Abstract
Actinobacillus pleuropneumoniae, the etiological agent of porcine pleuropneumonia, is able to persist on respiratory epithelia, in tonsils, and in the anaerobic environment of encapsulated lung sequesters. We have demonstrated previously that putative HlyX-regulated genes, coding for dimethyl sulfoxide (DMSO) reductase and aspartate ammonia lyase, are upregulated during infection and that deletions in these genes result in attenuation of the organism. The study presented here investigates the role of HlyX, the fumarate nitrate reductase regulator (FNR) homologue of A. pleuropneumoniae. By constructing an isogenic A. pleuropneumoniae hlyX mutant, the HlyX protein is shown to be responsible for upregulated expression of both DMSO reductase and aspartate ammonia lyase (AspA) under anaerobic conditions. In a challenge experiment the A. pleuropneumoniae hlyX mutant is shown to be highly attenuated, unable to persist in healthy lung epithelium and tonsils, and impaired in survival inside sequestered lung tissue. Further, using an A. pleuropneumoniae strain carrying the luxAB genes as transcriptional fusion to aspA on the chromosome, the airway antioxidant glutathione was identified as one factor potentially responsible for inducing HlyX-dependent gene expression of A. pleuropneumoniae in epithelial lining fluid.
Actinobacillus pleuropneumoniae, the etiological agent of porcine pleuropneumonia (10), is able to persist in host tissues, such as tonsillar crypts and sequestered necrotic lung, where the oxygen supply is scarce. The resulting carrier animals are the major source of infection for previously A. pleuropneumoniae-free herds (10) and, therefore, unraveling the mechanisms of persistence is highly relevant to effective vaccination and control of the infection.
In Escherichia coli a number of genes expressed under anaerobic conditions are regulated by the global regulator FNR (for fumarate nitrate reductase regulator) (24). An A. pleuropneumoniae FNR homologue, HlyX, has been found to induce hemolytic activity in E. coli under anoxic conditions and to be able to complement E. coli fnr deletions (18, 26). Like FNR, HlyX contains four iron-sulfur clusters responsible for the DNA-binding ability of the protein (12).
In A. pleuropneumoniae, anaerobically regulated genes appear to play a role in virulence and persistence. We have shown previously that A. pleuropneumoniae genes upregulated under anaerobic conditions in culture are not only upregulated in sequestered necrotic lung tissue where anoxic conditions are to be expected (1) but also upon the supplementation of culture medium with bronchoalveolar lavage fluid from A. pleuropneumoniae-infected pigs, which mimics conditions as they occur on respiratory epithelium (1-3, 15). Further, we have shown that isogenic mutants lacking aspartate ammonia lyase (aspartase) activity or both dimethyl sulfoxide (DMSO) reductase and aspartase activity were reduced in virulence and in their ability to persist on unaltered respiratory epithelium (2, 15).
Since both the DMSO reductase (dmsA) and the aspartase (aspA) genes contain putative HlyX-binding motifs (2, 15), we constructed an isogenic A. pleuropneumoniae mutant strain lacking the hlyX gene, examined it in vitro with respect to the regulation of DMSO reductase expression and aspartase activity, and used the strain in an aerosol infection model in order to examine the effect of the hlyX deletion on A. pleuropneumoniae virulence and persistence. Further, in order to investigate how genes controlled by the anaerobic regulator HlyX could be upregulated in the aerobic environment of the respiratory epithelium, we examined the influence of a common airway antioxidant, reduced glutathione (GSH), on aspartase expression in a luciferase reporter assay with an A. pleuropneumoniae strain carrying the luxAB genes in transcriptional fusion to the aspA gene on the chromosome (15).
MATERIALS AND METHODS
Bacterial strains, plasmids, and primers.
The strains, plasmids, and primers used in the present study are listed in Table 1.
TABLE 1.
