Abstract
Listeria monocytogenes is a gram-positive intracellular pathogen that can enter phagocytic and nonphagocytic cells and colonize their cytosols. Taking advantage of this property to generate an intracellular vaccine delivery vector, we previously described a mutant strain of L. monocytogenes, Δdal Δdat, which is unable to synthesize cell wall by virtue of deletions in two genes (dal and dat) required for d-alanine synthesis. This highly attenuated strain induced long-lived protective systemic and mucosal immune responses in mice when administered in the transient presence of d-alanine. We have now increased the usefulness of this organism as a vaccine vector by use of an inducible complementation system that obviates the need for exogenous d-alanine administration. The strain expresses a copy of the Bacillus subtilis racemase gene under the control of a tightly regulated isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter present on a multicopy plasmid. This bacterium demonstrates strict dose-dependent growth in the presence of IPTG. After removal of inducer, bacterial growth ceased within two replication cycles. Following infection of mice in the absence of IPTG or d-alanine, the bacterium survived in vivo for less than 3 days. Nevertheless, a single immunization elicited a state of long-lasting protective immunity against wild-type L. monocytogenes and induced a subset of effector listeriolysin O-specific CD11a+ CD8+ T cells in spleen and other tissues that was strongly enhanced after secondary immunization. This improved L. monocytogenes vector system may have potential use as a live vaccine against human immunodeficiency virus, other infectious diseases, and cancer.
Vaccinology is the most cost effective and efficacious of medical interventions for the eradication or management of many infectious diseases. The generation of a cellular immune response is key to survival against a variety of viral and intracellular bacterial pathogens and cancer. Therefore, the development of safe vaccines capable of inducing strong cellular immunity continues to be a pressing challenge for medicine. We have explored the use of an attenuated strain of Listeria monocytogenes as a novel live vaccine vector for this purpose. L. monocytogenes is a gram-positive intracellular bacterial pathogen whose genome has been fully sequenced (22), greatly facilitating detailed studies of its biochemistry, genetics, and physiology and contributing to the existing wealth of knowledge of its adjuvant properties and immunogenicity.
Since the early work of Mackaness in the 1960s (30), L. monocytogenes has been studied extensively as a paradigm for understanding innate and cell-mediated immunity. The organism can enter phagocytic cells through Fc receptors or type I macrophage scavenger receptors (12, 48) and can invade nonphagocytic cells by the interaction of the bacterial surface proteins InlA (16) and InlB (18) with their cognate host cell receptors. Following internalization by phagocytic cells, the majority of engulfed organisms are degraded in the phagosome (11). However, listeriolysin O (LLO), a pore-forming cytolysin, and PI-PLC, a phosphatidylinositol-specific phospholipase C, mediate permeabilization of the phagosomal membrane, enabling a fraction of the organisms to enter the host cell cytosol (9, 31, 38, 45). Subsequently, proteins secreted by the organism can be delivered directly into the major histocompatibility complex (MHC) class I pathway of antigen processing and presentation (2, 5). Mutants of L. monocytogenes unable to enter the cytosol are avirulent. These bacteria fail to stimulate protective immunity in mice, and cells infected by such mutants are not recognized by L. monocytogenes-immune CD8+ T cells (7, 34). Conversely, mice infected with sublethal doses of wild-type bacteria develop long-lasting immunity, mediated predominantly by CD8+ T cells (13, 29). This unique property of L. monocytogenes makes it attractive as a potential live vaccine vector, and recombinant strains expressing foreign antigens have successfully protected mice against lymphocytic choriomeningitis (23, 44), papillomavirus (26) and influenza virus (25) infections, and tumor challenge (24, 36, 37).
Because of the potential use of this organism as a vaccine vector for protection against infectious diseases and cancer, the safety of L. monocytogenes is a critical issue. While infections by L. monocytogenes are fairly rare and can readily be controlled by a number of antibiotics, the organism can cause meningitis and death, particularly in immunocompromised or pregnant patients. A vaccine strain of L. monocytogenes should therefore be avirulent but immunogenic. We previously sought to generate an attenuated strain that could enter the host cell cytosol but would have limited growth potential there. We reported the first description of a conditional lethal strain of L. monocytogenes with deletions of the dal and dat genes (Δdal Δdat), attenuated by virtue of deletions in the two genes necessary for the synthesis of d-alanine, a rare amino acid required for listerial peptidoglycan and lipoteichoic acid formation (15, 27, 40). This deletion bacterium has potential use as a vaccine vector because d-alanine is found only in trace quantities in vertebrate animals; therefore, the bacterium can survive only as long as d-alanine is supplied exogenously (49). Brief administration of the amino acid at the time of immunization is adequate to allow sufficient bacterial replication and the induction of an L. monocytogenes-specific immune response in mice. We demonstrated that immunization of mice in this way with a human immunodeficiency virus-gag recombinant of this novel attenuated strain of L. monocytogenes elicited a persistent systemic and mucosal anti-Gag CD8+ cytotoxic T lymphocyte response (39).
While the use of this strain as a vaccine vector allows complete control over the viability of the organism through the provision or withholding of d-alanine, it is not possible to be certain that the required amount of the amino acid is available at the precise location of the organism in the host in order to achieve optimal immunogenicity. Also, vaccination requires the dual administration of both the organism and the amino acid. We have now attempted to produce a second-generation strain from the d-alanine-deficient Δdal Δdat strain that would no longer require exogenous administration of the amino acid and yet would still retain a high degree of attenuation. A multicopy plasmid, in which expression of the Bacillus subtilis dal gene was placed under the inducible control of the lac repressor, was introduced into the Δdal Δdat strain to complement its dal deletion. Racemase, encoded by the dal gene, catalyzes the interconversion of l-alanine and d-alanine. This strategy allows the accumulation of racemase enzyme during preparative growth of the bacterium in the presence of inducer. However, immunization of the host in the absence of inducer is followed by cessation of racemase expression, rapid depletion of the enzyme during successive bacterial division cycles, and death of the organism. In this study, we test and demonstrate the validity of this concept and examine the degree of attenuation and immunogenicity of this new experimental strain.
MATERIALS AND METHODS
Bacterial strains and cell growth conditions.
Δdal Δdat, a hyperattenuated strain of Listeria monocytogenes, was grown in brain heart infusion broth (BHI; Difco, Detroit, MI) at 30°C or 37°C and supplemented with 200 μg/ml of d-alanine as described previously (49). In order to use standardized and known quantities of bacteria, many experiments were performed with thawed cultures prepared from 3- to 4-h growth of overnight cultures diluted to ratios of 1:20. Bacteria were stored at −80°C in BHI. Escherichia coli strains used for molecular cloning were grown in Luria-Bertani (LB) medium at 37°C with agitation. The following antibiotics were used at the indicated concentrations: ampicillin, 100 μg/ml; chloramphenicol (CAM), 10 μg/ml; and streptomycin (SM), 50 μg/ml. Tissue culture cells were grown in Dulbecco modified Eagle medium (DMEM; Mediatech, Herndon, VA) supplemented with 10% fetal bovine serum and 2 mM l-glutamine (Mediatech) at 37°C in a 5% CO2-air atmosphere. Plasmid pKSV7-Bsdal contains the B. subtilis racemase gene dal with 562 upstream base pairs.
Construction of Δdal Δdat/pLidal.
