters (1 or 2 cells per focus). A. Planktonic-phase clustering: aliquots were removed at the times indicated, viewed microscopically, assessed for cell-cell interactions, and plotted as percent of foci in clusters as a function of time (n = 1,492 foci). B. Surface clustering: cells attached to a polystyrene surface were microscopically assayed (n = 18,632 foci). The percent of foci in clusters are plotted as a function of time. C. Clustering without protein synthesis: a planktonic culture was treated as above except that chloramphenicol (30 μg/ml) was added to inhibit protein synthesis. A double inoculum of cells was added at the onset of the experiment to cultures exposed to chloramphenicol to control for the difference in cell number that could affect the frequency of cell-cell interactions. At 180 min, samples from four wells per category were assessed microscopically for cell-cell interactions (n = 2,025 foci).
One potential caveat to the cell-cell interaction experiments is the possibility that the clustered foci we observed were from cells that failed to separate after division rather than derived from cells that came in contact with each other and then adhered. To address this possibility, two strains were mixed in TSB plus 0.2% glucose with heparin (1,000 U/ml) and plasmid-selective antibiotic, one with a plasmid-borne copy of dsred (which codes for a red fluorescent protein) under the control of a strong promoter and the other with a vector control. Using epifluorescent microscopy, we observed at 3 h that more than 40% of clusters of three cells or more contained both red-fluorescent and nonfluorescent cells, indicating that newly formed cell-cell interactions do occur under these conditions, in clusters of six or more cells more than 60% of clusters were mixed (n > 100, data not shown).
The cell-clustering phenotype was assessed in similar experiments on a polystyrene surface. We found that heparin mediates cell-cell adherence on the surface in a manner similar to that observed for planktonic cells (Fig. 3B). We also determined surface coverage by S. aureus in the presence and absence of heparin as described in Materials and Methods and found that heparin was associated with more surface coverage at 2 h (22% coverage with heparin, 11% without, P < 0.05) and 3 h (29% with heparin, 19% without, P < 0.05) but not at 30 min (2.7% with heparin, 3.6% without, P > 0.05).
Effect of a protein synthesis inhibitor on heparin-dependent cell-cell interactions.
The observed delay in elevated levels of cell-cell interactions and surface coverage in the presence of heparin raised the question of whether the effect of heparin in this system is dependent on protein synthesis. Chloramphenicol at 30 μg/ml was added to cells in the presence or absence of heparin (1,000 U/ml heparin or saline in TSB plus 0.2% glucose) to inhibit protein synthesis. It is formally possible that the kinetics of cell-cell interactions would be different in the experimental samples because cell number would not increase over time. Therefore, a double inoculum of cells (≈4 × 107 cells) was added to the samples containing chloramphenicol as a control for final cell number, because we had previously determined this was the approximate change in cell number at 3 h when samples were taken to assess cell clustering. We found that cells in which protein synthesis was inhibited by chloramphenicol did not demonstrate a heparin-dependent increase in cell-cell interactions at 180 min (Fig. 3C). The lack of this increase in cell-cell interactions suggests that heparin does not directly mediate cell-cell interactions.
Influence of sodium heparin on sigma B activity.
One possible mechanism for the effect of heparin in stimulating biofilm formation is that cells treated with heparin could be stressed without an obvious change in growth rate. Such stress might lead to stimulation of biofilm formation as has been demonstrated for high levels of ethanol and sodium chloride. Sigma B is a sigma factor that activates an array of genes in response to cellular stress (6). Sigma B activity correlates with cellular stress and has been implicated in staphylococcal biofilm formation (34, 35, 53).
A transcriptional fusion of gfp to a sigma B-dependent promoter (asp23) was employed to investigate whether heparin stimulates a stress response (20). For this experiment we utilized strain SH1000, known to be wild type for sigB regulation (34). Strain MZ100 is known to have a deletion in the sigB regulatory gene rsbU so it is inappropriate for studying sigB expression and activity (20, 34). SH1000 and an isogenic sigB mutant strain were transformed with a plasmid-borne asp23-gfp reporter construct (27, 34). Cells were grown in TSB plus 0.2% glucose with sodium heparin (1 to 1,000 U/ml) or saline. Expression of asp23 was then determined using a fluorometer. No change in asp23 transcription with respect to heparin was observed in SH1000 (3,857 RFU without heparin and 3,889 RFU with heparin at 100 U/ml, data not shown). Similarly, heparin caused no change in sigma B activity in two clinical isolates transformed with the asp23-gfp reporter construct (data not shown). sigB mutants of SH1000 displayed low GFP levels compared to the wild-type SH1000 (for example, at 1 U/ml of heparin, the sigB mutant had 61 RFU and the wild type had 3,533 RFU).