Characteristics of bacterial strains and primers used in this study
| Strain, plasmid, or primers | Characteristicsa | Source and/or references |
|---|---|---|
| Strains | ||
| E. coli β2155 | thrB1004 pro thi strA hsdS lacZΔM15 (F′ lacZΔM15 laqIqtraD36 proA+proB+) ΔdapA::erm (Ermr)recA::RPA-2-tet (Tcr)::Mu-km (Kmr) λpir | 9 |
| E. coli DH5αF′ | F′ endA1 hsdR17 (rK− mK−) supE44 thi-1 recA1 gyrA (Nalr) relA1 Δ(lacZYA-argF)U169 deoR [φ80dlacΔ(lacZ)M15] | 21 |
| A. pleuropneumoniae AP76 | A. pleuropneumoniae serotype 7, field strain | 19 |
| A. pleuropneumoniae ΔhlyX | Unmarked hlyX-negative knockout mutant of A. pleuropneumoniae AP76 | This study |
| A. pleuropneumoniae ΔaspA::luxAB | Unmarked A. pleuropneumoniae mutant carrying luxAB as a transcriptional fusion in the aspA gene | 15 |
| Plasmids | ||
| pCR2.1-TOPO | Topoisomerase I enhanced E. coli cloning vector | TOPO TA Cloning, Invitrogen, Groningen, The Netherlands (25) |
| pBluescript SK(+) | 3.0-kb cloning vector; Apr | Stratagene, La Jolla, CA |
| pEMOC2 | Transconjugation vector based on pBluescript SK with mobRP4, polycloning site, Cmr, and transcriptional fusion of the omlA promoter with the sacB gene | Accession no. AJ868288 (5) |
| pHLYX810 | 2,338-bp PCR product of primers oHLYX5 and oHLYX6, containing the hlyX ORF, cloned into pCR 2.1-TOPO | This study |
| pHLYX110 | ApaI/NotI fragment carrying the hlyX ORF from pHLYX810, ligated into pBluescript SK(+) | This study |
| pHLYX111 | Deletion of an 883-bp fragment between the BgIII and XcmI restriction sites of pHLYX110 | This study |
| pHLYX701 | ApaI/NotI fragment from pHLYX111, ligated into pEMOC2 | This study |
| pLS88 | Broad-host-range shuttle vector from Haemophilus ducreyi; Strr Smr Kmr | 33 |
| pHLYX1300 | PCR product generated from primers oHLYX9 and oHLYX10 containing hlyX ORF, ligated into plasmid pLS88 after MfeI restriction in a 5′-3′ orientation with respect to the vector-derived sulII promoter | This study |
| pHLYX1301 | PCR product generated from primers oHLYX9 and oHLYX10 containing hlyX ORF ligated into plasmid pLS88 after MfeI restriction in a 3′-5′ orientation with respect to the vector-derived sulII promoter | This study |
| Primers | ||
| oHLYX5 | 5′-TGG GGC CCT CGG TAC AAC GGT ATG TCC TT-3′; forward primer containing an ApaI restriction site, situated 986 bp upstream of the hlyX start codon | This study |
| oHLYX6 | 5′-TCG CGG CCG CCA ACG TGA GAG CTT CGT TCA-3′; reverse primer containing a NotI restriction site, situated 626 bp downstream of the hlyX ORF | This study |
| oHLYX7 | 5′-TCC GAA ACC GGA TAA TTC AC-3′; forward primer situated 507 bp upstream of the hlyX start codon | This study |
| oHLYX8 | 5′-AGC GAA AGG GTT AAT CAG CA-3′; reverse primer situated 228 bp downstream of the hlyX ORF | This study |
| oHLYX9 | 5′-ATG ACA ATT GTT TTA AAA GAC GGT AGC CCT TAT G-3′; forward primer containing an MfeI restriction site, situated 20 bp upstream of the hlyX start codon | This study |
| oHLYX10 | 5′-CTT ACA ATT GCG CCT ATA CGG TCA GTT CGT-3′; | This study |
| reverse primer containing an MfeI restriction site situated 4 bp downstream of the hlyX ORF |
Ermr, erythromycin resistance; Tcr, tetracycline resistance; Kmr, kanamycin resistance; Strr, streptomycin resistance; Smr, sulfonamide resistance; Kmr, kanamycin resistance.
Media and growth conditions.
E. coli strains were cultured in LB medium supplemented with the appropriate antibiotics (ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml); for cultivation of E. coli β2155 (Δdap), diaminopimelic acid (1 mM; Sigma-Aldrich, Munich, Germany) was added. A. pleuropneumoniae strains were cultured in PPLO medium (Difco GmbH, Augsburg, Germany) supplemented with nicotinamide dinucleotide (10 μg/ml; Merck, Darmstadt, Germany), l-glutamine (100 μg/ml; Serva, Heidelberg, Germany), l-cysteine-hydrochloride (260 μg/ml; Sigma-Aldrich), l-cystine-dihydrochloride (10 μg/ml; Sigma-Aldrich), dextrose (1 mg/ml), and Tween 80 (0.1%) on Columbia Sheep Blood agar (Oxoid GmbH. Wesel, Germany) or on selective meat-blood agar (16). A. pleuropneumoniae transconjugants (single crossovers) and transformants were grown in PPLO medium containing chloramphenicol (5 μg/ml), and the medium for counterselection was prepared as described previously (28).
Anaerobic cultures used for the determination of aspartase activity and DmsA expression were prepared as described previously (2, 15). To compare growth of the different strains under anaerobic conditions, 100 ml of supplemented PPLO medium was preincubated in an anaerobic chamber for at least 48 h, inoculated with a single colony, and further incubated at 37°C for 16 h. Since all A. pleuropneumoniae strains used in the present study showed severe clumping under anaerobic conditions, bacterial growth was determined as dry pellet weight in triplicate. Bacteria were harvested by centrifugation, and the pellets were dried at 80°C for 24 h and then weighed. Statistical analysis of pellet weights was performed by using Student’s t test.
Manipulation of DNA.
DNA-modifying enzymes were purchased from New England Biolabs (Bad Schwalbach, Germany) and used according to the manufacturer's instructions. Taq polymerase was purchased from Gibco-BR Life Technologies (Karlsruhe, Germany). Chromosomal DNA for PCR and Southern blotting, as well as plasmid DNA, was prepared by standard protocols (22). PCR, Southern blotting, transformation, and gel electrophoresis were done by standard procedures (22), and pulsed-field gel electrophoresis was performed as described previously (19).
Electrotransformation.