The plasmid pLIV1 (10) contains a SPAC/lacOid promoter/operator, so genes cloned into its unique downstream XbaI site will be expressed under the transcriptional control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter. Also present is the lac repressor gene (lacI) constitutively regulated by the L. monocytogenes p60 gene promoter. Primer BsubUp2 (5′-GCGTCTAGAAAGCTAGGAAGTGTCGTAATG-3′), containing the upstream XbaI site (underlined), and primer BsubHALow (5′-CGTCTAGATTATTATGCATAATCTGGAACATCATATGGATAATTGCTTATATTTACCTGCAATAAAGG-3′), containing the downstream XbaI site (underlined) and a hemagglutinin (HA) tag, were used to PCR amplify a promoterless B. subtilis dal gene from pKSV7-Bsdal. The 1,253-bp PCR fragment contained an 18-bp translation initiation signal and dal coding sequence, an HA tag, a downstream translation termination sequence, and XbaI sites at the 5′ and 3′ termini. pLIV1/idal (pLidal) was generated by ligating the XbaI-digested B. subtilis racemase gene fragment into the XbaI site of pLIV1. The Δdal Δdat strain was transformed with pLidal by electroporation. The transformants were selected on BHI-plus-d-alanine-plus-CAM agar plates at 30°C. The plasmid-containing transformants were further screened on BHI-plus-CAM-plus-IPTG and BHI-plus-CAM agar plates. The resulting IPTG-inducible strain of Δdal Δdat was named Δdal Δdat/pLidal.
Construction of Δdal Δdat/pAidal.
A KpnI fragment harboring the idal-expressing cassette from pLidal was cloned into pAM401, generating pAidal, as shown in Fig. 1. The KpnI fragment was digested with T4 DNA polymerase to form blunt ends and ligated with EcoRV-digested pAM401 vector. The new recombinants were transformed into E. coli XL1-Blue and confirmed by enzyme digestion and then transferred into Δdal Δdat and characterized on differential plates. This strain was named Δdal Δdat/pAidal.
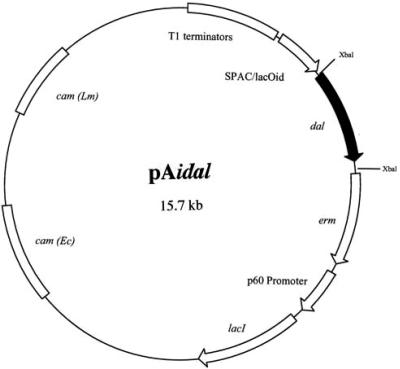
FIG. 1.
An inducible expression vector for the B. subtilis dal gene. pAidal is a plasmid containing a hybrid SPAC/lacOid IPTG-inducible promoter (10) followed downstream by a unique XbaI restriction site into which we cloned a promoterless B. subtilis dal gene containing 18 upstream bases to provide a Shine-Dalgarno ribosome binding site. A 27-bp HA tag was inserted prior to the termination codon of the dal gene. The lacI repressor is expressed constitutively by the L. monocytogenes p60 gene promoter. Upstream of the SPAC/lacOid region are tandem copies of the rrnB T1 transcription terminator (T1 terminators) to ensure that the transcription of the dal gene initiates only at the SPAC promoter. Downstream of dal is an erythromycin resistance determinant (erm). In addition, the plasmid has a chloramphenicol resistance gene [cam (Lm)] for plasmid selection in L. monocytogenes and a chloramphenicol resistance gene [cam (Ec)] that allows plasmid selection in E. coli.
The inducible expression of alanine racemase: dependence on IPTG for growth.
The idal strains were grown overnight in BHI plus CAM plus IPTG (1 mM). One milliliter of overnight culture was added to 19 ml of BHI-plus-CAM broth, aliquoted into multiple 1-ml cultures, and grown at 30°C (Δdal Δdat/pLidal) or 37°C (Δdal Δdat/pAidal) in the presence or absence of various concentrations of IPTG. Wild-type L. monocytogenes and the Δdal Δdat strain in the presence of d-alanine (200 μg/ml) were used as controls. At various times, samples were taken for measurement of their optical densities at 600 nm..
Dependence on IPTG for intracellular growth.
The intracellular growth of Δdal Δdat/pLidal and Δdal Δdat/pAidal in J774 cells was studied as previously described (38), with minor modification. Approximately 1 × 106 washed bacteria from a frozen stock culture, previously grown in BHI plus CAM plus IPTG (1 mM), were used to infect 1.5 × 106 J774 cells seeded 18 h previously onto 12-mm-diameter glass coverslips in a 60-mm-diameter petri dish. Infection was performed in DMEM without IPTG. At 30 min postinfection, the J774 cells were washed with phosphate-buffered saline (PBS) three times, and DMEM with or without 1 mM IPTG was added. At 1 h postinfection, gentamicin was added to a final concentration of 50 μg per ml. The control was Δdal Δdat in the presence of d-alanine (200 μg/ml). Three coverslips were taken from the cultures at 2, 4, 6, 8, and 10 h, separately placed in sterile distilled H2O, and vortexed for 15 s to lyse J774 cells. Dilutions were plated on BHI with 200 μg/ml of d-alanine to determine the total numbers of viable intracellular bacteria. At 6 h postinfection, the J774 cells were washed with PBS to remove gentamicin from the medium, which was replaced with fresh DMEM medium in the presence or absence of IPTG (1 mM).
Analysis of plaque formation in L2 fibroblasts.
Assays of plaque formation in L2 cell monolayers were performed as previously described (47), with some modification. Briefly, L2 cell monolayers were grown to confluence in six-well tissue culture plates. Approximately 2 × 105 bacteria from a frozen stock culture were used to infect the monolayers for 1 h in DMEM in the presence of various concentrations of IPTG. Monolayers were washed three times with PBS, and a DMEM-0.7% agarose overlay containing 10 μg/ml of gentamicin and various concentrations of IPTG was added. Plates were incubated for 3 days to allow plaque formation. At day 4, the overlay was removed, and the cells were fixed in PBS-10% paraformaldehyde for 60 min, stained with 0.1% crystal violet-20% ethanol for 5 min, washed, and air dried. The diameters of plaques in the monolayers were measured after ×10 amplification.
Plasmid stability in vitro and in vivo.
(i) In vitro stability was assessed as follows. Δdal Δdat/pLidal and Δdal Δdat/pAidal were cultured for approximately 125 generations in BHI broth in the presence of d-alanine or IPTG and then plated on BHI-plus-d-alanine or BHI-plus-IPTG agar plates. The resulting colonies were further tested on BHI-plus-d-alanine and BHI-plus-IPTG duplicate plates to confirm their phenotypes and subsequently analyzed by PCR to determine the presence of plasmid. (ii) In vivo stability was assessed as follows. Five- to six-week-old mice were inoculated intravenously (i.v.) with 2 × 107 bacteria and sacrificed 24 h later. Splenocytes were lysed in water and plated on BHI-plus-d-alanine-plus-SM or BHI-plus-IPTG-plus-CAM-plus-SM agar plates. The ratios of bacterial colonies on these plates were determined, and selected colonies were analyzed by PCR.
Bacterial proliferation in spleen and liver.
To examine the virulence of the L. monocytogenes constructs in mice, animals were infected i.v. with 2 × 107 Δdal Δdat cells in the presence of d-alanine, Δdal Δdat/pLidal cells, or Δdal Δdat/pAidal cells or with 2 × 103 wild-type L. monocytogenes cells. The numbers of viable bacteria in spleen and liver were determined at days 1, 2, 3, and 7 postinfection. Spleen and liver cells were lysed in H2O and H2O containing 0.1% NP-40 detergent (Sigma, St. Louis, MO), respectively, and dilutions were plated on BHI plus d-alanine plus SM.
Immunogenicity: LLO-tetramer-specific T cells.