A role for sigma B in heparin-dependent biofilm formation was further tested genetically. We hypothesized that if components of the sigma B regulon were responsible for increased bacterial attachment in the presence of heparin, then sigB mutants would not display increased attachment with the addition of heparin. Consistent with another report (63), a sigB mutation in MZ100 did not adversely affect biofilm formation, nor did it reduce PIA levels (data not shown). However, we observed that a sigB mutation in the SH1000 background conferred a 70 to 80% reduction in biofilm formation that was partially rescued by heparin. In one representative experiment where biofilms were allowed to form for 8 h, heparin stimulated a 92% increase in biofilm formation in the sigB mutant and a similar increase for SH1000 (68%, P < 0.05).
Sodium heparin enhances biofilm formation of known biofilm mutants.
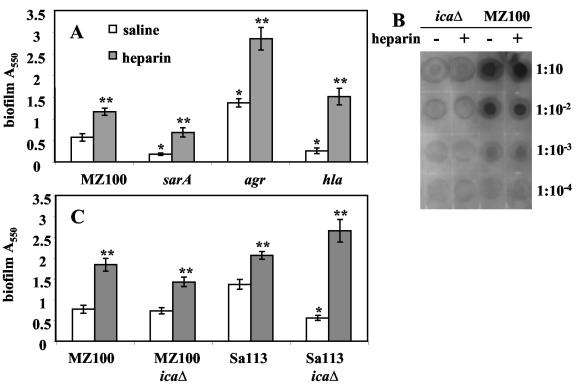
We hypothesized that if known biofilm-related factors were responsible for increased bacterial attachment in the presence of heparin, strains with mutations in these factors would not display increased attachment with the addition of heparin. To this end we tested agr, hla, and sarA mutants for biofilm formation in the presence and absence of heparin using the microtiter dish assay (Fig. 5A). In the absence of heparin, these mutants behaved as previously described when we tested them in the MZ100 background (3, 8, 63, 65, 66). Mutation of the gene that codes for the staphylococcal accessory regulator, sarA (9), severely reduced biofilm formation (P < 0.01) and was almost completely rescued by the addition of exogenous heparin at 1,000 U/ml to wild-type levels without heparin (P < 0.01, Fig. 5A). We found that an agr mutation in the MZ100 background confers a hyperbiofilm phenotype that was further enhanced by heparin (P < 0.01). Mutation of the hla gene, which codes for alpha-toxin, confers a biofilm-defective phenotype (P < 0.01) in MZ100 that was completely rescued by heparin (P < 0.01).
FIG. 5.
Effect of heparin on known biofilm formation mutants. A. Mutants (gene names noted) were assessed for biofilm formation in the microtiter dish assay (8-h biofilms) with heparin at 1,000 U/ml (shaded bars) or with saline (white bars). B. Relative PIA levels were determined in response to heparin at 1,000 U/ml. Tenfold serial dilutions of protease-treated whole-cell lysates were immunoblotted with anti-PIA antibodies. C. Isogenic pairs of wild-type and ica mutants in the MZ100 and Sa113 backgrounds were assessed for biofilm formation in the microtiter dish assay (9-h biofilms). One asterisk signifies statistical significance (P < 0.05) between a wild-type strain and its isogenic mutant strain without heparin; two asterisks signify statistical significance (P < 0.05) between strains with and without heparin.
In addition to the microtiter dish assay we directly observed 3- to 5-hour biofilms with phase microscopy and fluorescent microscopy. To quantify the microscopic data we stained cells with Syto-9, a quantitative fluorescent stain that binds to DNA, and measured the relative fluorescence of several fields of view for each strain. These results closely mirror these found with the microtiter plate assay (data not shown).
