Abstract
The adherence of pathogenic Escherichia coli strains to intestinal epithelium is essential for initiation of infection. The cad operon encodes the lysine decarboxylase (LDC) system responsible for metabolizing lysine, and this operon has been proposed as an antivirulence mechanism in enteroinvasive E. coli and Shigella flexneri and as a factor mediating E. coli O157:H7 adherence. We sought to determine whether the LDC activity was present in a phylogenetically characterized collection of diarrheagenic E. coli (DEC) strains and to establish whether its expression was associated with their adherence to tissue culture cells. LDC activity was found in most of the pathogenic E. coli strains tested and was absent from Shiga toxin-producing E. coli (STEC) O111 strains (DEC pathotype 8). Analysis of the cad region in these O111 strains indicates that the operon has been rearranged and some of the genes are either missing or disrupted. A similar rearrangement was found in an E. coli O111:H8 strain recently isolated from an outbreak in Texas. Complementation of the LDC-negative strains with the cad operon in trans restored the LDC activity and resulted in a reduction in adherence to tissue culture cells. Initial analysis of the protein profiles on the surface of the O111 strains indicates that the LDC activity has an effect on the expression of the adhesin intimin. Cadaverine had a slight effect on LDC-negative strain adhesion but none on intimin expression. Our data suggest that this pathoadaptive mutation is an important mechanism to control functions potentially implicated in the pathogenesis of these organisms.
Shiga toxin-producing Escherichia coli (STEC) strains are a group of zoonotic enteric pathogens which include multiple serotypes associated with a broad spectrum of human illness ranging from uncomplicated diarrhea to hemorrhagic colitis and hemolytic uremic syndrome (27, 38). In the United States alone, STEC cause an estimated 110,000 illnesses and 90 deaths annually (34). While many of the outbreaks associated with STEC infections have been related to serotype O157:H7, recent data indicated that the incidence of STEC O157 cases has declined substantially (15). In contrast, several non-O157 STEC have been isolated more frequently from stool samples and from patients suffering from hemolytic uremic syndrome (2, 23, 26), including those from serogroups O26, O91, O103, O111, and O113 (38, 46).
Two distinct mechanisms are involved in the evolution of commensal E. coli toward pathogenic phenotypes: the acquisition of additional genes encoding virulence determinants (generally carried on plasmids, pathogenicity islands [PAIs], or phages) and the loss or modification of existing genetic material (pathogenicity-adaptive or pathoadaptive mutations) (43, 53). In the case of STEC strains, the production of Shiga toxins Stx1 and Stx2 and the expression of the PAI-encoded intimin adhesin or the plasmid-encoded enterohemolysin are some examples of acquisition of virulence determinants which are in straight association with human disease (25, 38). In contrast, the functional modification or elimination of an original gene(s) associated with a pathoadaptive mutation has not been clearly established in STEC strains.
A pathoadaptive mutation improves survival within host tissues, increases the pathogenic potential of the bacteria, and drives the evolution of bacteria toward an enhanced pathogenic phenotype (53). One of the best examples of pathoadaptive mutation in enteric pathogens was described by Maurelli et al., who proposed that cadA acts as an antivirulence gene in enteroinvasive E. coli and Shigella spp. (32). The cadA gene encodes the lysine decarboxylase enzyme, responsible for metabolizing lysine, and these investigators found that loss to lysine decarboxylase (LDC) activity was due to a large chromosomal deletion comprising the cadA region (16). Further studies indicated that introduction of cadA in S. flexneri affects the activity of two enterotoxins and the presence of the by-product cadaverine, generated from the decarboxylation of lysine, caused attenuation in S. flexneri virulence (18, 33).
We recently found that the product of the cadA gene is one of multiple factors mediating STEC O157:H7 adherence to tissue culture cells (58). Our data showed that a transposon insertion inactivating cadA produced a hyperadherent STEC O157:H7 strain and suggested that CadA or the by-products from LDC metabolism have a role in STEC adhesion. In the case of STEC strains, adherence to intestinal epithelium is essential for initiation of infection. Intimin (the product of the eae gene) is an outer membrane adhesin that mediates intimate attachment to the host cell and is essential for the establishment of the attaching and effacing lesion (24). Intimin has been associated with the pattern of colonization and tissue tropism in the host (19, 48) and in the case of STEC O157:H7, intimin expression correlates with colonization of the human large intestine follicle-associated epithelium (19, 49).
