Abstract
A novel DNA assay demonstrating sensitive and accurate detection of Helicobacter pylori from stool samples is reported. Moreover, in three individuals tested for therapeutic response, the assay showed the disappearance of H. pylori DNA during treatment. Thus, this noninvasive molecular biology-based assay has the potential to be a powerful diagnostic tool given its ability to specifically identify H. pylori DNA.
Helicobacter pylori infection is recognized as a major causative agent of gastritis and peptic ulcer disease and as a carcinogen responsible for gastric carcinoma and lymphoma (6, 8, 11). To date, none of the available clinical tests for H. pylori infection has been ideal, with each of the tests observed to have limitations (2). At present, only the PCR assay of gastric biopsy specimens, the retrieval of which is invasive for the patient, has sensitivity and specificity that approaches 100%.
Novel, efficient methods of stool specimen processing and specific DNA extraction have recently been developed by the Exact Sciences Corporation (Maynard, Mass.) (1). Herein, we report on the use of this method to optimize detection of H. pylori DNA shed in the stool. The importance of H. pylori DNA detection in diagnostic tests is supported by the capability to genotype infecting strains and to identify virulence factors and markers of resistance to specific antibiotic treatments (3, 5, 7, 9).
Stool samples from 25 patients who underwent upper endoscopy were collected and stored frozen for molecular biology-based analyses. Twenty-two of these stool samples were obtained from patients (patients 1 through 22) for whom tests were being performed to detect H. pylori infection prior to H. pylori treatment (4). Stool samples were collected before and after treatment from three additional patients with histologically documented infection (patients 23 through 25).
Four grams of frozen stool from each patient were homogenized in 28 ml of EXACT Buffer A (Exact Sciences Corporation). Following homogenization, each sample was centrifuged twice to remove particulate matter and the supernatant was incubated with DNase-free RNase. The DNA was then precipitated with 300 mM sodium acetate and isopropanol, centrifuged, and resuspended in TE buffer (10 mM Tris, 1 mM EDTA [pH 7.4]).
Next, the human APC (adenomatous polyposis coli) and the Helicobacter 16S rRNA genes were captured with oligonucleotide capture probes specific for each target gene (H. pylori-specific probe, biotin-GGGGAGTACGGTCGCAAGATTAAAACTCAAAGGAATA; APC-specific probe, biotin-CAGATAGCCCTGGACAAACCATGCCACCAAGCAGAAG). The extracted DNA was combined with an equal volume of 6 M guanidinium isothiocyanate and 20 nmol of each sequence-specific biotinylated oligonucleotide capture probe for incubation and subsequent magnetic separation of target DNA complex sequences with paramagnetic polystyrene beads coated with streptavidin (Dynabeads M-280 Streptavidin; Dynal ASA, Oslo, Norway), as described previously (1).
H. pylori- and human APC-specific PCR amplifications were performed with primers designed to be complementary to 16S rRNA sequences unique to H. pylori and sequences within exon 15 of the human APC gene, respectively, as published previously (1, 4, 10). As positive controls, gene sequences were amplified from commercial sources of H. pylori genomic DNA (catalog no. 43504D; American Type Culture Collection, Manassas, Va.) and human genomic DNA (catalog no. 1691112; Roche Molecular Biochemicals, Indianapolis, Ind.). PCR amplifications were performed under standard PCR conditions (Perkin-Elmer) with 50-μl reaction mixtures, which included 10 μl of sample eluant and the PCR primers at concentrations of 500 nM. The conditions used with the thermocycler instrument (MJ Research, Waltham, Mass.) consisted of an initial denaturation (94°C for 5 min), followed by 40 cycles of denaturation (94°C for 1 min), annealing (60°C for 1 min), and extension (72°C for 1 min) and a final extension at 72°C for 5 min. The PCR products (8 μl) were resolved by agarose (4% NuSieve 3:1 agarose; FMC Corporation) gel electrophoresis. Following electrophoresis, the gels were stained with ethidium bromide and the reactions were scored as positive or negative by visual analysis and image analysis (Eagle Eye II still video system; Stratagene, La Jolla, Calif.).