Six- to eight-week-old BALB/c (H-2d haplotype) female mice were infected i.v. with 2 × 103 wild-type L. monocytogenes cells, 2 × 107 Δdal Δdat cells in the presence of d-alanine, or 2 × 107 Δdal Δdat/pLidal or Δdal Δdat/pAidal cells. At day 21 postinfection, some of these animals were boosted at a fivefold-higher dose. Groups of three mice each were sacrificed at day 9 after primary infection or at day 6 after secondary infection. Peritoneal exudate cells, peripheral blood lymphocytes, and splenocytes were collected, the red blood cells were removed with ACK lysis buffer, and the washed lymphocytes were stained with anti-CD8a-fluorescein isothiocyanate, anti-CD11a-phycoerythrin (BD Biosciences, San Diego, CA) and LLO91-99-MHC I tetramers labeled with allophycocyanin (NIH). Flow cytometry was performed using a FACSCalibur flow cytometer (Becton Dickinson, Mountain View, CA), and the data were analyzed using FlowJo software (Tree Star, Inc, Ashland, OR).
Immunogenicity: protection.
For protection studies, mice were infected with 2 × 107 Δdal Δdat cells in the presence of d-alanine, Δdal Δdat/pLidal cells, or Δdal Δdat/pAidal cells and challenged 21 days later with 2 ×104 wild-type L. monocytogenes cells. The mice were sacrificed at day 3 postchallenge. Spleens were removed for the enumeration of bacteria in these organs.
RESULTS
The inducible expression of alanine racemase in Δdal Δdat/pLidal and Δdal Δdat/pAidal: dependence on IPTG for growth and expression of enzyme.
The growth and viability of Δdal Δdat, the double-deletion strain of L. monocytogenes, is absolutely dependent on an exogenous supply of d-alanine, both in vitro and in vivo (49). While this provides a high degree of safety and control when these organisms are used as a vaccine, we wanted to explore the possibility of generating a strain based on the attenuation of Δdal Δdat that would nevertheless obviate the need for exogenous administration of the amino acid. We reasoned that this might be achieved if a gene encoding one of the d-alanine-synthesizing enzymes could be introduced into the bacteria under conditions in which its expression would be tightly regulated, allowing, when desired, a supply of enzyme sufficient to provide for the abundant need for d-alanine for cell wall synthesis, growth, and division or for its complete shutdown at other times.
Gene expression systems for gram-positive bacteria based on ligand-activated repressors, including the mercury resistance operon, activated by free Hg2+ (8), tetracycline resistance genes, activated by that antibiotic (19), and the lac repressor/operator system (53), have been described previously. Recently, plasmid pLIV1, with a lac regulatory cassette that shows stringent regulation and strong dose dependence on the inducer IPTG, has been described for use with L. monocytogenes (10). Important features of the cassette include multiple tandem copies of a transcriptional terminator that prevents aberrant read-through to the experimental gene, strong constitutive expression of the lac repressor mediated by the L. monocytogenes p60 promoter, and the presence of a modified SPAC/lacOid promoter/operator sequence that shows affinity for the lac repressor that is six- to eightfold higher than that of the native operator (35). We have introduced into this expression cassette a promoterless copy of the B. subtilis alanine racemase (dal) gene downstream of the SPAC/lacOid promoter/operator.
The B. subtilis dal gene with an intact 562-bp upstream promoter region, when expressed from the low-copy-number pKSV7 plasmid (pKSV7-Bsdal), fully complements the d-alanine deficiency of Δdal Δdat during growth in vitro (unpublished data and reference 49). However, the B. subtilis gene, with only 18 upstream base pairs containing a Shine-Dalgarno sequence for ribosome binding, failed to allow any growth of Δdal Δdat (data not shown). This promoterless dal gene was introduced into the temperature-sensitive pLIV1 construct. The resulting SPAC/lacOid:idal cassette was also transferred to the non-temperature-sensitive plasmid pAM401. We will refer to the double-deletion bacterium containing the pLIV1 plasmid as Δdal Δdat/pLidal and to that carrying the pAM401 plasmid as Δdal Δdat/pAidal (Fig. 1).
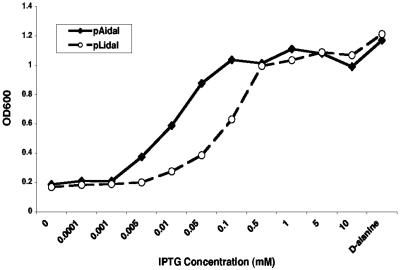
Growth of the idal-expressing bacteria, on either plasmid backbone, was found to be absolutely dependent either on d-alanine or, if d-alanine was absent, on IPTG (Fig. 2). Δdal Δdat/pAidal and Δdal Δdat/pLidal began to grow at concentrations of about 0.003 and 0.01 mM IPTG and showed maximal growth rates at 0.1- and 0.5-mM concentrations of IPTG and higher, respectively, which are close to the growth rate of the Δdal Δdat strain supplemented with d-alanine or of wild-type L. monocytogenes. The more efficient use of inducer by Δdal Δdat/pAidal may result from its higher copy number. Bacteria from which IPTG had been removed divided once or twice before replication ceased and death of the culture ensued (Fig. 3).
FIG. 2.
IPTG effect on growth is dose dependent. Overnight cultures of Δdal Δdat/pLidal (pLidal) and Δdal Δdat/pAidal (pAidal) were diluted at ratios of 1:20 into BHI plus CAM and grown for 5 h at 30°C (Δdal Δdat/pLidal) or 37°C (Δdal Δdat/pAidal) in the presence of the indicated concentrations of IPTG or with 200 μg/ml d-alanine. OD600, optical density at 600 nm.
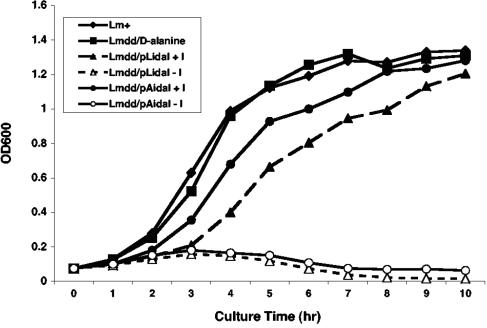
FIG. 3.
Growth kinetics of Δdal Δdat/pLidal and Δdal Δdat/pAidal in the presence or absence of IPTG. Overnight cultures were diluted at ratios of 1:20 and grown in BHI medium at the following temperatures: wild-type L. monocytogenes (Lm+), 37oC; Δdal Δdat (Lmdd) plus 200 μg/ml d-alanine, 37oC; Δdal Δdat/pLidal ± 1 mM IPTG (I), 30oC; and Δdal Δdat/pAidal ± 1 mM IPTG, 37oC. OD600, optical density at 600 nm.
In the presence of IPTG, the idal-expressing bacteria should accumulate alanine racemase enzyme. However, attempts to detect the enzyme by Western blotting, using antibody directed against an HA tag incorporated at the C terminus of the dal gene, yielded no band when lysates of Δdal Δdat/pLidal were analyzed and only a very faint band from lysates of Δdal Δdat/pAidal, whereas analysis of lysates of the Δdal Δdat strain containing the B. subtilis dal gene with its native promoter on pKSV7 (pKSV7-Bsdal) resulted in an easily detected strong band (Fig. 4, lane 4). Nevertheless, the SPAC/lacOid promoter was shown to be effective for the inducible expression of racemase in E. coli. Fig. 4 shows a Western blot of E. coli/pLidal containing the HA-tagged dal gene grown in the presence or absence of IPTG. The HA-tagged protein is clearly strongly induced. The ability to detect the racemase protein in E. coli, where it is superfluous, but not in Δdal Δdat, where it is required, suggests either that significantly more protein is induced in E. coli than in L. monocytogenes, perhaps due to higher plasmid copy number or more favorable synthesis/turnover rates or to the fact that when the enzyme is functional, it may be consumed or altered.