Electrotransformation was performed by using a GenePulser (Bio-Rad, Munich, Germany) according to a published protocol (30) adapted to A. pleuropneumoniae. In brief, 250 ml of supplemented PPLO medium was inoculated with a 1/10 volume of an overnight culture, followed by incubation with shaking to an optical density at 600 nm (OD600) of 0.3. The culture was chilled on ice for 15 min, and bacteria were harvested by centrifugation at 4,000 × g and 4°C. Bacteria were washed three times by centrifugation in 150 ml of ice-cold GYTT medium (10% glycerol, 0.125% Bacto yeast [Difco], 0.25% Bacto tryptone [Difco], 0.02% Tween 80). Finally, bacteria were resuspended in a final volume of 2.5 ml of GYTT medium. Then, 5 μg of salt-free plasmid DNA was added to a 0.4-ml aliquot of competent cells, and electrotransformation was performed in 0.2-cm cuvettes with the settings 2.5 kV, 25 μFa, and 800 W. After electropulsing, bacteria were immediately added to 1 ml of prewarmed, supplemented PPLO medium, followed by incubation for 4 h at 37°C in a 5% CO2 incubator. Transformants were then plated on PPLO agar with chloramphenicol and incubated for 24 h.
Cloning of the A. pleuropneumoniae hlyX gene and construction of an isogenic deletion mutant.
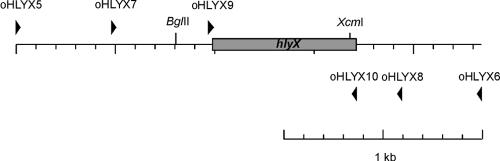
The hlyX gene (GenBank accession no. M80712) of A. pleuropneumoniae was amplified on a 2,338-bp fragment from genomic A. pleuropneumoniae AP76 DNA by using primers oHLYX5 and oHLYX6 (Table 1 and Fig. 1), which contained ApaI or NotI restriction endonuclease sites, respectively. After restriction with enzymes ApaI and NotI, the PCR product was ligated into pBluescript SK(+) to obtain plasmid pHLYX100. An 883-bp fragment was then deleted from the hlyX gene by using enzymes BglII and XcmI, and the plasmid was religated after fill-in with Klenow fragment to yield plasmid pHLYX101. The truncated hlyX gene was removed from pHLYX101 on an ApaI-NotI fragment and ligated into transconjugation plasmid pEMOC2, resulting in plasmid pHLYX701, which was then used to introduce the hlyX deletion into A. pleuropneumoniae via the single-step transconjugation system as described previously (5, 20) resulting in A. pleuropneumoniae ΔhlyX.
FIG. 1.
Map of the A. pleuropneumoniae ΔhlyX locus with primers and restriction sites used in the present study. Primer binding sites are indicated by arrowheads; restriction sites used are indicated by upward vertical lines.
Complementation of A. pleuropneumoniae ΔhlyX.
Plasmids pHLYX1300 and pHLYX1301 were constructed by cloning the 746-bp MfeI-restricted PCR product generated by primers oHLYX9 and oHLYX10 (Table 1 and Fig. 1), containing the entire hlyX open reading frame (ORF) without the putative promoter region, into EcoRI-restricted plasmid pLS88 (33) and electroporated into A. pleuropneumoniae ΔhlyX. In plasmid pHLYX1300 the orientation of the hlyX gene is in the orientation of transcription initiated by the vector-derived sulII promoter; in pHLYX1301 it is located in the opposite orientation.
Determination of aspartase and urease activity of A. pleuropneumoniae.
The aspartase test was performed by determination of fumarate formation as described previously (15). For testing urease activity individual colonies grown on supplemented PPLO agar were overlaid with 0.5% agarose containing 0.3 M urea and 0.01% phenol red (Sigma). The color of the colonies was assessed after 1 min. Urease-positive colonies turned red, whereas urease-negative colonies turned yellow.
Western blot analysis.
A. pleuropneumoniae whole-cell lysates were analyzed by discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10.8% acrylamide and 0.29% bisacrylamide) and Western blotting by using a Protean II Minigel system (Bio-Rad) as described previously (11). The serum directed against DmsA had been raised in rabbits as described previously (2).
Virulence studies.
Virulence of A. pleuropneumoniae ΔhlyX was assessed in an aerosol infection model previously described (4); A. pleuropneumoniae free and clinically healthy pigs (German Landrace) 7 to 9 weeks of age were randomly assigned to the different groups and cared for in accordance with the principles outlined in the European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes [European Treaty Series, no. 123: http://conventions.coe.int/treaty/EN/Menuprincipal.htm; permit no. 009i-(neu)42502-98/45] Clinical examinations were performed daily or as needed. Body temperature and clinical symptoms were recorded daily for each individual pig. A clinical scoring system based on the directive in the European Pharmacopoeia for testing A. pleuropneumoniae vaccines (porcine actinobacillosis vaccine [inactivated]) was used to assess the clinical condition of each individual animal as follows: a score of 1 was given for each occurrence of coughing, dyspnea, and vomitus, resulting in a minimum clinical score of 0 and a maximum score of 3 per day; the added daily clinical scores of days 1 to 7 were designated as the total clinical score. Statistical analysis of the total clinical score was performed by using Student’s t test. In order to confirm the absence of A. pleuropneumoniae-specific antibodies, blood samples were taken 1 week prior to infection; blood samples on days 7 and 21 postinfection were taken to determine the serological response to challenge with the different A. pleuropneumoniae strains by using the ApxII-ELISA (17). Postmortem analysis, as well as bacteriological, serological, and histological examinations, was performed as described previously (4). Briefly, lung lesion scores were determined as described by Hannan et al. (14) and statistically analyzed by using the Mann-Whitney test. The bacteriological examination included surface swabs of affected and unaffected lung tissue, palatine tonsils, bronchial lymph nodes, and heart muscle on Columbia sheep blood agar and selective meat-blood agar (16). Several individual A. pleuropneumoniae-like colonies were subcultured on supplemented PPLO agar and confirmed by urease assay and PCR analysis with primers oHLYX7 and oHLYX8.