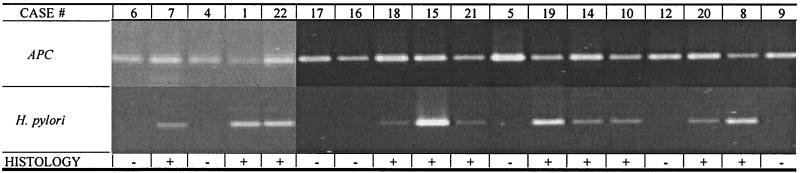
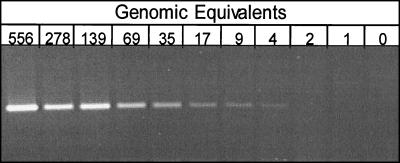
DNA was first extracted from stool samples collected from 22 patients prior to treatment and previously characterized histologically for H. pylori infection. PCR amplification of these DNA templates with the human APC-specific primers was initially performed, and the expected 184-bp PCR product and amplifiable DNA were demonstrated in all 22 samples (Fig. 1). Amplification PCRs were then performed with the H. pylori-specific primers. The expected 204-bp PCR product was clearly seen in samples from all 11 patients who had tested positive for H. pylori infection by clinical tests, including histologic and DNA examinations of gastric biopsy specimens (100% sensitivity) (Fig. 1). Additionally, stool samples from all 11 patients with negative test results for H. pylori infection, as determined by histologic and PCR analyses of gastric biopsy specimens, also had negative results by the DNA-based assay (specificity, 100%). These findings were reproducible in multiple independent analyses. Control H. pylori DNA samples were diluted severalfold and were analyzed as described above to confirm the sensitivity of the assay, which demonstrated that as few as four bacterial cells or genomic equivalents of H. pylori DNA could be detected (Fig. 2).
FIG. 1.
Specific detection of H. pylori DNA in stool specimens. Illustrated are representative results for samples displaying human APC- and H. pylori-specific amplification products. DNA was captured from stool samples with sequence-specific probes and was subsequently amplified by PCR with either human APC- or H. pylori-specific primers. The top panel demonstrates the presence of amplifiable DNA in each of the samples with human APC-specific primers. The bottom panel demonstrates amplification of the expected 204-bp PCR product with H. pylori-specific primers for samples with H. pylori infection documented by positive histologic results.
FIG. 2.
Determination of assay sensitivity. Control H. pylori DNA was serially diluted from 556 to 0 genomic equivalents. Each dilution was amplified with H. pylori sequence-specific primers, followed by electrophoresis to detect DNA amplification. The presence of a band at 4 genomic equivalents demonstrates that the detection of H. pylori DNA was possible down to 4 genomic equivalents.
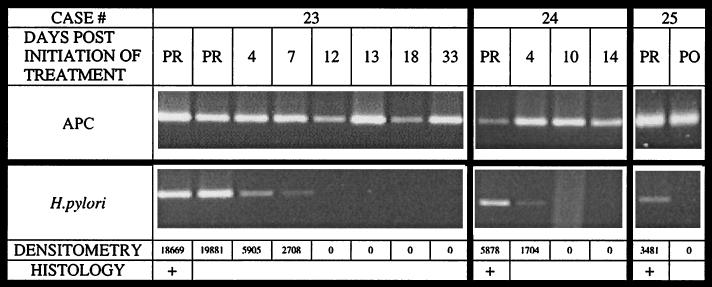
Next, multiple stool samples collected at different time points before, during, and after treatment in three patients with histologically documented infection were analyzed. Treatment for these patients was administered for 10 days by a Prevpac (TAP Pharmaceuticals). In PCRs with human APC-specific primers, amplifiable DNA was found in all samples from these three patients (Fig. 3). Amplification with H. pylori-specific primers demonstrated the presence of H. pylori DNA in stool samples collected before treatment and the absence of H. pylori DNA after the termination of treatment (Fig. 3). Moreover, amplification with H. pylori-specific primers demonstrated the disappearance of bacterial DNA during therapy between days 7 and 12 of treatment for patient 23 and days 4 and 10 of treatment for patient 24. Densitometry readings confirmed the gradual disappearance of the H. pylori-specific PCR product as treatment progressed, with the relative intensity of the PCR product decreasing by day 4 of treatment for both patients 23 and 24.
FIG. 3.
H. pylori DNA in stool before and after treatment. The results show the amplification of stool samples collected at multiple time points from patients 23 to 25 with human APC- and H. pylori-specific primers. PR, pretreatment; PO, posttreatment. DNA was captured from stool samples with sequence-specific probes and was subsequently amplified by PCR with either APC- or H. pylori-specific primers as described in the legend to Fig. 1. The top panel demonstrates the presence of amplifiable DNA in each of the samples. The second panel demonstrates the disappearance of H. pylori DNA during treatment. The gradual decrease begins by day 4 postinitiation of treatment for both patients 23 and 24. The complete eradication of bacterial DNA occurs between days 7 and 12 and between days 4 and 10 for patients 23 and 24, respectively. These results are confirmed by the densitometry readings determined for these samples, as indicated in the bottom panel.
We have reported herein on the development of a novel, noninvasive, molecular biology-based assay capable of accurately detecting H. pylori infection. Further studies with large numbers of subjects are planned to confirm these high rates of sensitivity and specificity of the assay in practice. Moreover, this assay can be automated at many steps to help make it minimally labor-intensive for clinical use.