S. aureus has adhesins on its surface that enable it to bind to a number of proteins, such as fibrinogen and fibronectin. These adhesins may have a role in heparin-stimulated biofilm formation. We tested whether some of these surface adhesins are necessary for heparin-dependent biofilm stimulation after 24 h of incubation. ClfA and ClfB are surface molecules that help S. aureus bind to fibrinogen (40, 43). We determined whether clfA clfB double mutants were stimulated in biofilm formation by heparin using the microtiter dish assay. In the 8325-4 background, the wild type had an A550 reading at 24 h of 0.26 ± 0.017 with saline and 2.67 ± 0.225 with heparin, and the clfA clfB double mutant was also stimulated by heparin (0.15 ± 0.21 with saline and 1.58 ± 0.18 with heparin). We observed a similar pattern with a clfA clfB double mutant in the Newman strain background (0.16 ± 0.22 with saline and 1.34 ± 0.22 with heparin).
S. aureus binds to fibronectin via the large surface proteins FnbA and FnbB (18, 32). We found that fnbA and fnbB mutants were stimulated in adherence by heparin. SH1000 strains with fnbA and fnbB mutations exhibited an increase in A550 reading from 0.18 ± 0.02 with saline to 2.16 ± 0.2 with heparin for fnbA and from 0.2 ± 0.03 in saline to 1.4 ± 0.15 with heparin for fnbB.
Heparin stimulates biofilms independently of polysaccharide intercellular adhesin.
Polysaccharide intercellular adhesin is produced by the icaADBC gene products and is a well-characterized factor involved in staphylococcal biofilm formation (22, 38, 41, 45). Staphylococcal strains with ica mutations are reported to form poor biofilms in which primary attachment is unaffected but later stages of biofilm formation are impeded (39). In addition, cultures of S. aureus containing episomal copies of the ica locus exhibit enhanced biofilm formation, suggesting that elevated levels of PIA are sufficient to stimulate biofilm formation (12). Therefore, we tested whether heparin enhances biofilm formation through increased PIA levels.
Levels of PIA from cells grown to the stationary phase in the presence or absence of heparin (1,000 U/ml) were assayed biochemically (Fig. 5B). PIA antibodies were used to probe a dilution series of cell lysates. An isogenic ica deletion strain was generated for use as a negative control. PIA levels were indistinguishable for cells grown in the presence and absence of heparin (Fig. 5B).
The dependence of biofilm formation on PIA was also tested genetically. Biofilm formation of isogenic icaΔ and wild-type strains was tested in the presence and absence of heparin. We found that deletion of the ica operon in MZ100 conferred no significant reduction in biofilm formation (P = 0.43) as has been reported in some strain backgrounds (4) (Fig. 5C). Heparin stimulated biofilm formation by both MZ100 and its ica derivative. Therefore, we performed the analysis in strain Sa113, where the ica locus has been shown to be important in biofilm formation (12). icaΔ mutants in the Sa113 background were significantly reduced in biofilm formation (P < 0.01) and were completely rescued by heparin (Fig. 5C). The observations that an icaΔ mutant strain can form robust biofilms in the presence of heparin and that PIA levels are not altered are consistent with heparin stimulating biofilm formation independently of PIA.
Heparin affects biofilms formation of other S. aureus strains and other staphylococcal species.
Since there can be major phenotypic differences between S. aureus strains, the effect of heparin on several strains was tested. We found that heparin stimulated biofilm formation in seven of seven laboratory strains tested (Col, Newman, MZ100, RN4220, SH1000, 8325-4, and Sa113; the last five are closely related). The increase in biofilm formed in the presence of heparin versus an equal volume of saline ranged from 80 to 800% when assessed by the 96-well plate biofilm assay (data not shown). Eight out of eight S. aureus clinical isolates also exhibited increased biofilm formation in the presence of heparin (data not shown).
Staphylococcus saprophyticus is a source of urinary tract infections. Heparin had no discernible effect upon biofilm formation in the one strain of S. saprophyticus tested. One of three Staphylococcus epidermidis strains tested (strain O-47) (25) had a biofilm more than 200% greater with heparin than with saline when assessed by the microtiter plate biofilm assay. Strain ATCC R97-03 exhibited less than half of the biofilm formed in saline, and ATCC R94-10 had a slight reduction in biofilm formation in the presence of heparin (data not shown).