FIG. 4.
IPTG induction of B. subtilis racemase protein in E. coli. E. coli XL1-Blue containing pLidal (tagged with HA) was grown in LB medium for 5 h at 30°C in the presence or absence of 1 mM IPTG. Culture pellets were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting by use of a mouse monoclonal anti-HA tag antibody. Lanes: 1, E. coli XL1-Blue; 2, E. coli XL1-Blue/pLidal-HA grown without IPTG; 3, E. coli XL1-Blue/pLidal-HA grown with IPTG; 4, Δdal Δdat/pKSV7-Bsdal-HA grown in BHI medium for 5 h at 30°C.
Dependence of Δdal Δdat/pLidal and Δdal Δdat/pAidal on IPTG for intracellular growth in J774 cells.
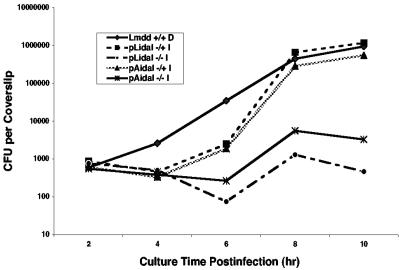
Because the growth of Δdal Δdat/pLidal and Δdal Δdat/pAidal in BHI medium showed strict dependence on IPTG, we next determined whether their growth within a eukaryotic host cell also depended on IPTG induction. Previous data had shown that Δdal Δdat in the presence of, but not in the absence of, d-alanine was able to replicate in J774 cells, a macrophage-like cell line (49). Therefore, these cells were infected with Δdal Δdat/pLidal and Δdal Δdat/pAidal for 30 min (in the absence of IPTG) and then subsequently cultured in the presence or the absence of IPTG and gentamicin (50 μg/ml). As shown in Fig. 5, the number of bacteria in the idal-infected cells decreased somewhat during the first 6 h of culture in the presence of gentamicin. However, after removal of the drug at 6 h, these cells grew well if IPTG was present but not if it was absent. The cells attained levels comparable to that of Δdal Δdat with the continuous presence of d-alanine. In the absence of inducer, only one or two cell divisions occurred. It appears that the idal-expressing bacteria may be sensitive to the high concentration of gentamicin, perhaps because the growth of these cells necessitates the additional burden of synthesizing an amount of racemase enzyme sufficient to satisfy their abundant need for d-alanine.
FIG. 5.
Intracellular growth of Δdal Δdat/pLidal and Δdal Δdat/pAidal in J774 cells. J774 cells were infected for 30 min in the absence of IPTG, washed with PBS, and subsequently cultured in the presence or absence of 1 mM IPTG and in 50 μg/ml gentamicin. At 6 h postinfection, gentamicin was removed, and cell growth continued in the presence or absence of 1 mM IPTG. The control was Δdal Δdat (Lmdd) in the presence of 200 μg/ml d-alanine (+/+ D). At 2-h intervals postinfection, coverslips were taken for the determination of bacterial numbers. −/− indicates the absence of IPTG during and after the 30-min infection period, whereas −/+ indicates the absence of IPTG during the 30-min infection period but its presence thereafter.
Racemase induction by IPTG is required for growth and cell-to-cell spread of Δdal Δdat/pLidal and Δdal Δdat/pAidal in L2 fibroblasts.
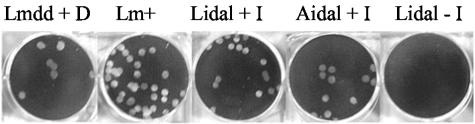
To evaluate the racemase requirement for cell-to-cell spread, we used the idal-expressing strains in a plaque formation assay in murine L2 fibroblasts, a nonprofessional phagocytic cell line. As shown in Fig. 6, no plaques were formed in L2 cell monolayers infected with either Δdal Δdat/pLidal or Δdal Δdat/pAidal in the absence of IPTG. Only when IPTG was present throughout the entire infection period were large plaques visible in the monolayers. The average diameter of Δdal Δdat/pLidal plaques in the presence of 1 mM of IPTG was less than that of plaques following infection with Δdal Δdat/pAidal or infection in the presence of d-alanine. Table 1 shows that plaque diameter was dependent on the concentration of IPTG and that Δdal Δdat/pAidal made plaques that were larger and detectable at lower concentrations than those seen with Δdal Δdat/pLidal. At very high IPTG concentrations, plaque diameter decreased, likely due to the toxicity of IPTG (10, 52).
FIG. 6.
Induction of racemase in Δdal Δdat/pLidal and Δdal Δdat/pAidal is required for cell-to-cell spread in mouse L2 fibroblasts. Wild-type L. monocytogenes (Lm+), Δdal Δdat plus 200 μg/ml d-alanine (Lmdd + D), Δdal Δdat/pLidal (Lidal), and Δdal Δdat/pAidal (Aidal), the latter two in the presence or absence of 1 mM IPTG (I), were added to monolayers of L2 fibroblasts and incubated for 1 h (the result for Δdal Δdat/pAidal in the absence of IPTG is not shown). Following washing with PBS, the infected cell monolayers were overlaid with agarose containing BHI medium plus gentamicin ± IPTG. Plaque formation following intracellular growth and spread of bacteria was visualized at 96 h postinfection.
TABLE 1.
IPTG dose-dependent plaque formation in mouse L2 fibroblastsa
| IPTG concentration (mM) | % Plaque size for:
|
|
|---|---|---|
| Δdal Δdat/pLidal | Δdal Δdat/pAidal | |
| 0 | 0 | 0 |
| 0.0001 | 0 | 0 |
| 0.001 | 0 | 26.1 |
| 0.005 | 0 | 45.3 |
| 0.01 | 15.8 | 85.4 |
| 0.05 | 49.2 | 89.2 |
| 0.1 | 61.8 | 91.6 |
| 0.5 | 71.8 | 92.0 |
| 1.0 | 78.3 | 97.3 |
| 5.0 | 80.6 | 94.7 |
| 10.0 | 80.3 | 92.7 |
Δdal Δdat/pLidal or Δdal Δdat/pAidal, grown in BHI broth containing 1 mM IPTG, were washed and used to infect monolayers of mouse L2 fibroblasts for 1 h at the indicated concentrations of IPTG. After washing the monolayers, an agarose overlay containing medium, gentamicin, and IPTG was added. Plaque formation was visualized after a 96-h incubation at 37°C. Plaque diameters are given as percentages of plaques produced by Δdal Δdat plus d-alanine (200 μg/ml).
Plasmid stability in vitro and in vivo.
The survival of the Δdal Δdat strain in the absence of d-alanine depends on retention of the idal plasmids. We therefore wondered how stable the idal plasmids were in the absence of selective antibiotics. The issue of stability is especially important in vivo, since the bacteria have been designed for use as vaccines. As shown in Table 2, both Δdal Δdat/pLidal and Δdal Δdat/pAidal lost all plasmid after about 125 generations of in vitro culture in BHI medium in the presence of d-alanine, a condition that exerts no selective pressure for plasmid retention. However, in the presence of IPTG but absence of d-alanine, all viable bacteria contained plasmid after 125 generations. Thus, bacteria that must rely on endogenous racemase for d-alanine production retain their plasmids. If any bacteria had lost plasmid, the resulting racemase-deficient bacteria would have quickly expired and been replaced by plasmid-containing organisms.
TABLE 2.