Quantitative analysis of A. pleuropneumoniae in tonsils and sequestered lung tissue.
To determine the number of CFU of A. pleuropneumoniae still present 3 weeks after infection, 100 mg of tissue was processed in a FastPrep FP120 instrument (QBiogene, Heidelberg, Germany) with six sterile 3-mm glass beads (Roth, Karlsruhe, Germany) in 1 ml of NaCl (150 mM) twice for 40 s each time at a setting of 5.5. The number of A. pleuropneumoniae CFU was assessed by serial 10-fold dilutions and plating on selective meat-blood agar.
Luciferase assay.
The A. pleuropneumoniae ΔaspA::luxAB mutant, carrying a transcriptional fusion of aspA and luxAB, was used in a luciferase assay as described previously (15). To investigate the effect of reduced GSH on transcription of the aspA gene, A. pleuropneumoniae ΔaspA::luxAB was grown in liquid culture with shaking at 200 rpm to an OD600 of 0.5 to 0.6. This culture was split into 5-ml aliquots, and an equal volume of NaCl (150 mM) containing 1.2 μM, 12 μM, 120 μM, 600 μM, 1200 μM, or 12 mM GSH, respectively, was added to the culture medium. To one aliquot, NaCl without GSH was added to serve as a negative control. The cultures were further incubated with shaking for 1 h, reaching an OD600 of ca. 0.7 to 0.8. For the luciferase assay, 2.5 ml of each culture was immediately transferred to a Lumox 24-well plate (In Vitro Systems & Services GmbH, Göttingen, Germany). After the addition of 5 μl of 1% N-decyl-aldehyde (Sigma-Aldrich), the plate was exposed to X-ray film for 3 min. Signals were quantified by using Multi-Analyst/PC software (Bio-Rad).
RESULTS
Construction and functional characterization of the isogenic mutant A. pleuropneumoniae ΔhlyX in vitro.
A 883-bp deletion eliminating the 5′ end of the hlyX gene (Fig. 1) was introduced into A. pleuropneumoniae AP76 via conjugation with plasmid pHLYX701, followed by sucrose counterselection as described previously (28). The resulting A. pleuropneumoniae hlyX mutant was verified by using PCR, Southern blotting, and pulsed-field gel electrophoresis analyses (data not shown).
Due to the tendency of A. pleuropneumoniae to form clumps under anaerobic conditions, the difference in growth rate was assessed by comparison of dry pellet weights. Under anaerobic conditions, growth of the A. pleuropneumoniae hlyX mutant was decreased by 37.3% over a 16-h incubation period compared to the A. pleuropneumoniae parent strain (A. pleuropneumoniae AP76, 21.7 ± 1.15 mg; A. pleuropneumoniae ΔhlyX 13.6 ± 0.5 mg; P < 0.01).
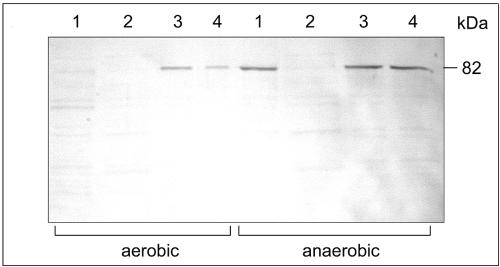
The mutant strain lacked the significant increase in aspartase activity under anaerobic conditions seen in the parent strain (Table 2). Aspartase activity was restored by introducing the hlyX gene on plasmid pHLYX1300 but not plasmid pHLYX1301 (Table 2). A. pleuropneumoniae ΔhlyX showed no expression of DmsA in Western blot analyses; expression was restored by either plasmid pHLYX1300 or pHLYX1301 under anaerobic conditions (Fig. 2).
TABLE 2.
Aspartase assay
| A. pleuropneumoniae strain | Mean increase in aspartase activity under anaerobic conditions ± SDa |
|---|---|
| Wild type | 191 ± 34 |
| Mutant ΔhlyX | 106 ± 21 |
| Mutant ΔhlyX + pHLYX 1300 | 202 ± 25 |
| Mutant ΔhlyX + pHLYX 1301 | 104 ± 13 |
Arithmetic mean of results from three independent experiments. The relative activity under aerobic conditions is 100%.
FIG. 2.
Expression of the A. pleuropneumoniae DmsA protein as assessed by Western blot analysis. Lanes: 1, A. pleuropneumoniae AP76; 2, A. pleuropneumoniae ΔhlyX; 3, A. pleuropneumoniae ΔhlyX transformed with pHLYX1300; 4, A. pleuropneumoniae ΔhlyX transformed with pHLYX1301.
Virulence studies.