DISCUSSION
The data presented here demonstrates that sodium heparin, a common catheter lock solution, stimulates S. aureus biofilm formation on abiotic surfaces when present in growth medium. We show that heparin enhances adherence to polystyrene in a dose-dependent manner and that the kinetics of adherence are positively affected. Although heparin is a heterogeneous polysaccharide, the stimulation of biofilm formation is not a simple consequence of more biomass, as the growth rate is not accelerated in the presence of this compound. The stage in biofilm formation at which heparin acts appears to be after primary attachment and is likely a result of increased cell-cell interactions. However, heparin was able to bypass the biofilm defect of mutations in genes reported to negatively impact the early stage (sarA and ica) and later stages (hla) of biofilm formation (8, 39, 63). These data suggest that S. aureus has multiple routes to build a biofilm and are consistent with but do not prove that heparin stimulates biofilm formation through a novel pathway or by activating an uncharacterized mechanism downstream of sarA/agr.
Strains with mutations in the agr quorum-sensing system form very robust biofilms compared to the wild type. We hypothesized that heparin may stimulate biofilm formation by inhibition of agr expression or Agr activity. The result that agr mutants are further stimulated by heparin indicates that the heparin-associated biofilm stimulation effect does not act through inhibiting agr expression or function.
Heparin increased the frequency of clumped cells in the planktonic phase and on polystyrene. The loss of clustering with the addition of chloramphenicol implies that this phenotype is dependent upon protein synthesis. Moreover, the addition of exogenous heparin does not act immediately as a cross-bridge between cocci. These data suggests that heparin stimulates the formation of adhesion molecules that make S. aureus better able to adhere to one another in either a heparin-dependent (where heparin acts as a cross-bridge) or heparin-independent manner. Consistent with this hypothesis, several groups have evidence suggesting that S. aureus codes for a heparin binding protein (14, 16, 37). Pascu and colleagues found that 30 of 38 coagulase-negative staphylococci were able to aggregate to heparin-coated beads and this binding was inhibited by several sulfated molecules (48). Another report showed the presence of heparin binding proteins on the surface of Neisseria gonorrhoeae, Helicobacter pylori, S. epidermidis, S. aureus, Staphylococcus pyogenes, and two species of Yersinia (14). The identity of the gene(s) that codes for the factor responsible for heparin binding activity has not been reported.
We show that heparin stimulates biofilm formation on abiotic surfaces, but do these biofilms display the characteristics reported for S. aureus biofilms? The observation that heparin-exposed biofilms form microcolonies and macrocolonies that are stained by calcofluor is consistent with typical biofilm formation. Additionally, we found that nonstimulated biofilms and heparin-stimulated biofilms had high levels of resistance to the antibiotic vancomycin; overnight biofilms from both exhibited a 64-fold increase in minimum bactericidal concentration compared to planktonic cells (data not shown).
The increase in cell-cell interactions and adherence to surfaces of S. aureus exposed to heparin may be part of a normal physiological response this pathogenic species has evolved to respond to the host environment. When an S. aureus cell contacts the epithelial cell matrix, it would naturally be exposed to glycosaminoglycans, and it could benefit the bacteria to respond to this signal by making adhesive molecules allowing it to maintain itself in a potentially beneficial niche. Consistent with this idea, several pathogenic microorganisms (bacteria, viruses, and eukaryotic parasites) have evolved ways to bind to heparin to mediate attachment to or internalization into mammalian cells (14, 15, 49). Other organisms, including gram-positive bacteria such as Listeria monocytogenes and Streptococcus pyogenes, utilize heparin and related glycosaminoglycans to facilitate interaction with mammalian cells (15, 26, 57).
Future studies more closely mimicking the clinical setting as well as clinical experiments should be performed to determine whether heparin promotes persistent bacterial infections. Additionally, alternative lock solutions should be and are being tested both in the laboratory and in clinical settings (7, 52, 55, 59, 64).