Additional adhesins have been proposed to play an important role in STEC intestinal colonization. One of these is the product of the efa-1 (EHEC factor for adherence) gene found in an STEC O111:H− isolate which mediates adherence to cultured Chinese hamster ovary (CHO) cells (39). When a mutation is introduced in the efa-1 gene of other STEC non-O157 strains (serotypes O5 and O111) and its role in colonization of the intestinal tract of calves is tested, the results have shown that Efa-1 is required for colonization (54). In the current study, we first investigated whether the LDC activity was present in all members of a phylogenetically characterized collection of diarrheagenic E. coli (DEC) strains (61) and then established whether LDC expression was a pathoadaptive mutation associated with STEC O111 adhesion to tissue culture cells.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains used in this study are listed in Table 1. Diarrheagenic E. coli (DEC) strains were obtained from the STEC Center at Michigan State University (61; http://www.shigatox.net/stec/). E. coli strain O111:H8 isolated from an outbreak in Texas was obtained from the Texas Department of Health. Strains were routinely grown in Luria-Bertani (LB) broth or on L agar at 37°C (30). pKER65 (Mu d5005 cad+ mel+ adi+) (35) was obtained from T. Maurelli.
TABLE 1.
Bacterial strains used in this study and their lysine decarboxylase activity
| Strain | Serotype | DEC no.a | LDC test | Strain | Serotype | DEC no.a | LDC test | |
|---|---|---|---|---|---|---|---|---|
| EPEC strains | ||||||||
| 572-56 | O55:H6 | 1A | + | |||||
| C54-58 | O55:H6 | 1B | + | |||||
| F196-5 | O55:H6 | 1C | + | |||||
| F563-55 | O55:H6 | 1D | + | |||||
| AC-C2 | O55:H6 | 1E | + | |||||
| 3787-62 | O55:H6 | 2A | + | |||||
| 5513-56 | O55:NM | 2B | + | |||||
| 607-54 | O55:H6 | 2C | + | |||||
| F60-51 | O55:H6 | 2D | + | |||||
| 2087-77 | O55:H6 | 2E | + | |||||
| 5624-50 | O55:H7 | 5A | + | |||||
| 660-79 | O55:H7 | 5B | + | |||||
| 5380-66 | O55:H7 | 5C | + | |||||
| C586-65 | O55:H7 | 5D | + | |||||
| C997-63 | O55:H7 | 5E | + | |||||
| 5338-66 | O111:H21 | 6A | + | |||||
| C142-54 | O111:H12 | 6B | + | |||||
| 2277-67 | O111:H12 | 6C | + | |||||
| F436-51 | O111:H12 | 6D | + | |||||
| 184-83 | O111:NM | 6E | + | |||||
| 2254-75 | O128a:H2 | 11A | + | |||||
| 3733-71 | O128a:H2 | 11B | + | |||||
| A9619-C2 | O45:H2 | 11C | + | |||||
| EHEC strains | ||||||||
| 3299-85 | O157:H7 | 3A | + | |||||
| 46240 | O157:H7 | 3B | + | |||||
| 3104-88 | O157:H7 | 3C | + | |||||
| 3009-88 | O157:H7 | 3D | + | |||||
| 3077-88 | O157:H7 | 3E | + | |||||
| 493/89 | O157:H- | + | ||||||
| C1520-77 | O157:H7 | 4A | + | |||||
| C999-87 | O157:H7 | 4B | + | |||||
| C374-83 | O157:H7 | 4C | + | |||||
| C681-87 | O157:H7 | 4D | + | |||||
| C7-88 | O157:H7 | 4E | + | |||||
| EDL-933 | O157:H7 | + | ||||||
| 2198-77 | O111a:NM | 8A | − | |||||
| 3030A-86 | O111:H8 | 8B | − | |||||
| 86-10049 | O111:NM | 8C | + | |||||
| C130-53 | O111:H11 | 8D | + | |||||
| C194-65 | O111:H8 | 8E | − | |||||
| Texas outbreakb | O111:H8 | − | ||||||