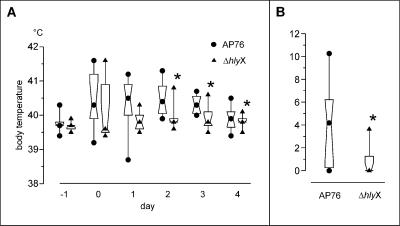
A. pleuropneumoniae ΔhlyX was used in an aerosol infection model and compared to the parent strain in an experiment with 18 German Landrace pigs that were randomly assigned to two groups. The cultures used for infection were grown to an OD600 of 0.39 (A. pleuropneumoniae parent strain, group 1) and to an OD600 of 0.44 (A. pleuropneumoniae ΔhlyX, group 2) and then diluted 1:30,000 as described previously (4). Pigs were infected in an aerosol chamber that holds four or five animals as described previously (4). Total infection doses aerosolized in 13 ml of diluted liquid culture as calculated from plate counts were 9.6 × 104 CFU for group 1 and 8.1 × 104 CFU for group 2. Aerosol infection led to an increase in body temperature to 40°C or higher in six out of nine animals in group 1 and in four out of nine animals in group 2 on day 1 postinfection. On days 2, 3, and 4 postinfection, the body temperatures in group 2 were significantly lower than in group 1 (P < 0.05, Fig. 3A). One animal in group 1 died on day 2 postinfection due to respiratory failure. All remaining animals were sacrificed on day 21 postinfection. At necropsy, six out of nine pigs in group 2 had no macroscopically visible lung lesions compared to two out of eight animals in group 1. The difference in lung lesion scores between the two groups was statistically significant (P < 0.05, Fig. 3B). Histological examination revealed no differences in the organization of lesions in both groups.
FIG. 3.
Virulence studies of A. pleuropneumoniae ΔhlyX in an aerosol infection model. Symbols: •, A. pleuropneumoniae AP76 wild-type strain (AP76); ▴, A. pleuropneumoniae ΔhlyX (ΔdmsA). The central symbol within the hourglass shape represents the geometric mean, the hinges present the values in the middle of each half of data, and the top and bottom symbols mark the maximum and minimum value. Asterisks denote statistical significance (P < 0.05) in the Wilcoxon signed-rank test. (A) Body temperatures of pigs over the course of 6 days, with day 0 marking the day of infection; (B) lung lesion scores assessed according to the method of Hannan et al. (14). Statistical significance (P < 0.05) in the Mann-Whitney Test is denoted by an asterisk.
In group 1, A. pleuropneumoniae was isolated as dense to confluent growth from sequesters in all six animals that exhibited lung lesions; however, due to contamination of two samples, quantitative analysis of bacterial counts in lung tissue was successful only in four animals (4.6 × 106 to 4 × 107 per g of tissue). In group 2, A. pleuropneumoniae was present in sequestered lung material in only two of the three pigs with lung lesions, at 2.6 × 105 and 3.4 × 105 per g of tissue, respectively. In animals from group 1, A. pleuropneumoniae could be isolated from tonsils (six of eight pigs, 6.2 × 104 to 3 × 107 CFU/g of tissue), tracheobronchial lymph nodes (four of eight pigs), and macroscopically unaltered lung tissue (seven of eight pigs). In contrast, no A. pleuropneumoniae could be isolated from these tissues in any of the animals in group 2.
Serum samples were obtained 1 week before and 3 weeks after experimental infection. Before the infection, all animals were seronegative. In group 1, seven of eight animals were seropositive; in contrast, in group 2, only the three animals that had lung lesions were serologically positive in the ApxII-ELISA (17; data not shown).
Influence of GSH on aspartase activity.
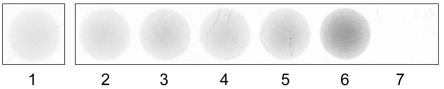
The reporter strain A. pleuropneumoniae Δ aspA::luxAB (15) was used to investigate the influence of GSH on aspartase activity. Liquid cultures were split at an OD600 of 0.53. Cultures were diluted 1:2 with 150 mM NaCl, resulting in an OD600 of 0.275. After 1 h of incubation, all cultures had reached an OD600 of 0.7 to 0.75 with the exception of the culture that had received 12 mM GSH, which only grew to an OD600 of 0.36. A gradual increase in luminescence as a result of increased aspartase activity was observed up to a GSH concentration of 1,200 μM, at which a threefold increase in activity was seen, whereas the culture that received 12 mM GSH showed no luciferase activity (Fig. 4). Upon addition of 80 μM oxidized GSH, which is the maximum physiological concentration that was measured in the murine lung model (8), no increase in luciferase activity could be induced (data not shown).
FIG. 4.
Influence of GSH on HlyX-induced activation of aspA transcription in a luciferase assay using A. pleuropneumoniae ΔaspA::luxAB. Lanes: 1, NaCl control; 2, 1.2 μM GSH; 3, 12 μM GSH; 4, 120 μM GSH; 5, 600 μM GSH; 6, 1,200 μM GSH; 7, 12 mM GSH.