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| MZ100 | Wild type | Microbia, Inc. |
| ALC2713 | MZ100 agr::tetM | This study |
| SMC2714 | MZ100 icaADBC::tetK | This study |
| SMC2715 | MZ100 hla::ermC | This study |
| ALC4007 | MZ100 sarA::aph3-A | This study |
| ALC3741 | MZ100 sigB::ermC | This study |
| 8325-4 | Wild type | 58 |
| SMC1017 | 8325-4 hla::ermC | 46 |
| RN4220 | Wild type, DNA acceptor strain | 44 |
| Sa113 | Wild type, DNA acceptor strain | 29 |
| SMC1441 | Sa113 ica::tetK | 12 |
| SH1000 | Wild type | 27 |
| ALC3085 | SH1000 sigB::ermC | This study |
| Plasmids | ||
| pALC2201 | Promoter of asp23 driving gfpuv | 56 |
| pALC1743 | RNAIII promoter regulating gfpuvr | 2 |
| pALC2111 | dsred replacing gfpuvr on pALC1743 | This study |
| pSC23 | pBT5 with icaADBC::tetK | 12 |
Acknowledgments
We thank members of the O'Toole lab for thoughtful discussion, Nicholas Jacobs for kindly sharing laboratory space with us as well as feedback on this project, Nicholas Shworak for the kind gift of several of the heparin-related compounds used in this study and for insight on studying heparin, Marybeth Maloney and Todd Jarry for sharing protocols, Ron Taylor for the use of the microplate reader, and Microbia, Inc., for the kind gift of strain MZ100. We also thank C. Daghlian and the Ripple Electron Microscopy facility at Dartmouth College for assistance with SEM.
This work was supported by NIH training grants T32 AI07363 and F32 GM66658-01A1 to R.M.Q.S. and grants from NIH (R21-AI055774), Microbia, Inc., and the Pew Charitable Trusts to G.A.O. G.A.O. is a Pew Scholar in the Biomedical Sciences.
Editor: J. T. Barbieri
REFERENCES
- 1.Arnow, P. M., E. M. Quimosing, and M. Beach. 1993. Consequences of intravascular catheter sepsis. Clin. Infect. Dis. 16:778-784. [DOI] [PubMed] [Google Scholar]
- 2.Bateman, B. T., N. P. Donegan, T. M. Jarry, M. Palma, and A. L. Cheung. 2001. Evaluation of a tetracycline-inducible promoter in Staphylococcus aureus in vitro and in vivo and its application in demonstrating the role of sigB in microcolony formation. Infect. Immun. 69:7851-7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beenken, K. E., J. S. Blevins, and M. S. Smeltzer. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 71:4206-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beenken, K. E., P. M. Dunman, F. McAleese, D. Macapagal, E. Murphy, S. J. Projan, J. S. Blevins, and M. S. Smeltzer. 2004. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 186:4665-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bischoff, M., P. Dunman, J. Kormanec, D. Macapagal, E. Murphy, W. Mounts, B. Berger-Bachi, and S. Projan. 2004. Microarray-based analysis of the Staphylococcus aureus sigB regulon. J. Bacteriol. 186:4085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buturovic, J., R. Ponikvar, A. Kandus, M. Boh, J. Klinkmann, and P. Ivanovich. 1998. Filling hemodialysis catheters in the interdialytic period: heparin versus citrate versus polygeline: a prospective randomized study. Artif. Organs 22:945-947. [DOI] [PubMed] [Google Scholar]
- 8.Caiazza, N. C., and G. A. O'Toole. 2003. Alpha-toxin is required for biofilm formation by Staphylococcus aureus. J. Bacteriol. 185:3214-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung, A. L., and S. J. Projan. 1994. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J. Bacteriol. 176:4168-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen, G. D., W. A. Simpson, J. J. Younger, L. M. Baddour, F. F. Barrett, D. M. Melton, and E. H. Beachey. 1985. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22:996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa, G. M., C. Pizzi, C. Leone, A. Borghi, E. Cordioli, and R. Bugiardini. 1999. Thrombosis of a mitral valve prosthesis resulting from Staphylococcus epidermidis endocarditis. Cardiologia 44:675-678. [PubMed] [Google Scholar]
- 12.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Gotz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeAngelis, P. L. 2002. Evolution of glycosaminoglycans and their glycosyltransferases: implication for the extracellular matrices of animals and the capsules of pathogenic bacteria. Anat. Rec. 268:317-326. [DOI] [PubMed] [Google Scholar]
- 14.Duensing, T. D., J. S. Wing, and J. P. van Putten. 1999. Sulfated polysaccharide-directed recruitment of mammalian host proteins: a novel strategy in microbial pathogenesis. Infect. Immun. 67:4463-4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fallgren, C., A. Andersson, and A. Ljungh. 2001. The role of glycosaminoglycan binding of staphylococci in attachment to eukaryotic host cells. Curr. Microbiol. 43:57-63. [DOI] [PubMed] [Google Scholar]