| 3323-61 | O26:H11 | 9A | + | |||||
| 2262-79 | O26:N | 9B | − | |||||
| C240-52 | O26:N | 9C | + | |||||
| C814-67 | O26:H11 | 9D | + | |||||
| 45 | O26:H11 | 9E | + | |||||
| E335021/OF | O128:H2 | 11D | ||||||
| WM63 | O128:H2 | 11E | + | |||||
| F1-50 | O111:H2 | 12A | ||||||
| 2966-56 | O111:H2 | 12B | + | |||||
| 3942-67 | O111:NM | 12C | + | |||||
| 9101-83 | O111:H2 | 12D | ||||||
| 3291-86 | O111:N | 12E | + | |||||
| C916-70 | O128:H21 | 14A | + | |||||
| C691-71 | O128:H21 | 14B | + | |||||
| 9088-83 | O128a:H21 | 14C | + | |||||
| 1791-79 | O128:NM | 14D | + | |||||
| C639-77 | O128:H21 | 14E | + | |||||
| 5430-66 | O111:H21 | 15A | + | |||||
| 448-71 | O111:H21 | 15B | + | |||||
| 2660-77 | O111:H21 | 15C | + | |||||
| 2708-78 | O111:H21 | 15D | + | |||||
| 2394-80 | O111:H21 | 15E | + | |||||
| RDEC strain | ||||||||
| RDEC-1 | O15:NM | + | ||||||
| H30 | O26:H11 | + | ||||||
| 3047-86 | O26:H11 | 10B | + | |||||
| 1557-77 | O26:H11 | 10C | + | |||||
| C12-52 | O26:H11 | 10D | + | |||||
| 900105 | O26:H11 | 10E | + | |||||
| ETEC strains | ||||||||
| 75001 | O157:H43 | 7A | + | |||||
| 902034 | O149:NM | 7B | + | |||||
| 820691 | O157:H43 | 7C | + | |||||
| 831015 | O157:H43 | 7D | + | |||||
| 861575 | O157:NM | 7E | + | |||||
| 3350-73 | O128:H7 | 13A | + | |||||
| 5024-71 | O128:H7 | 13B | + | |||||
| C500-74 | O128:H7 | 13C | + | |||||
| C1083-79B | O128:H7 | 13D | + | |||||
| 2384-81 | O128:H47 | 13E | + | |||||
| E. coli K-12 strains | ||||||||
| MG1655c | + |
To construct plasmid pCadABC, the cad operon was obtained by restriction digestion from plasmid pSM10 (36) and cloned as an EcoRI/BamHI fragment in pBR322. The cadA gene was amplified by PCR from EHEC O157:H7 strain EDL933 using primer pair P7 and P8 (Table 2) and the 3.8-kb fragment was cloned into the pGEMT-Easy vector (Promega) to create pCadA. Plasmids were introduced into E. coli by electroporation as described by Dower et al. (17). When indicated, the bacterial strains were grown in Dulbecco's minimal Eagle's medium (DMEM) (Gibco/Invitrogen cat. no. 11885-084) at 37°C. Antibiotics were added to media at the following concentrations: kanamycin, 50 μg/ml; ampicillin, 100 μg/ml; chloramphenicol, 30 μg/ml; and streptomycin, 100 μg/ml.
TABLE 2.
Primers used in this study.
| Primer | Sequence |
|---|---|
| P1 | 5′-TTTACGGCAGGAAACACGCG-3′ |
| P2 | 5′-GACCGATTTAGGCCATACGCTG-3′ |
| P3 | 5′-CGCGTCATATCACTGATTTGCG-3′ |
| P4 | 5′-ACTTGGGACAAAAAAACGCGC-3′ |
| P5 | 5′-AGTTGAATACGGCCACCG-3′ |
| P6 | 5′-ATCGGCGCTCACTATCCG-3′ |
| P7 | 5′-CCGGATCCGGACCAGTCGTTCTC-3′ |
| P8 | 5′-CCTCTAGAATACTGCGGATACCACTG-3′ |
| P9 | 5′-GCGATGAAGTTCACGGAT-3′ |
| P10 | 5′-CCATCAGTGGTATCCGCAGTAT-3′ |
| P11 | 5′-GAGTCTTACCAGGCGATA-3′ |
| P12 | 5′-TATCGCCTGGTAAGACTC-3′ |
| P13 | 5′-AAGATATCCTGCGCTTCG-3′ |
| P14 | 5′-GCGGTTGTTTTTAACGCACTCC-3′ |
| P15 | 5′-AACCGCTTGCTGCCGAAAAG-3′ |
| P16 | 5′-GCTGCACGTCGGCAATTTTC-3′ |
| P17 | 5′-AGTGATTCAGGGTCATCCGTGG-3′ |
| P18 | 5′-AGCAGATGGAGGTAGCTGATGC-3′ |
| P19 | 5′-TGCAGTTGAACGTTATGGAGCC-3′ |
| P20 | 5′-CTGGAGAACGACTGGTCC-3′ |
| P21 | 5′-ACCCATCCGTGAACTTCATCGC-3′ |
Lysine decarboxylase assay.