DISCUSSION
In E. coli the FNR protein, a global response regulator containing four iron-sulfur clusters, upregulates the expression of a number of genes involved in anaerobic metabolism, such as the gene coding for DMSO reductase, and simultaneously represses genes involved in aerobic metabolism, such as the gene coding for cytochrome oxidase (6). In A. pleuropneumoniae the FNR homologue HlyX, which is able to complement fnr deletions in E. coli, has been identified (18). As we had shown in previous studies, genes containing putative HlyX binding motifs are expressed by A. pleuropneumoniae under the influence of bronchoalveolar lavage fluid from infected pigs, and deletion of these genes can cause attenuation of the organism (2, 15). In addition, an analysis of genes expressed in necrotic porcine lung tissue revealed the expression of DMSO reductase and several other genes involved in anaerobic metabolism under these conditions (1). This repeated isolation of presumably HlyX-regulated genes led to the hypothesis that deletion of this global anaerobic regulator would inhibit colonization and/or persistence in A. pleuropneumoniae if it was indeed involved in the regulation of the genes we identified. Since neither the regulation of A. pleuropneumoniae genes by HlyX nor the impact of the deletion of a global anaerobic regulator on the virulence of a respiratory tract pathogen had been investigated to date, we constructed a hlyX deletion mutant and examined it both in vitro and in vivo in an aerosol infection experiment.
The isogenic mutant A. pleuropneumoniae ΔhlyX shows a markedly reduced growth rate under anaerobic conditions in vitro, which implies that HlyX regulation is important but not essential for adaptation of A. pleuropneumoniae to anaerobiosis in culture. Our finding that both DMSO reductase expression and induction of aspartase activity are abolished in the hlyX-negative mutant supported our hypothesis that expression of these genes is regulated by HlyX (Fig. 2 and Table 2). In order to confirm that the effects we observed were specific for hlyX, we set up a complementation experiment reintroducing a plasmid-encoded hlyX gene into A. pleuropneumoniae ΔhlyX; in plasmid pHLYX1300 the hlyX gene is positioned in the orientation of transcription initiated by the plasmid-encoded sulII promoter, and in pHLYX1301 it is in the opposite orientation. Both DMSO expression and induction of aspartase activity under anaerobic conditions were restored in pHLYX1300 transformants. The finding that pHLYX1301 transformants also had a detectable DmsA expression could be explained by the incidental formation of a fusion promoter in plasmid pHLYX1301 that is able to initiate transcription. Sequence differences between the HlyX binding sites of dmsA (TTGAT—ATCAG) and aspA (GTGAT—ATCAC) could be responsible for the absence of this effect in the ΔaspA mutant. In addition, Western blot analysis may be more sensitive than the aspartase assay in the detection of the fusion promoter activity. The faint bands that are visible under aerobic conditions for the complemented strains may be due to the plasmid-encoded constitutive expression of HlyX.
In the challenge experiment with the A. pleuropneumoniae parent strain (group 1) and A. pleuropneumoniae ΔhlyX (group 2), we observed a significantly reduced virulence of the mutant strain. The finding that six of nine animals in group 2 had no macroscopically visible lung lesions and no detectable antibody titers implied that A. pleuropneumoniae ΔhlyX was clearly reduced in its ability to colonize, a prerequisite for inflicting lung damage. In contrast to the findings in group 2, six of the surviving eight animals in group 1 had severe lesions; additionally, one animal in this group died on day 2 postinfection.
The reduced colonizing and persisting ability of A. pleuropneumoniae ΔhlyX was confirmed by the complete lack of reisolation from tonsils and healthy lung epithelium 21 days postinfection; in contrast, the majority of animals challenged with the parent strain (group 1) still harbored the organisms in these tissues. Further, an hlyX deletion appears to also hamper long-term survival inside sequestered lung tissue, since A. pleuropneumoniae ΔhlyX was completely absent from the lung lesions of one animal in group 2, and the number of CFU per gram of material from the other two animals was 1 to 2 logs below the numbers found in animals in group 1. This difference likely is the result of ineffective adaptation of A. pleuropneumoniae ΔhlyX to anaerobic conditions in sequestered lung tissue and tonsils, and derepression of FNR-regulated genes involved in aerobic metabolism may add to this effect by causing the bacterium to waste energy. Together, these results confirm and extend the observations we made with mutant strains lacking DMSO reductase and aspartase activity (2, 15) and clearly show that genes under transcriptional control of the anaerobic regulator HlyX encode virulence-associated proteins required for bacterial survival in the presumably aerobic environment on the respiratory epithelium.
In an initial attempt to solve this apparent contradiction, we hypothesized that an airway antioxidant such as GSH or nitrogen species might be responsible. GSH is secreted by respiratory epithelial cells of a number of mammals including pigs and humans (7, 8, 34) as a primary line of defense against reactive oxygen or nitrogen species might be responsible. This hypothesis was particularly intriguing since GSH has recently been shown to be upregulated in the murine lung during infection with Pseudomonas aeruginosa (7, 8, 34). Further, GSH, on the one hand, reduces the redox potential of the epithelial lining fluid (ELF), a signal which has been shown to induce expression of FNR-regulated genes in E. coli (31). On the other hand, GSH has been shown to stabilize the iron-sulfur clusters required for the DNA-binding of FNR (29). Our finding that a concentration of 1,200 μM GSH induces luciferase expression in A. pleuropneumoniae ΔaspA::luxAB in culture supports this hypothesis, since this concentration has been calculated to be present in the murine lung (8); in addition, oxidized glutathione at a concentration of 80 μM, the maximum concentration observed in the murine lung (8), failed to achieve the same effect, which demonstrates that the effect we observed is specific for the reduced form of glutathione. The observed bactericidal activity of a GSH concentration of 1.2 mM is supported by reports of other groups investigating the role of GSH in the respiratory tract (13, 32). Therefore, these initial results imply that GSH is one of the factors in ELF that is sensed by A. pleuropneumoniae, thereby causing an HlyX-induced expression of virulence-associated proteins. Whether GSH acts by reducing oxygen tension or by direct interaction with the iron-sulfur clusters of HlyX and, likewise, which other factors in ELF might be recognized by the pathogen remains to be determined.