- 16.Fallgren, C., M. Utt, and A. Ljungh. 2001. Isolation and characterisation of a 17-kDa staphylococcal heparin-binding protein with broad specificity. J. Med. Microbiol. 50:547-557. [DOI] [PubMed] [Google Scholar]
- 17.Fedtke, I., F. Gotz, and A. Peschel. 2004. Bacterial evasion of innate host defenses: the Staphylococcus aureus lesson. Int. J. Med. Microbiol. 294:189-194. [DOI] [PubMed] [Google Scholar]
- 18.Flock, J. I., G. Froman, K. Jonsson, B. Guss, C. Signas, B. Nilsson, G. Raucci, M. Hook, T. Wadstrom, and M. Lindberg. 1987. Cloning and expression of the gene for a fibronectin-binding protein from Staphylococcus aureus. EMBO J. 6:2351-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frick, I. M., A. Schmidtchen, and U. Sjobring. 2003. Interactions between M proteins of Streptococcus pyogenes and glycosaminoglycans promote bacterial adhesions to host cells. Eur. J. Biochem. 270:2303-2311. [DOI] [PubMed] [Google Scholar]
- 20.Giachino, P., S. 2001. Engelmann, and M. Bischoff. sigma(B) activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 183:1843-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorman, S. P., W. M. Mawhinney, C. G. Adair, and M. Issouckis. 1993. Confocal laser scanning microscopy of peritoneal catheter surfaces. J. Med. Microbiol. 38:411-417. [DOI] [PubMed] [Google Scholar]
- 22.Gotz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367-1378. [DOI] [PubMed] [Google Scholar]
- 23.Greenberg, D. P., A. S. Bayer, A. L. Cheung, and J. I. Ward. 1989. Protective efficacy of protein A-specific antibody against bacteremic infection due to Staphylococcus aureus in an infant rat model. Infect. Immun. 57:1113-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamon, M. A., and B. A. Lazazzera. 2001. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol. Microbiol. 42:1199-1209. [DOI] [PubMed] [Google Scholar]
- 25.Heilmann, C., C. Gerke, F. Perdreau-Remington, and F. Gotz. 1996. Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect. Immun. 64:277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henry-Stanley, M. J., D. J. Hess, E. A. Erickson, R. M. Garni, and C. L. Wells. 2003. Role of heparan sulfate in interactions of Listeria monocytogenes with enterocytes. Med. Microbiol. Immunol. 192:107-115. [DOI] [PubMed] [Google Scholar]
- 27.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. SigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hussain, M., A. Haggar, C. Heilmann, G. Peters, J. I. Flock, and M. Herrmann. 2002. Insertional inactivation of eap in Staphylococcus aureus strain Newman confers reduced staphylococcal binding to fibroblasts. Infect. Immun. 70:2933-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iordanescu, S., and M. Surdeanu. 1976. Two restriction and modification systems in Staphylococcus aureus NTC 8325. J. Gen. Microbiol. 96:2777-2781. [DOI] [PubMed] [Google Scholar]
- 30.Jackson, R. L., S. J. Busch, and A. D. Cardin. 1991. Glycosaminoglycans: molecular properties, protein interactions, and role in physiological processes. Physiol. Rev. 71:481-539. [DOI] [PubMed] [Google Scholar]
- 31.Joh, D., E. R. Wann, B. Kreikemeyer, P. Speziale, and M. Hook. 1999. Role of fibronectin-binding MSCRAMMs in bacterial adherence and entry into mammalian cells. Matrix Biol. 18:211-223. [DOI] [PubMed] [Google Scholar]
- 32.Jonsson, K., C. Signas, H. P. Muller, and M. Lindberg. 1991. Two different genes encode fibronectin binding proteins in Staphylococcus aureus. The complete nucleotide sequence and characterization of the second gene. Eur. J. Biochem. 202:1041-1048. [DOI] [PubMed] [Google Scholar]
- 33.Kadouri, D., and G. A. O'Toole. Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 34.Kies, S., M. Otto, C. Vuong, and F. Gotz. 2001. Identification of the sigB operon in Staphylococcus epidermidis: construction and characterization of a sigB deletion mutant. Infect. Immun. 69:7933-7936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knobloch, J. K., K. Bartscht, A. Sabottke, H. Rohde, H. H. Feucht, and D. Mack. 2001. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 183:2624-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kornblum, J., B. Kreiswirth, S. J. Projan, H. Ross, and R. P. Novick. 1990. agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus, p. 373-402. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH Publishers, New York, N.Y.