To determine the lysine decarboxylase (LDC) activity, strains were grown in Moeller decarboxylase broth with lysine (BD BBL, Fisher Scientific). This medium contained decarboxylase basal medium supplemented with 1% l-lysine and bromocresol purple indicator. An inoculated control made up of the above ingredients minus l-lysine was included with each set of samples tested. Glycerol was added to the top of the inoculated medium, and the results were read after incubation for 24 or 36 h at 37°C.
Analysis of the cad region.
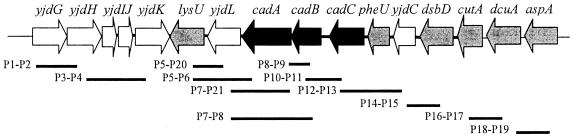
The region containing the cad operon in LDC-negative E. coli strains was screened by PCR for the presence of the genes encoding the cad system or those genes flanking the operon, using the E. coli K-12 and EHEC O157:H7 genome sequences as templates (6, 47). The cad region and flanking genes were amplified using Taq polymerase and the primer pairs listed in Table 2. The locations of the primer pairs within the DNA sequence of E. coli K-12 and the predicted amplicon sizes are also presented in Fig. 1. The amplification was performed for 30 cycles, adjusting the annealing temperature and the extension times based on the primer sequence and the length of the expected product. PCR products of interest were sequenced at the Protein Chemistry Laboratory, Biomolecular Resource Facility Core at University of Texas Medical Branch.
FIG. 1.
E. coli K-12 region containing the cad operon and predicted PCR products. Strain MG1655 was used as the template.
Bacterial adhesion to epithelial cells.
For both qualitative and quantitative adhesion assays, E. coli strains were evaluated for their ability to adhere to HeLa cell monolayers by our standard protocol as previously described (57). Briefly, the strains were grown in LB broth overnight at 37°C and added to tissue culture cells replenished with fresh DMEM at a concentration of 107 bacteria per well for 3 h at 37°C. The monolayers were washed, fixed and stained with Giemsa solution for microscopic evaluation or bacteria were recovered with 0.1% Triton X-100 in phosphate-buffered saline (PBS, pH 7.4) and plated on L agar plates for quantification.
To determine the effect of cadaverine (1,5-pentanediamine dihydrochloride) on the adherence of the different LDC-negative E. coli strains to HeLa cells, each strain was grown in LB broth overnight at 37°C. The next day, bacteria and HeLa cells were incubated for 3 h in DMEM only, or in the presence of 100, 300 or 500 μM of cadaverine (Sigma-Aldrich). The concentrations of cadaverine were similar to those reported by Maurelli et al. (32) and represented physiological levels. One set of experiments were fixed and stained with Giemsa solution for microscopic evaluation, and the other three sets were used for quantitative studies. Adherence data are expressed as CFU per ml of bacterial inoculum recovered from triplicate wells and are the means of the experiments. Statistical difference is expressed as the P value determined by a t test analysis.
Growth curves.
LDC-negative strains were grown for 18 h in LB at 37°C, diluted 1:500 in either fresh DMEM or DMEM supplemented with 500 μM of cadaverine, and grown at 37°C with shaking at 250 rpm. Optical density at 600 nm measurements were taken every hour, and 100 μl of the cultures was diluted and plated on LB agar plates to obtain CFU counts.
Preparation of whole-cell lysates.
Bacterial cultures were grown overnight at 37°C in DMEM, LB, or LB supplemented with 300, 500, or 900 μM cadaverine and aliquoted into portions of approximately 4.0 × 1010 bacteria. The cells were harvested by centrifugation at 12,000 × g for 5 min at room temperature, washed in 1 ml 1X phosphate-buffered saline (pH 7.4), centrifuged again, resuspended in 100 μl of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) solubilization buffer, and lysed at 100°C for 10 min. Cell debris was removed by centrifugation prior to separation of supernatant proteins by 10% SDS-PAGE minigels according to the method of Laemmli (29). Proteins were either stained with Coomassie brilliant blue or prepared for Western blot analysis.