The fact that A. pleuropneumoniae ΔhlyX is still virulent and able to persist in sequestered lung tissue further demonstrates that HlyX is not essential for virulence or adaptation to oxygen-reduced conditions and therefore implies that other, as-yet-unidentified regulators are able to partially compensate for the loss of HlyX function. One possible candidate, the global regulator ArcA has recently been linked to virulence in Haemophilus influenzae and Vibrio cholerae (23, 27). ArcA has not been characterized in A. pleuropneumoniae; however, genomic sequence data available for A. pleuropneumoniae at http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi? under the sequence ID NZ_AACK01000005 reveal a putative response regulator protein that is 73% identical to the ArcA protein of H. influenzae (accession number NP_439045). Whether this putative ArcA protein plays a role in A. pleuropneumoniae virulence and, if so, how coordination of HlyX and ArcA regulation of virulence-associated genes occurs in the different compartments affected by an A. pleuropneumoniae infection (tonsils, sequestered lung, and intact respiratory epithelium) remains to be determined.
Acknowledgments
This study was supported by Sonderforschungsbereich 587 (Project A4) from the Deutsche Forschungsgemeinschaft, Bonn, Germany. I.D.J. is a fellow of the Graduiertenkolleg (GRK 745) of the Deutsche Forschungsgemeinschaft, and M.N. is a fellow of the Programme d'Appui aux Services Agricoles et aux Organisations Paysannes of the Institut d'Economie Rurale du Mali. Vector sequencing was supported by a grant given by the Bundesministerium fuer Bildung und Forschung within the BioProfile programme (project PTJ-BIO/0313037).
Editor: F. C. Fang
REFERENCES
- 1.Baltes, N., and G. F. Gerlach. 2004. Identification of genes transcribed by Actinobacillus pleuropneumoniae in necrotic porcine lung tissue by using selective capture of transcribed sequences. Infect. Immun. 72:6711-6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baltes, N., I. Hennig-Pauka, I. Jacobsen, A. D. Gruber, and G. F. Gerlach. 2003. Identification of dimethyl sulfoxide reductase in Actinobacillus pleuropneumoniae and its role in infection. Infect. Immun. 71:6784-6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baltes, N., S. Kyaw, I. Hennig-Pauka, and G. F. Gerlach. 2004. Lack of influence of the anaerobic [NiFe] hydrogenase and l-1,2 propanediol oxidoreductase on the outcome of Actinobacillus pleuropneumoniae serotype 7 infection. Vet. Microbiol. 102:67-72. [DOI] [PubMed] [Google Scholar]
- 4.Baltes, N., W. Tonpitak, G. F. Gerlach, I. Hennig-Pauka, A. Hoffmann-Moujahid, M. Ganter, and H. J. Rothkotter. 2001. Actinobacillus pleuropneumoniae iron transport and urease activity: effects on bacterial virulence and host immune response. Infect. Immun. 69:472-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baltes, N., W. Tonpitak, I. Hennig-Pauka, A. D. Gruber, and G. F. Gerlach. 2003. Actinobacillus pleuropneumoniae serotype 7 siderophore receptor FhuA is not required for virulence. FEMS Microbiol. Lett. 220:41-48. [DOI] [PubMed] [Google Scholar]
- 6.Bauer, C. E., S. Elsen, and T. H. Bird. 1999. Mechanisms for redox control of gene expression. Annu. Rev. Microbiol. 53:495-523. [DOI] [PubMed] [Google Scholar]
- 7.Cantin, A. M., S. L. North, R. C. Hubbard, and R. G. Crystal. 1987. Normal alveolar epithelial lining fluid contains high levels of glutathione. J. Appl. Physiol. 63:152-157. [DOI] [PubMed] [Google Scholar]
- 8.Day, B. J., A. M. van Heeckeren, E. Min, and L. W. Velsor. 2004. Role for cystic fibrosis transmembrane conductance regulator protein in a glutathione response to bronchopulmonary Pseudomonas infection. Infect. Immun. 72:2045-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dehio, C., and M. Meyer. 1997. Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J. Bacteriol. 179:538-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fenwick, B., and S. Henry. 1994. Porcine pleuropneumonia. J. Am. Vet. Med. Assoc. 204:1334-1340. [PubMed] [Google Scholar]
- 11.Gerlach, G. F., C. Anderson, A. A. Potter, S. Klashinsky, and P. J. Willson. 1992. Cloning and expression of a transferrin-binding protein from Actinobacillus pleuropneumoniae. Infect. Immun. 60:892-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green, J., and M. L. Baldwin. 1997. HlyX, the FNR homologue of Actinobacillus pleuropneumoniae, is a [4Fe-4S]-containing oxygen-responsive transcription regulator that anaerobically activates FNR-dependent class I promoters via an enhanced AR1 contact. Mol. Microbiol. 24:593-605. [DOI] [PubMed] [Google Scholar]