Plasmid stability in vitro and in vivo
| Colony-forming fraction (%) ina:
| |||||
|---|---|---|---|---|---|
| BHI + d-Ala in vitro
|
BHI + IPTG in vitro
|
Spleen of mice at 24 h after i.v. infection
|
|||
| Δdal Δdat/pLidal | Δdal Δdat/pAidal | Δdal Δdat/pLidal | Δdal Δdat/pAidal | Δdal Δdat/pLidal | Δdal Δdat/pAidal |
| 0 | 0 | 100 | 100 | 25 | 66 |
Bacteria were grown in BHI broth in the presence of either d-alanine or IPTG, cultured for approximately 125 generations, and then plated on BHI-plus-d-alanine or BHI-plus-IPTG agar plates. The resulting colonies were further tested in duplicate on BHI-plus-d-alanine and BHI-plus-IPTG plates. Mice were infected i.v. with 2 × 107 bacteria and sacrificed at 24 h postinfection. Splenocytes were lysed in water, and aliquots plated on BHI-plus-d-alanine-plus-SM or BHI-plus-IPTG-plus-CAM-plus-SM agar plates. The numbers shown represent the fractions of d-alanine-dependent bacteria able to form colonies in the presence of IPTG but absence of d-alanine. The presence of plasmid in colonies was confirmed by PCR.
We examined the in vivo retention of plasmid as well for mice at 24 h postinfection, when only a fraction of the bacteria were found to still survive in the spleen (see Fig. 7). As shown in Table 2, only 25% of the surviving organisms retained the pLidal plasmid, while 66% retained the pAidal plasmid. The surviving non-plasmid-containing bacteria may temporarily be sustained by accumulated racemase. These in vivo data indicate that Δdal Δdat/pAidal is more stable in vivo.
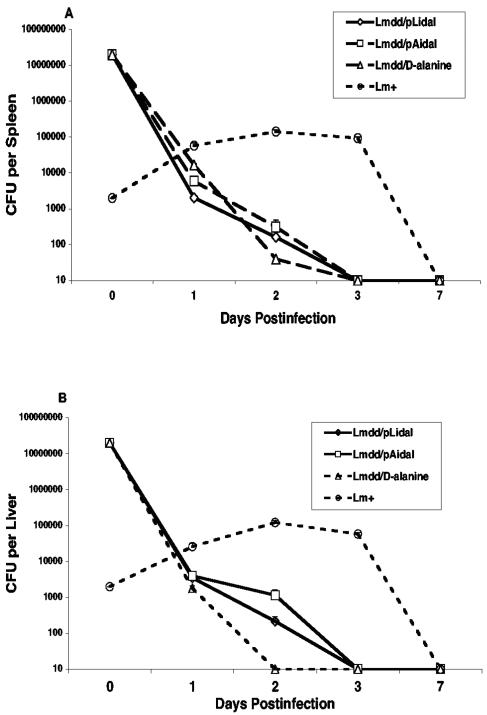
FIG. 7.
Proliferation of bacteria in spleen (A) and liver (B). Mice were infected i.v. with 2 × 107 Δdal Δdat/pLidal cells (Lmdd/pLidal), Δdal Δdat/pAidal cells (Lmdd/pAidal), or Δdal Δdat cells plus d-alanine or with 2 × 103 wild-type L. monocytogenes (Lm+) cells. Viable bacteria in spleen and liver were determined at days 1, 2, 3, and 7. Spleen and liver cells were lysed in H2O and H2O containing 0.1% NP-40, respectively, and dilutions were plated on BHI-plus-d-alanine-plus-SM or BHI-plus-CAM-plus-IPTG-plus-SM agar plates. The values at each time point represent the means and standard errors of the means from at least six mice per curve in two individual experiments.
Accelerated clearance of Δdal Δdat/pLidal and Δdal Δdat/pAidal in mice.
To assess the safety of the organisms, the number of bacteria found in tissues of infected host animals was examined at various times after infection. Groups of mice were infected intravenously with 2 × 107 Δdal Δdat/pLidal cells, Δdal Δdat/pAidal cells, or Δdal Δdat cells plus d-alanine or with 2 × 103 wild-type L. monocytogenes cells, and the total number of viable bacteria in spleens and livers was assessed on BHI-plus-d-alanine-plus-SM plates at various times up to 7 days postinfection. As indicated in Fig. 7, wild-type L. monocytogenes multiplied in these organs and reached a maximum at days 2 to 3 and were then eliminated by the host immune response. In contrast, Δdal Δdat plus d-alanine, Δdal Δdat/pLidal, and Δdal Δdat/pAidal failed to replicate significantly and were cleared from spleen and liver within 2 to 3 days. This rapid clearance agrees with preliminary tests that indicate that the 50% lethal doses of both of these organisms are approximately 108 or higher (data not shown).
Immunogenicity: LLO-specific CD8 T cells.
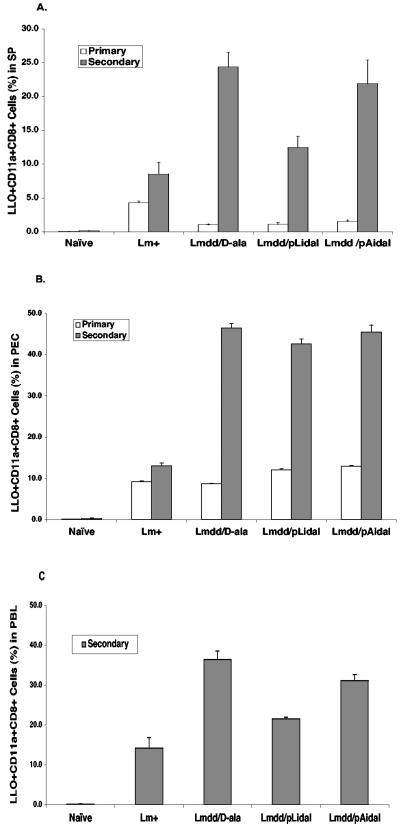
LLO, the major virulence factor for L. monocytogenes pathogenesis, is responsible for lysing the phagocytic vacuole to allow bacterial access to the cytosol (17, 28). This protein also contains the dominant CD8+ T cell epitope of L. monocytogenes (51). To assess the level of induction of activated LLO-specific CD11a+ CD8+ effector T cells following immunization of mice with Δdal Δdat/pLidal and Δdal Δdat/pAidal, lymphocytes from spleen, peritoneal exudate, and peripheral blood were analyzed during primary and secondary responses by flow cytometry. As shown in Fig. 8, wild-type L. monocytogenes, the positive control, elicited the highest percentage of LLO-specific CD11a+ CD8+ T cells in spleen during the primary response relative to the other strains tested. However, during a secondary response, the attenuated strains, i.e., Δdal Δdat plus d-alanine, Δdal Δdat/pLidal, and Δdal Δdat/pAidal, showed responses significantly higher than that of wild-type L. monocytogenes in all the tissues tested.
FIG. 8.
LLO tetramer-positive CD11a+ CD8+ T-cell populations during primary and secondary responses. BALB/c mice were infected i.v. with 2 × 107 Δdal Δdat/pLidal cells (Lmdd/pLidal), Δdal Δdat/pAidal cells (Lmdd/pAidal), or Δdal Δdat cells plus d-alanine (Lmdd/D-ala) or with 2 × 103 wild-type L. monocytogenes cells (Lm+) and reinfected at fivefold-higher doses 3 weeks postinfection. At day 9 after primary infection or day 6 after secondary infection, splenocytes (SP) (A), peritoneal exudate cells (PEC) (B), and peripheral blood lymphocytes (PBL) (C) were analyzed by use of a fluorescence-activated cell sorter after staining with anti-CD11a and CD8a monoclonal antibodies and LLO91-99 tetramers.
Immunogenicity: protection against challenge by wild-type L. monocytogenes.