Western blot assay.
Proteins separated by 10% SDS-PAGE minigels were transferred to Immobilon-P (polyvinylidene difluoride) membranes (Millipore) using a Trans-Blot SD transfer cell (Bio-Rad) at 15 V for 22 min. The membrane was blocked with a solution containing PBS (pH 7.4), 0.5% Triton X-100, and 5% nonfat milk. Incubations with anti-intimin (1:2,000 dilution) or anti-Efa-1 serum (1:5,000) and secondary antibody (1:30,000) were carried out for 1 h at room temperature. The blot was developed with the ECL Western blotting analysis system (Amersham Biosciences).
RESULTS
Ability of DEC strains to decarboxylate lysine.
We recently observed that a transposon insertion in the cadA gene of EHEC O157:H7 caused the strain to express a hyperadherence phenotype on tissue culture cells (58). Although the ability to decarboxylate lysine has never been associated with this phenotype, the role of the Cad system as an antivirulence factor has been previously proposed (32). Therefore, the ability to decarboxylate lysine was evaluated by the colorimetric change in Moeller decarboxylase broth with lysine in a collection of phylogenetically well characterized diarrheagenic E. coli strains representing 16 serotypes associated with enteric disease, including samples from patients with hemorrhagic colitis and hemolytic uremic syndrome and those isolated originally from infants with diarrhea (61).
These strains were originally isolated from diverse geographical locations between 1950 and 1990, belonging to three categories: enteropathogenic E. coli, enterohemorrhagic E. coli (EHEC), and enterotoxigenic E. coli (ETEC), and including the serogroups O55, O111, O128, O45, O157, O26, O149, and O15 (Table 1). Four of the 78 DEC strains were unable to decarboxylate lysine even after 48 h incubation at 37°C. These LDC-negative strains belong to serotypes O111:NM (DEC8A), O111:H8 (DECs 8B and 8E) and O26:N (DEC9B). Recently, an E. coli O111:H8 strain has been isolated from an outbreak in the United States (7), and we were also interested in determining the LDC activity of this isolate. As previously observed with the other O111:H8 strains, the Texas outbreak strain was unable to decarboxylate lysine. Our results indicate that some DEC strains, particularly those from serotype O111:H8, lack LDC activity. Therefore, we mapped the region containing the cad operon encoding the LDC system to detect mutations within this region leading to the LDC-negative phenotype.
Mapping the cad operon in LDC-negative E. coli strains.
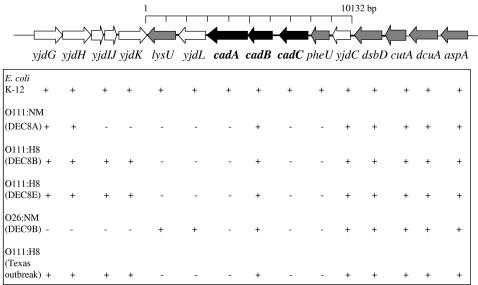
To determine whether the cad operon was present in the chromosome of the LDC-negative E. coli strains, we examined the cad operon and the sequences flanking the 5-prime and 3-prime ends of this region by PCR. Based on E. coli K-12 strain MG1655 (6), we designed primers downstream and upstream of the cadA gene (Table 2 and Fig. 1). As shown in Table 3, E. coli O111 (DECs 8A, 8B, and 8E and the Texas outbreak isolate) and E. coli O26 (DEC9B) strains yielded the same size PCR products as the E. coli K-12 control strain using primer pairs P14-P15, P16-P17, and P18-P19 (0.97, 0.99, and 0.98 kb, respectively). This result indicates that yidC, dsbD, cutA, dcuA, and aspA, genes located upstream from the cad operon, are present in these strains (Fig. 2). Similarly, amplification of the E. coli O111 DECs 8B and 8E and the Texas outbreak isolate with primers annealing downstream of the cad operon (primer pairs P1-P2 and P3-P4) yielded the expected PCR products (Table 3), suggesting that this region, including the yjdG, yjdH, yjdI, yjdJ, and yjdK genes, is intact in these strains. The E. coli O111:NM (DEC 8A) failed to produce a PCR product with primer pair P3-P4 and the E. coli O26:NM (DEC9B) strain was negative with primer pairs P1-P2 and P3-P4, suggesting that the region containing the yjdI, yjdJ, and yjdK genes is different in these two strains compared to the E. coli K-12 DNA sequence.