In analyzing stool samples collected from patients at different time points before, during, and after H. pylori treatment, the absence of detectable H. pylori DNA could be observed at least 12 days after treatment initiation, illustrating the potential ability of this test to document the efficacy of H. pylori treatment. Confirmation of H. pylori clearance in those with complicated peptic ulcer disease or mucosa-associated lymphoid tissue lymphomas might be accomplished by this test with stool specimens, which can be recovered by noninvasive means. Further studies with more subjects who are treated for H. pylori infection and who receive follow-up tests are planned to determine the clinical utility of this assay in the documentation of eradication. Finally, the potential of the DNA-based assay described here to genotype infecting strains of H. pylori by noninvasive means has important clinical implications. Certain H. pylori genotypes have been found to be associated with more virulent infections (e.g., genotypes that include the cagA gene) and resistance to certain antibiotic treatments (e.g., genotypes with mutations in domain V of the 23S rRNA gene or the rdxA gene), and these might be detectable by a DNA-based test such as the one described here (3, 5, 7, 9). Thus, our molecular biology- and DNA-based assay might ultimately possess clinical utility for the efficient identification of infected individuals, help guide their antibiotic treatment, and document eradication soon after therapy.
Acknowledgments
Support for this study was received through NCI grant CA67900-06.
REFERENCES
- 1.Ahlquist, D. A., J. E. Skoletsky, K. A. Boynton, J. J. Harrington, D. W. Mahoney, W. E. Pierceall, S. N. Thibodeau, and A. P. Shuber. 2000. Colorectal cancer screening by detection of altered human DNA in stool: feasibility of a multitarget assay panel. Gastroenterology 119:1219–1227. [DOI] [PubMed] [Google Scholar]
- 2.Bravos, E. D., and R. H. Gilman. 2000. Accurate diagnosis of Helicobacter pylori—other tests. Gastroenterol. Clin. N. Am. 29:925–929. [DOI] [PubMed] [Google Scholar]
- 3.Gibson, J. R., N. A. Saunders, B. Burke, and R. J. Owen. 1999. Novel method for rapid determination of clarithromycin sensitivity in Helicobacter pylori. J. Clin. Microbiol. 37:3746–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gramley, W. A., A. Asghar, H. F. Frierson, Jr., and S. M. Powell. 1999. Detection of Helicobacter pylori DNA in fecal samples from infected individuals. J. Clin. Microbiol. 37:2236–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeong, J. Y., A. K. Mukhopadhyay, D. Dailidiene, Y. Wang, B. Velapatino, R. H. Gilman, A. J. Parkinson, G. B. Nair, B. C. Wong, S. K. Lam, R. Mistry, I. Segal, Y. Yuan, H. Gao, T. Alarcon, M. L. Brea, Y. Ito, D. Kersulyte, H. K. Lee, Y. Gong, A. Goodwin, P. S. Hoffman, and D. E. Berg. 2000. Sequential inactivation of rdxA (HP0954) and frxA (HP0642) nitroreductase genes causes moderate and high-level metronidazole resistance in Helicobacter pylori. J. Bacteriol. 182:5082–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marshall, B., and J. Armstrong. 1995. Attempt to prove Koch’s postulates for pylori campylobacter. Med. J. Aust. 142:436–439. [DOI] [PubMed] [Google Scholar]
- 7.Megraud, F., N. Lehn, T. Lind, E. Bayerdorffer, C. O’Morain, R. Spiller, P. Unge, S. V. van Zanten, M. Wrangstadh, and C. F. Burman. 1999. Antimicrobial susceptibility testing of Helicobacter pylori in a large multicenter trial: the MACH 2 study. Antimicrob. Agents Chemother. 43:2747–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parsonnet, J., G. Friedman, D. Vandersteen, Y. Chang, J. H. Vogelman, N. Orentreich, and R. K. Sibley. 1991. Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med. 325:1127–1131. [DOI] [PubMed] [Google Scholar]
- 9.Tankovic, J., D. Lamarque, J. C. Delchier, C. J. Soussy, A. Labigne, and P. J. Jenks. 2000. Frequent association between alteration of the rdxA gene and metronidazole resistance in French and North African isolates of Helicobacter pylori. Antimicrob. Agents Chemother. 44:608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiss, J., J. Mecca, E. Silva, and D. Gassner. 1994. Comparison of PCR and other diagnostic techniques for detection of Helicobacter pylori infection in dyspeptic patients. J. Clin. Microbiol. 32:1663–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wotherspoon, A. C., C. Ortiz-Hidalgo, M. F. Falzon, et al. 1993. Helicobacter pylori-associated gastritis and primary B-cell lymphoma. Lancet 338:1175–1176. [DOI] [PubMed] [Google Scholar]