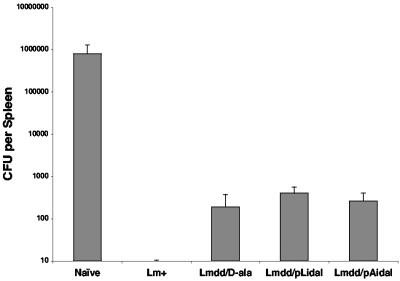
To determine whether infection of mice with sublethal doses of Δdal Δdat/pLidal and Δdal Δdat/pAidal could induce long-lasting protective immunity, we injected mice intravenously with 2 × 107 Δdal Δdat/pLidal cells, Δdal Δdat/pAidal cells, or Δdal Δdat cells plus d-alanine or with 2 × 103 wild-type L. monocytogenes cells and subsequently challenged the mice with 2 × 104 wild-type L. monocytogenes cells 3 weeks later. As shown in Fig. 9, the levels of viable bacteria remaining in the spleens of mice immunized by Δdal Δdat/pLidal and Δdal Δdat/pAidal were similar to that following Δdal Δdat-plus-d-alanine immunization, with more than a 2-log10-unit reduction compared to the naïve group. However, immunization with 2 × 103 wild-type L. monocytogenes cells provided the greatest protection.
FIG. 9.
Protection of immunized BALB/c mice against challenge with 2 × 104 (approximately 1 50% lethal dose) wild-type L. monocytogenes cells. Groups of three mice were immunized with 2 × 103 wild-type L. monocytogenes cells (Lm+) or with 2 × 107 Δdal Δdat cells plus d-alanine (Lmdd/D-ala), Δdal Δdat/pLidal cells (Lmdd/pLidal), or Δdal Δdat/ pAidal cells (Lmdd/pAidal). Mice were challenged 3 weeks later, and the total numbers of viable bacteria in spleen were determined by growth on BHI-plus-d-alanine-plus-SM plates. The detection limit was 10 CFU/spleen.
DISCUSSION
L. monocytogenes has long served as a model for the study of innate and cellular immunity and recently has been viewed as a potential live vaccine vehicle for infectious diseases and cancer (4, 7, 14, 21, 30, 41, 43, 50). However, the organism can cause listeriosis, a serious food-borne illness for certain populations at risk, such as neonates and infants, pregnant women, elderly persons, and immunocompromised individuals (20, 32, 42). The most common clinical findings include meningitis, septicemia, encephalitis, and intrauterine/cervical infections. Even though the incidence of natural infection is low, it is of significant concern if L. monocytogenes strains are to be used as human vaccine vectors. The organism must not only be attenuated sufficiently so as to pose little risk for highly susceptible hosts but must also retain the ability to elicit the desired immune response.
A candidate safe vaccine vector, an L. monocytogenes strain with deletions of the dal and dat genes (Δdal Δdat) is a highly attenuated strain, with its growth fully controlled by the exogenous administration of d-alanine. It was shown to effectively induce protective immunity against challenge either by the wild-type organism itself or by other organisms expressing a common antigen when infection was initiated with a small dose of d-alanine (39, 49).
In the present report, we have devised conditional lethal forms of the Δdal Δdat vector, called Δdal Δdat/pLidal and Δdal Δdat/pAidal, designed to overcome the shortcoming of the need for external administration of d-alanine. In these new constructs, we chose to use the homologous B. subtilis racemase gene to complement the racemase deletion in the Δdal Δdat strain. The deduced protein sequence of L. monocytogenes alanine racemase shares only 50% identity with that of the B. subtilis racemase, yet the protein of B. subtilis, regulated by its natural 562-bp upstream promoter region, fully complemented the d-alanine deficiency of Δdal Δdat during growth in vitro when expressed from a low-copy-number plasmid (unpublished data and reference 49). However, the restricted homology of the two genes substantially reduced the possibility of recombination and the potential regeneration of dal+ on the L. monocytogenes chromosome. In the new constructs, the B. subtilis racemase (dal) gene, with only a short Shine-Dalgarno-sequence-containing 18-bp upstream region, was placed downstream of the SPAC/lacOid IPTG-inducible promoter to assure tight control of the gene by IPTG. This construct completely failed to grow without d-alanine, while the presence of IPTG allowed uninhibited growth.
Use of the pLIV1 plasmid (10), harboring regions of homology with the L. monocytogenes chromosome, allows integration of the IPTG-regulated cassette into the chromosome. We chose not to use this approach to achieve d-alanine independence of the Δdal Δdat strain. First, the synthesis of cell wall components in a growing population of L. monocytogenes cells requires abundant d-alanine, both for peptidoglycan formation (27) and for lipoteichoic acid synthesis (1, 40). We surmised that a single copy of the B. subtilis dal gene controlled by the fairly weak SPAC/lacOid promoter would not satisfy this need. Even the requirement for the actA gene product by Listeria is not satisfied with a single copy of the pLIV1 cassette (unpublished observation). Conversely, multiple copies of the plasmid should assure an abundant supply of enzyme to support bacterial growth. This in fact did allow full growth of Δdal Δdat, which has a deletion of racemase. Nevertheless it still proved difficult to detect racemase by Western blotting. Multiple copies of the lac repressor gene also assured that mutations on any individual plasmid leading to inducer-independent gene expression would have negligible effects on the control of the system.
The two new constructs that we generated, Δdal Δdat/pLidal and Δdal Δdat/pAidal, both contain a promoterless B. subtilis dal gene under the control of the SPAC/lacOid promoter, complementing the dal gene defect of Δdal Δdat when expression was induced by IPTG. Both constructs had the desired property of complete dependence on the inducer for growth with no requirement for exogenous d-alanine. Thus, growth in laboratory medium and intercellular spread in culture were stringently dependent upon IPTG. These organisms, unlike Δdal Δdat, carry their own transient in situ supply of racemase. Therefore, following mouse infection, regardless of the route of infection or the site of in vivo replication, they may satisfy their need for d-alanine more efficiently than Δdal Δdat. In the absence of IPTG, the strains were very rapidly depleted from the spleens and livers of infected animals, days before a sublethal dose of wild-type L. monocytogenes reached maximal growth and almost a week before wild-type L. monocytogenes was eliminated. Despite this short survival time in mice, the strains were able to induce effective immune responses. They elicited strong LLO-tetramer-specific CD8+ T cell responses and provided protection against challenge by wild-type L. monocytogenes at levels similar to that provided by Δdal Δdat-plus-d-alanine administration. The CD8+ T cell response was weaker than that elicited by wild-type L. monocytogenes after primary immunization but significantly stronger than that of wild-type L. monocytogenes after a boost, perhaps as a result of the greater amount of antigen that could be introduced with these attenuated organisms. It is therefore possible that after a boost, these new strains might provide stronger protection against challenge than could be provided by wild-type L. monocytogenes. Thus, despite the rapid death of the organisms in infected animals, they nevertheless elicited strong immune responses that were able to protect against subsequent challenge by the wild-type organism.
Since we did not utilize the integrative property of pLidal, the pAidal plasmid showed multiple advantages over pLidal. First, the electroporation of pAidal into competent L. monocytogenes was considerably more efficient than that of pLidal. In addition, the fact that it is not temperature sensitive and is a higher-copy-number plasmid may account for the better retention of pAidal by Δdal Δdat both in culture and in vivo. These characteristics may have led to the somewhat better induction of immune CD8+ T cells by pAidal, as the duration of in vivo bacterial growth during the first day after infection may determine the magnitude of the LLO-specific T-cell response (33). Furthermore, it has been observed that during growth at low temperatures the pLIV1 plasmid can integrate into the chromosome at its homology regions, thus allowing secondary integration events to occur more readily (unpublished data). This suggests that some uncertainty is associated with the use of pLIV1-derived vectors as nonintegrative plasmids.