TABLE 3.
PCR analysis of E. coli regions containing the cad operon and the predicted amplicon products
| Strain | Size (kb) of PRC product generated with target sequence (primer pair):
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| yjdG-yjdH (P1-P2) | yjdlJ-yjdK (P3-P4) | lysU-yjdL (P5-P20) | lysU-cadA (P5-P6) | yjdL-cadA (P7-P21) | yjdL-cadB (P7-P8) | cadB (P8-P9) | cadB-cadC (P10-P11) | cadC-pheU (P12-P13) | yjdC-dsbD (P14-P15) | cutA-dcuA (P16-P17) | dcuA-aspA (P18-P19) | |
| E. coli K-12a | 0.99 | 0.96 | 0.87 | 1.3 | 2.83 | 3.8 | 0.95 | 1.14 | 2.17 | 0.97 | 0.99 | 0.98 |
| O111:NM (DEC8A) | 0.99 | NAb | NA | NA | NA | 2.5 | 0.95 | NA | NA | 0.97 | 0.99 | 0.98 |
| O111:H8 (DEC8B) | 0.99 | 0.96 | NA | NA | NA | 2.5 | 0.95 | NA | NA | 0.97 | 0.99 | 0.98 |
| O111:H8 (DEC8E) | 0.99 | 0.96 | NA | NA | NA | 2.5 | 0.95 | NA | NA | 0.97 | 0.99 | 0.98 |
| O26:NM (DEC9B) | NA | NA | 0.87 | 1.3 | NA | NA | 0.95 | NA | NA | 0.97 | 0.99 | 0.98 |
| O111:H8 (Texas outbreak) | 0.99 | 0.96 | NA | NA | NA | 2.5 | 0.95 | NA | NA | 0.97 | 0.99 | 0.98 |
Strain MG1655 was the template.
NA, no amplicon obtained.
FIG. 2.
Distribution of genes within and flanking the cad region among E. coli K-12, O111, and O26 strains based on PCR experiments carried out as described in Materials and Methods and in Table 3. +, presence of a positive PCR with a product of the expected size based on data obtained from the E. coli K-12 strain MG1655 genome sequence (6). DEC nomenclature is based on the phylogenetic distribution proposed by Whittam (61).
We then performed an extensive PCR analysis of the 10.1-kb region comprising the yjdC, pheU, cadC, cadB, cadA, yjdL, and lysU genes as described for E. coli K-12 strain MG1655 and compared it with the LDC-negative strains. No PCR products were obtained for E. coli O111 or O26 strains when primer pairs targeting the cadA, cadC, and pheU genes were used (Table 3 and Fig. 2). These results suggest that these genes in the chromosome of LDC-negative strains either are not present or have been rearranged. The presence of lysU and yjdL in the E. coli O26:NM (DEC9B) strain, which are absent from the E. coli O111 strains, suggest a different molecular arrangement of this region in the genome of these strains. Interestingly, a PCR product of 0.9 kb (primers P8 and P9) corresponding to the cadB gene was obtained in all the strains tested (Table 3 and Fig. 2).
Based on this analysis, we were able to map the putative limits of this deletion as being yjdK and yjdC for E. coli DEC strains 8B and 8E and the Texas outbreak strain, and yjdH and yjdC for E. coli strain DEC8A. We were unable to determine one of the ends of this putative deletion for E. coli strain DEC9B. Furthermore, we were unable to obtain a PCR product when primers flanking the yjdK (primer P3) and yjdC (primer P15) genes were used, even at extended amplification times (to amplify a 5.0- to 6.0-kb fragment), suggesting that an intergenic region larger than 5.0 kb is probably present within these two genes. A PCR product of ≈2.5 kb was obtained with the E. coli O111:H8 strains DEC8A, 8B, and 8E and the Texas outbreak strain when primer pairs P7-P8 (comprising the cadA gene, the 3′ end of cadB, and the 5′ end of yjdL) were used, instead of the 3,790-bp product obtained with E. coli K-12 strain MG1655. In contrast, no PCR product was obtained with the DEC9B strain. The PCR product obtained from DEC8A was sequenced and compared with the cad region of E. coli K-12. The DNA sequence indicated that the cadA gene is disrupted and only portions of the gene are remaining in the chromosome of this E. coli O111:H8 strain (data not shown). This result is in agreement with the LDC-negative activity of these strains and confirmed that the cad region is disrupted.