- 13.Green, R. M., A. Seth, and N. D. Connell. 2000. A peptide permease mutant of Mycobacterium bovis BCG resistant to the toxic peptides glutathione and S-nitrosoglutathione. Infect. Immun. 68:429-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hannan, P. C., B. S. Bhogal, and J. P. Fish. 1982. Tylosin tartrate and tiamutilin effects on experimental piglet pneumonia induced with pneumonic pig lung homogenate containing mycoplasmas, bacteria, and viruses. Res. Vet. Sci. 33:76-88. [PubMed] [Google Scholar]
- 15.Jacobsen, I., I. Hennig-Pauka, N. Baltes, M. Trost, and G. F. Gerlach. 2005. Enzymes involved in anaerobic respiration appear to play a role in Actinobacillus pleuropneumoniae virulence. Infect. Immun. 73:226-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobsen, M. J., and J. P. Nielsen. 1995. Development and evaluation of a selective and indicative medium for isolation of Actinobacillus pleuropneumoniae from tonsils. Vet. Microbiol. 47:191-197. [DOI] [PubMed] [Google Scholar]
- 17.Leiner, G., B. Franz, K. Strutzberg, and G. F. Gerlach. 1999. A novel enzyme-linked immunosorbent assay using the recombinant Actinobacillus pleuropneumoniae ApxII antigen for diagnosis of pleuropneumonia in pig herds. Clin. Diagn. Lab. Immunol. 6:630-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacInnes, J. I., J. E. Kim, C. J. Lian, and G. A. Soltes. 1990. Actinobacillus pleuropneumoniae hlyX gene homology with the fnr gene of Escherichia coli. J. Bacteriol. 172:4587-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oswald, W., D. V. Konine, J. Rohde, and G. F. Gerlach. 1999. First chromosomal restriction map of Actinobacillus pleuropneumoniae and localization of putative virulence-associated genes. J. Bacteriol. 181:4161-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oswald, W., W. Tonpitak, G. Ohrt, and G. Gerlach. 1999. A single-step transconjugation system for the introduction of unmarked deletions into Actinobacillus pleuropneumoniae serotype 7 using a sucrose sensitivity marker. FEMS Microbiol. Lett. 179:153-160. [DOI] [PubMed] [Google Scholar]
- 21.Raleigh, F. A., K. Lech, and R. Brent. 1989. Selected topics from classical bacterial genetics, p. 1.4.1-1.4.14. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, et al. (ed.), Current protocols in molecular biology. Wiley Interscience, New York, N. Y. [DOI] [PubMed]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Sengupta, N., K. Paul, and R. Chowdhury. 2003. The global regulator ArcA modulates expression of virulence factors in Vibrio cholerae. Infect. Immun. 71:5583-5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaw, D. J., and J. R. Guest. 1982. Nucleotide sequence of the fnr gene and primary structure of the Enr protein of Escherichia coli. Nucleic Acids Res. 10:6119-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shuman, S. 1994. Novel approach to molecular cloning and polynucleotide synthesis using vaccinia DNA topoisomerase. J. Biol. Chem. 269:32678-32684. [PubMed] [Google Scholar]
- 26.Soltes, G. A., and J. I. MacInnes. 1994. Regulation of gene expression by the HlyX protein of Actinobacillus pleuropneumoniae. Microbiology 140:839-845. [DOI] [PubMed] [Google Scholar]
- 27.Souza-Hart, J. A., W. Blackstock, M. Di, V., I. B. Holland, and M. Kok. 2003. Two-component systems in Haemophilus influenzae: a regulatory role for ArcA in serum resistance. Infect. Immun. 71:163-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tonpitak, W., N. Baltes, I. Hennig-Pauka, and G. F. Gerlach. 2002. Construction of an Actinobacillus pleuropneumoniae serotype 2 prototype live negative-marker vaccine. Infect. Immun. 70:7120-7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tran, Q. H., T. Arras, S. Becker, G. Holighaus, G. Ohlberger, and G. Unden. 2000. Role of glutathione in the formation of the active form of the oxygen sensor FNR ([4Fe-4S] · FNR) and in the control of FNR function. Eur. J. Biochem. 267:4817-4824. [DOI] [PubMed] [Google Scholar]
- 30.Tung, W. L., and K. C. Chow. 1995. A modified medium for efficient electrotransformation of Escherichia coli. Trends Genet. 11:128-129. [DOI] [PubMed] [Google Scholar]
- 31.Unden, G., S. Achebach, G. Holighaus, H. G. Tran, B. Wackwitz, and Y. Zeuner. 2002. Control of FNR function of Escherichia coli by O2 and reducing conditions. J. Mol. Microbiol. Biotechnol. 4:263-268. [PubMed] [Google Scholar]
- 32.Venketaraman, V., Y. K. Dayaram, A. G. Amin, R. Ngo, R. M. Green, M. T. Talaue, J. Mann, and N. D. Connell. 2003. Role of glutathione in macrophage control of mycobacteria. Infect. Immun. 71:1864-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willson, P. J., W. L. Albritton, L. Slaney, and J. K. Setlow. 1989. Characterization of a multiple antibiotic resistance plasmid from Haemophilus ducreyi. Antimicrob. Agents Chemother. 33:1627-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yam, J., and R. J. Roberts. 1980. Oxygen-induced lung injury in the newborn piglet. Early Hum. Dev. 4:411-424. [DOI] [PubMed] [Google Scholar]