In summary, we have characterized a new vaccine system for L. monocytogenes based on Δdal Δdat, a d-alanine-requiring auxotroph. This new bacterium satisfies its need for d-alanine through conditional expression of a racemase gene from an IPTG-inducible promoter. The bacterium shows complete dependence on IPTG, is avirulent in mice, and results in an effective immune response that leads to protection against subsequent challenge. Δdal Δdat/pAidal may be a good candidate for a live bacterial vaccine vector. In an attempt to satisfy the important medical need for vectors able to elicit cellular immunity, several potential new vaccine strains of L. monocytogenes have recently been reported (3, 6, 46). Further study of our idal system and comparative examination of all of these new strains should focus on their relative safety and explore the quality of the immunogenicity of passenger foreign genes expressed by the vectors.
Acknowledgments
We thank the NIH AIDS Research and Reference Reagent Program for the preparation of MHC tetramers.
This work was supported by Public Health Service grants AI-42509 (F.R.F.) and AI-53669 (D.E.H.) from the National Institutes of Health.
Editor: J. D. Clements
REFERENCES
- 1.Abachin, E., C. Poyart, E. Pellegrini, E. Milohanic, F. Fiedler, P. Berche, and P. Trieu-Cuot. 2002. Formation of d-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol. Microbiol. 43:1-14. [DOI] [PubMed] [Google Scholar]
- 2.Ada, G. L. 1990. The immunological principles of vaccination. Lancet 335:523-526. [DOI] [PubMed] [Google Scholar]
- 3.Angelakopoulos, H., K. Loock, D. M. Sisul, E. R. Jensen, J. F. Miller, and E. L. Hohmann. 2002. Safety and shedding of an attenuated strain of Listeria monocytogenes with a deletion of actA/plcB in adult volunteers: a dose escalation study of oral inoculation. Infect. Immun. 70:3592-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barry, R. A., H. G. A. Bouwer, D. A. Portnoy, and D. J. Hinrichs. 1992. Pathogenicity and immunogenicity of Listeria monocytogenes small-plaque mutants defective for intracellular growth and cell-to-cell spread. Infect. Immun. 60:1625-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braciale, T. J., L. A. Morrison, M. T. Sweetser, J. Sambrook, M. J. Gething, and V. L. Braciale. 1987. Antigen presentation pathways to class I and class II MHC-restricted T lymphocytes. Immunol. Rev. 98:95-114. [DOI] [PubMed] [Google Scholar]
- 6.Brockstedt, D. G., M. A. Giedlin, M. L. Leong, K. S. Bahjat, Y. Gao, W. Luckett, W. Liu, D. N. Cook, D. A. Portnoy, and T. W. Dubensky, Jr. 2004. Listeria-based cancer vaccines that segregate immunogenicity from toxicity. Proc. Natl. Acad. Sci. USA 101:13832-13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunt, L. M., D. A. Portnoy, and E. R. Unanue. 1990. Presentation of Listeria monocytogenes to CD8+ T cells requires secretion of hemolysin and intracellular bacterial growth. J. Immunol. 145:3540-3546. [PubMed] [Google Scholar]
- 8.Chu, L., D. Mukhopadhyay, H. Yu, K. S. Kim, and T. K. Misra. 1992. Regulation of the Staphylococcus aureus plasmid pI258 mercury resistance operon. J. Bacteriol. 174:7044-7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cossart, P., M. F. Vicente, J. Mengaud, F. Baquero, J. C. Perez-Diaz, and P. Berche. 1989. Listeriolysin O is essential for virulence of Listeria monocytogenes: direct evidence obtained by gene complementation. Infect. Immun. 57:3629-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dancz, C. E., A. Haraga, D. A. Portnoy, and D. E. Higgins. 2002. Inducible control of virulence gene expression in Listeria monocytogenes: temporal requirement of listeriolysin O during intracellular infection. J. Bacteriol. 184:5935-5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Chastellier, C., and P. Berche. 1994. Fate of Listeria monocytogenes in murine macrophages: evidence for simultaneous killing and survival of intracellular bacteria. Infect. Immun. 62:543-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunne, D. W., D. Resnick, J. Greenberg, M. Krieger, and K. A. Joiner. 1994. The type I macrophage scavenger receptor binds to gram-positive bacteria and recognizes lipoteichoic acid. Proc. Natl. Acad. Sci. USA 91:1863-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finelli, A., K. M. Kerksiek, S. E. Allen, N. Marshall, R. Mercado, I. Pilip, D. H. Busch, and E. G. Pamer. 1999. MHC class I restricted T cell responses to Listeria monocytogenes, an intracellular bacterial pathogen. Immunol. Res. 19:211-223. [DOI] [PubMed] [Google Scholar]
- 14.Frankel, F. R., S. Hedge, J. Lieberman, and Y. Paterson. 1995. Induction of cell-mediated immune responses to human immunodeficiency virus type 1 Gag protein by using Listeria monocytogenes as a live vaccine vector. J. Immunol. 155:4775-4782. [PubMed] [Google Scholar]
- 15.Fujii, H., K. Kamisango, M. Nagaoka, K. Uchikawa, I. Sekikawa, K. Yamamoto, and I. Azuma. 1985. Structural study on teichoic acids of Listeria monocytogenes types 4a and 4d. J. Biochem. 97:883-891. [DOI] [PubMed] [Google Scholar]
- 16.Gaillard, J., P. Berche, C. Frehel, E. Gouin, and P. Cossart. 1991. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell 65:1127-1141. [DOI] [PubMed] [Google Scholar]
- 17.Gaillard, J. L., P. Berche, and P. Sansonetti. 1986. Transposon mutagenesis as a tool to study the role of hemolysin in the virulence of Listeria monocytogenes. Infect. Immun. 52:50-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaillard, J. L., F. Jaubert, and P. Berche. 1996. The inlAB locus mediates the entry of Listeria monocytogenes into hepatocytes in vivo. J. Exp. Med. 183:359-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geissendorfer, M., and W. Hillen. 1990. Regulated expression of heterologous genes in Bacillus subtilis using the Tn10 encoded tet regulatory elements. Appl. Microbiol. Biotechnol. 33:657-663. [DOI] [PubMed] [Google Scholar]
- 20.Gellin, B. G., and C. V. Broome. 1989. Listeriosis. JAMA 261:1313-1320. [PubMed] [Google Scholar]
- 21.Gentschev, I., G. Dietrich, S. Spreng, A. Kolb-Maurer, V. Brinkmann, L. Grode, J. Hess, S. H. Kaufmann, and W. Goebel. 2001. Recombinant attenuated bacteria for the delivery of subunit vaccines. Vaccine 19:2621-2628. [DOI] [PubMed] [Google Scholar]
- 22.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 23.Goossens, P. L., G. Milon, P. Cossart, and M.-F. Saron. 1995. Attenuated Listeria monocytogenes as a live vector for induction of CD8+ T cells in vivo: a study with the nucleoprotein of the lymphocytic choriomeningitis virus. Int. Immunol. 7:797-802. [DOI] [PubMed] [Google Scholar]
- 24.Gunn, G. R., A. Zubair, C. Peters, Z. K. Pan, T. C. Wu, and Y. Paterson. 2001. Two Listeria monocytogenes vaccine vectors that express different molecular forms of human papilloma virus-16 (HPV-16) E7 induce qualitatively different T cell immunity that correlates with their ability to induce regression of established tumors immortalized by HPV-16. J. Immunol. 167:6471-6479. [DOI] [PubMed] [Google Scholar]