Effect of the cad operon on adhesion of LDC-negative strains to HeLa cells.
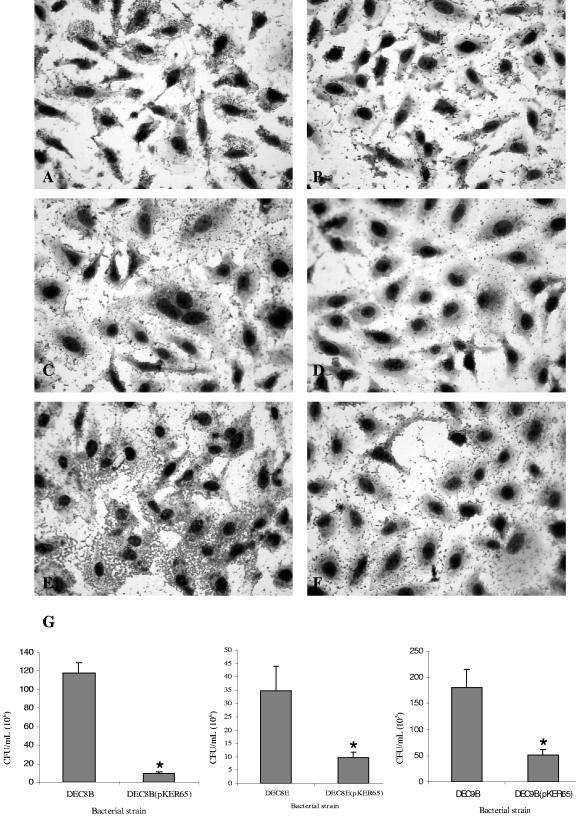
Our previous studies have shown that the cad operon had an effect in the adherence of EHEC O157:H7 to HeLa cells (58). We examined whether the introduction of the cad operon on a plasmid affects adhesion of LDC-negative strains to HeLa cells. As shown in Fig. 3, the wild-type DECs 8B, 8E, and 9B adhered to HeLa cells in distinct patterns, and in the case of DEC8B, the formation of microcolonies was observed (Fig. 3A, C, and E). The three DEC strains transformed with plasmid pKER65 (cad+) (35) were able to decarboxylate lysine (LDC positive activity) and showed reduced adhesion to HeLa cells compared to their corresponding wild-type strains (Fig. 3B, D, and F).
FIG. 3.
Adherence phenotypes of E. coli LDC-negative strains O111:H8 (DEC8B), O111:H8 (DEC8E), and O26:N (DEC9B) (panels A, C, and E, respectively), and their corresponding pKER65-carrying (cad+) (B, D, and F, respectively) strains on HeLa cells 3 h after infection. (G) Quantification of adherence. The data were obtained as described in Materials and Methods.
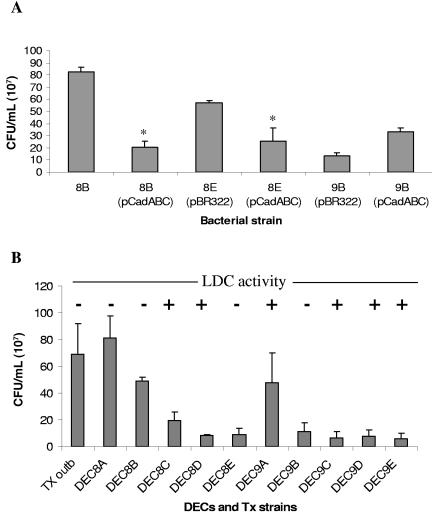
The adherence of the strains carrying pKER65 recovered after 3 h of incubation was reduced to 72 to 92% of that of wild-type strains (P < 0.05 in each case, Fig. 3G). Furthermore, we observed that the strains containing the pKER65 plasmid were unable to form microcolonies on HeLa cells. To confirm that the effect in adhesion was strictly associated with the presence of the cad operon, plasmid pCadABC was constructed and introduced into DECs 8B, 8E, and 9B. Bacterial strains carrying pCadABC became LDC-positive and in the case of DECs 8B(pCadABC) and 8E(pCadABC), they showed reduced adhesion to HeLa cells compared to their corresponding wild-type strains (Fig. 4A, a reduction of 55 to 75%, P < 0.05). In contrast, the DEC9B(pCadABC) strain did not display a reduction in adherence and instead, it became slightly more adhesive than the wild-type strain. Furthermore, transformation of a plasmid containing the cadA gene (pCadA) into DEC 8B, 8E, and 9B strains neither restored LDC activity nor affected the adherence phenotypes of the wild-type strains (data not shown).