- 37.Liang, O. D., F. Ascencio, L. A. Fransson, and T. Wadstrom. 1992. Binding of heparan sulfate to Staphylococcus aureus. Infect. Immun. 60:899-906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mack, D. 1999. Molecular mechanisms of Staphylococcus epidermidis biofilm formation. J. Hosp. Infect. 43(Suppl):S113-125. [DOI] [PubMed] [Google Scholar]
- 39.Mack, D., M. Nedelmann, A. Krokotsch, A. Schwarzkopf, J. Heesemann, and R. Laufs. 1994. Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide intercellular adhesin. Infect. Immun. 62:3244-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McDevitt, D., T. Nanavaty, K. House-Pompeo, E. Bell, N. Turner, L. McIntire, T. Foster, and M. Hook. 1997. Characterization of the interaction between the Staphylococcus aureus clumping factor (ClfA) and fibrinogen. Eur. J. Biochem. 247:416-424. [DOI] [PubMed] [Google Scholar]
- 41.McKenney, D., J. Hubner, E. Muller, Y. Wang, D. A. Goldmann, and G. B. Pier. 1998. The ica locus of Staphylococcus epidermidis encodes production of the capsular polysaccharide/adhesin. Infect. Immun. 66:4711-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyake, Y., S. Fujiwara, T. Usui, and H. Suginaka. 1992. Simple method for measuring the antibiotic concentration required to kill adherent bacteria. Chemotherapy 38:286-290. [DOI] [PubMed] [Google Scholar]
- 43.Ni Eidhin, D., S. Perkins, P. Francois, P. Vaudaux, M. Hook, and T. J. Foster. 1998. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol. Microbiol. 30:245-257. [DOI] [PubMed] [Google Scholar]
- 44.Novick, R. P. 1990. The staphylococcus as a molecular genetic system, p. 1-40. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH Publishers, New York, N.Y.