- 25.Ikonomidis, G., D. Portnoy, W. Gerhard, and Y. Paterson. 1997. Influenza-specific immunity induced by recombinant Listeria monocytogenes vaccines. Vaccine 15:433-440. [DOI] [PubMed] [Google Scholar]
- 26.Jensen, E. R., R. Selvakumar, H. Shen, R. Ahmed, F. O. Wettstein, and J. F. Miller. 1997. Recombinant Listeria monocytogenes vaccination eliminates papillomavirus-induced tumors and prevents papilloma formation from viral DNA. J. Virol. 71:8467-8474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamisango, K., I. Saiki, Y. Tanio, H. Okumura, Y. Araki, I. Sekikawa, I. Azuma, and Y. Yamamura. 1982. Structure and biological activities of peptidoglycans of Listeria monocytogenes and Propionibacterium acnes. J. Biochem. 92:23-33. [DOI] [PubMed] [Google Scholar]
- 28.Kathariou, S., P. Metz, H. Hof, and W. Goebel. 1987. Tn916-induced mutations in the hemolysin determinant affecting virulence of Listeria monocytogenes. J. Bacteriol. 169:1291-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaufmann, S. H. E. 1993. Immunity to intracellular bacteria. Annu. Rev. Immunol. 11:129-163. [DOI] [PubMed] [Google Scholar]
- 30.Mackaness, G. B. 1962. Cellular resistance to infection. J. Exp. Med. 116:381-406. [PubMed] [Google Scholar]
- 31.Marquis, H., V. Doshi, and D. A. Portnoy. 1995. The broad-range phospholipase C and a metalloprotease mediate listeriolysin O-independent escape of Listeria monocytogenes from a primary vacuole in human epithelial cells. Infect. Immun. 63:4531-4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meier, J., and L. Lopez. 2001. Listeriosis: an emerging food-borne disease. Clin. Lab. Sci. 14:187-192. [PubMed] [Google Scholar]
- 33.Mercado, R., S. Vijh, S. E. Allen, K. Kerksiek, I. M. Pilip, and E. G. Pamer. 2000. Early programming of T cell populations responding to bacterial infection. J. Immunol. 165:6833-6839. [DOI] [PubMed] [Google Scholar]
- 34.Michel, E., K. A. Reich, R. Favier, P. Berche, and P. Cossart. 1990. Attenuated mutants of the intracellular bacterium Listeria monocytogenes obtained by single amino acid substitutions in listeriolysin O. Mol. Microbiol. 4:2167-2178. [DOI] [PubMed] [Google Scholar]
- 35.Oehler, S., M. Amouyal, P. Kolkhof, B. von Wilcken-Bergmann, and B. Muller-Hill. 1994. Quality and position of the three lac operators of E. coli define efficiency of repression. EMBO J. 13:3348-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan, Z.-K., G. Ikonomidis, A. Lazenby, D. Pardoll, and Y. Paterson. 1995. A recombinant Listeria monocytogenes vaccine expressing a model tumour antigen protects mice against lethal tumour cell challenge and causes regression of established tumours. Nat. Med. 1:471-477. [DOI] [PubMed] [Google Scholar]
- 37.Paterson, Y., and G. Ikonomidis. 1996. Recombinant Listeria monocytogenes cancer vaccines. Curr. Opin. Immunol. 8:664-669. [DOI] [PubMed] [Google Scholar]
- 38.Portnoy, D. A., P. S. Jacks, and D. J. Hinrichs. 1988. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J. Exp. Med. 167:1459-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rayevskaya, M. V., and F. R. Frankel. 2001. Systemic immunity and mucosal immunity are induced against human immunodeficiency virus gag protein in mice by a new hyperattenuated strain of Listeria monocytogenes. J. Virol. 75:2786-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruhland, G. J., and F. Fiedler. 1987. Occurrence and biochemistry of lipoteichoic acids in the genus Listeria. Syst. Appl. Microbiol. 9:40-46. [Google Scholar]
- 41.Schafer, R., D. A. Portnoy, S. A. Brassell, and Y. Paterson. 1992. Induction of a cellular immune response to a foreign antigen by a recombinant Listeria monocytogenes vaccine. J. Immunol. 149:53-59. [PubMed] [Google Scholar]
- 42.Schlech, W. F., III. 1991. Lowbury lecture. Listeriosis: epidemiology, virulence and the significance of contaminated foodstuffs. J. Hosp. Infect. 19:211-224. [DOI] [PubMed] [Google Scholar]
- 43.Shen, H., J. F. Miller, X. Fan, D. Kolwyck, R. Ahmed, and J. T. Harty. 1998. Compartmentalization of bacterial antigens: differential effects on priming of CD8 T cells and protective immunity. Cell 92:535-545. [DOI] [PubMed] [Google Scholar]
- 44.Shen, H., M. K. Slifka, M. Matloubian, E. R. Jensen, R. Ahmed, and J. F. Miller. 1995. Recombinant Listeria monocytogenes as a live vaccine vehicle for the induction of protective anti-viral cell-mediated immunity. Proc. Natl. Acad. Sci. USA 92:3987-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith, G. A., H. Marquis, S. Jones, N. C. Johnston, D. A. Portnoy, and H. Goldfine. 1995. The two distinct phospholipases C of Listeria monocytogenes have overlapping roles in escape from a vacuole and cell-to-cell spread. Infect. Immun. 63:4231-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stritzker, J., J. Janda, C. Schoen, M. Taupp, S. Pilgrim, I. Gentschev, P. Schreier, G. Geginat, and W. Goebel. 2004. Growth, virulence, and immunogenicity of Listeria monocytogenes aro mutants. Infect. Immun. 72:5622-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun, A. N., A. Camilli, and D. A. Portnoy. 1990. Isolation of Listeria monocytogenes small-plaque mutants defective for intracellular growth and cell-to-cell spread. Infect. Immun. 58:3770-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzuki, H., Y. Kurihara, M. Takeya, N. Kamada, M. Kataoka, K. Jishage, O. Ueda, H. Sakaguchi, T. Higashi, T. Suzuki, Y. Takashima, Y. Kawabe, O. Cynshi, Y. Wada, M. Honda, H. Kurihara, H. Aburatani, T. Doi, A. Matsumoto, S. Azuma, T. Noda, Y. Toyoda, H. Itakura, Y. Yazaki, and T. Kodama. 1997. A role for macrophage scavenger receptors in atherosclerosis and susceptibility to infection. Nature 386:292-296. [DOI] [PubMed] [Google Scholar]
- 49.Thompson, R. J., H. G. A. Bouwer, D. A. Portnoy, and F. R. Frankel. 1998. Pathogenicity and immunogenicity of a Listeria monocytogenes strain that requires d-alanine for growth. Infect. Immun. 66:3552-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weiskirch, L. M., and Y. Paterson. 1997. Listeria monocytogenes: a potent vaccine vector for neoplastic and infectious disease. Immunol. Rev. 158:159-169. [DOI] [PubMed] [Google Scholar]
- 51.Wipke, B. T., S. C. Jameson, M. J. Bevan, and E. G. Pamer. 1993. Variable binding affinities of listeriolysin O peptides for the H-2Kd class I molecule. Eur. J. Immunol. 23:2005-2010. [DOI] [PubMed] [Google Scholar]
- 52.Wyborski, D. L., and J. M. Short. 1991. Analysis of inducers of the E. coli lac repressor system in mammalian cells and whole animals. Nucleic Acids Res. 19:4647-4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yansura, D. G., and D. J. Henner. 1984. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 81:439-443. [DOI] [PMC free article] [PubMed] [Google Scholar]