- 45.O'Gara, J. P., and H. Humphreys. 2001. Staphylococcus epidermidis biofilms: importance and implications. J. Med. Microbiol. 50:582-587. [DOI] [PubMed] [Google Scholar]
- 46.O'Reilly, M., A. L. de Araujo, S. Kennedy, and T. J. Foster. 1986. Inactivation of the alpha-haemolysin gene of Staphylococcus aureus 8325-4 by site-directed mutagenesis and studies on the expression of its haemolysins. Microb. Pathog. 1:125-138. [DOI] [PubMed] [Google Scholar]
- 47.Park, P. W., J. Rosenbloom, W. R. Abrams, and R. P. Mecham. 1996. Molecular cloning and expression of the gene for elastin-binding protein (ebpS) in Staphylococcus aureus. J. Biol. Chem. 271:15803-15809. [DOI] [PubMed] [Google Scholar]
- 48.Pascu, C., S. Hirmo, A. Ljungh, and T. Wadstrom. 1996. A particle agglutination assay for rapid identification of heparin binding to coagulase-negative staphylococci. J. Med. Microbiol. 45:263-269. [DOI] [PubMed] [Google Scholar]
- 49.Patti, J. M., B. L. Allen, M. J. McGavin, and M. Hook. 1994. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48:585-617. [DOI] [PubMed] [Google Scholar]
- 50.Paulsson, M., I. Gouda, O. Larm, and A. Ljungh. 1994. Adherence of coagulase-negative staphylococci to heparin and other glycosaminoglycans immobilized on polymer surfaces. J. Biomed. Mater. Res. 28:311-317. [DOI] [PubMed] [Google Scholar]
- 51.Raad, I. 1998. Intravascular-catheter-related infections. Lancet 351:893-898. [DOI] [PubMed] [Google Scholar]
- 52.Raad, I., I. Chatzinikolaou, G. Chaiban, H. Hanna, R. Hachem, T. Dvorak, G. Cook, and W. Costerton. 2003. In vitro and ex vivo activities of minocycline and EDTA against microorganisms embedded in biofilm on catheter surfaces. Antimicrob. Agents Chemother. 47:3580-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rachid, S., K. Ohlsen, U. Wallner, J. Hacker, M. Hecker, and W. Ziebuhr. 2000. Alternative transcription factor σB is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J. Bacteriol. 182:6824-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rajan, S., and L. Saiman. 2002. Pulmonary infections in patients with cystic fibrosis. Semin. Respir. Infect. 17:47-56. [DOI] [PubMed] [Google Scholar]
- 55.Root, J. L., O. R. McIntyre, N. J. Jacobs, and C. P. Daghlian. 1988. Inhibitory effect of disodium EDTA upon the growth of Staphylococcus epidermidis in vitro: relation to infection prophylaxis of Hickman catheters. Antimicrob. Agents Chemother. 32:1627-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmidt, K. A., N. P. Donegan, W. A. Kwan, Jr., and A. Cheung. 2004. Influences of sigmaB and agr on expression of staphylococcal enterotoxin B (seb) in Staphylococcus aureus. Can. J. Microbiol. 50:351-360. [DOI] [PubMed] [Google Scholar]
- 57.Schou, C., T. C. Bog-Hansen, and N. E. Fiehn. 1999. Bacterial binding to extracellular matrix proteins: in vitro adhesion. APMIS 107:493-504. [DOI] [PubMed] [Google Scholar]
- 58.Schroeder, C. J., and P. A. Pattee. 1984. Transduction analysis of transposon Tn551 insertions in the trp-thy region of the Staphylococcus aureus chromosome. J. Bacteriol. 157:533-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shah, C. B., M. W. Mittelman, J. W. Costerton, S. Parenteau, M. Pelak, R. Arsenault, and L. A. Mermel. 2002. Antimicrobial activity of a novel catheter lock solution. Antimicrob. Agents Chemother. 46:1674-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shirtliff, M. E., and J. T. Mader. 2002. Acute septic arthritis. Clin. Microbiol. Rev. 15:527-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stephens, R. S., F. S. Fawaz, K. A. Kennedy, K. Koshiyama, B. Nichols, C. Van Ooij, and J. N. Engel. 2000. Eukaryotic cell uptake of heparin-coated microspheres: a model of host cell invasion by Chlamydia trachomatis. Infect. Immun. 68:1080-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stewart, P. S. 2002. Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 292:107-113. [DOI] [PubMed] [Google Scholar]
- 63.Valle, J., A. Toledo-Arana, C. Berasain, J. M. Ghigo, B. Amorena, J. R. Penades, and I. Lasa. 2003. SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 48:1075-1087. [DOI] [PubMed] [Google Scholar]
- 64.Vercaigne, L. M., S. A. Zelenitsky, I. Findlay, K. Bernstein, and S. B. Penner. 2002. An in vitro evaluation of the antibiotic/heparin lock to sterilize central venous haemodialysis catheters. J. Antimicrob. Chemother. 49:693-696. [DOI] [PubMed] [Google Scholar]
- 65.Vuong, C., H. L. Saenz, F. Gotz, and M. Otto. 2000. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 182:1688-1693. [DOI] [PubMed] [Google Scholar]
- 66.Yarwood, J. M., D. J. Bartels, E. M. Volper, and E. P. Greenberg. 2004. Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 186:1838-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]