FIG. 4.
(A) Quantification of the adherence phenotypes of E. coli LDC-negative DEC strains 8B, 8E, and 9B and their corresponding strains complemented with plasmid pCadABC on HeLa cells 3 h after infection (asterisks indicate P < 0.05). (B) Quantification of the adherence phenotypes of E. coli DEC 8 and 9 strains and the O111:H8 Texas outbreak isolate on HeLa cells 3 h after infection.
Overall, our data indicate that introduction of the cad operon in trans into DEC8 LDC-negative strains had a significant effect on their adhesion to tissue culture cells and presumably in the ability of these LDC-negative organisms to form microcolonies. Furthermore, our results suggest that the cad operon or by-products produced by decarboxylation of lysine negatively regulate adhesion of DEC8 LDC-negative strains to HeLa cells. These statements are only applicable to DEC8 LDC-negative strains, since introduction of pCadABC into DEC9B did not have a significant effect on adhesion, suggesting that an additional protein product encoded in pKER65 may be necessary for the reduction in adherence observed when DEC9B strain carries this plasmid, because the introduction of only the cad operon is not sufficient to restore this phenotype.
cad+-complemented DEC strains differentially express the protein intimin.
Since some of the adherence factors in DEC8 strains are known, namely intimin and Efa-1, we determined the effect of the products encoded in the cad operon on the expression of these adherence factors. Bacterial whole-cell extracts of strains DEC8B, DEC8B(pKER65), DEC8E, DEC8E(pKER65), DEC9B, and DEC9B(pKER65) were fractionated, their proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes, and Western blots were performed with anti-intimin serum (kindly provided by G. Frankel) and anti-Efa-1 serum (kindly provided by E. Hartland).
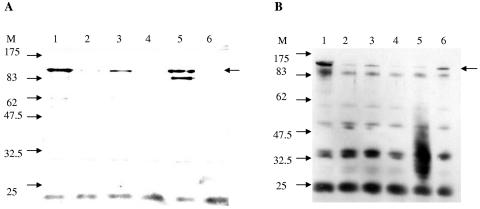
As shown in Fig. 5, the antiserum recognized the intimin protein in strains DEC8B, DEC8E, and DEC9B (Fig. 5A, lanes 1, 3, and 5). In contrast, the expression of intimin was diminished in the strains containing pKER65, suggesting that differences in adhesion of the complemented strains to HeLa cells were probably due to deficient intimin expression. The intimin protein appears as a doublet in strain DEC9B, which could represent isoforms of the protein or degradation products (Fig. 5A, lane 5).
FIG. 5.
(A and B) Western blotting with anti-intimin serum of whole-cell lysates fractionated by SDS-10% PAGE. Panel A lanes: 1, DEC8B; 2, DEC8B(pKER65); 3, DEC8E; 4, DEC8E(pKER65); 5, DEC9B; 6, DEC9B(pKER65). Panel B lanes: 1, DEC8B; 2, DEC8B(pCadABC); 3, DEC8E; 4, DEC8E(pCadABC); 5, DEC9B; 6, DEC9B(pCadABC); M, molecular size standards in kilodaltons. The intimin protein is identified with a black arrow.
To further confirm that the cad operon was associated with the reduction in intimin expression, a Western blot experiment with anti-intimin serum was performed with strains DEC8B, DEC8B(pCadABC), DEC8E, DEC8E(pCadABC), DEC9B, and DEC9B(pCadABC) (Fig. 5B). Again, we observed that the expression of intimin was reduced when the DEC8 strains carried the cad operon on a plasmid. In the case of the DEC9B(pCadABC) strain, we observed a slight increase in intimin expression, a result that goes in accordance with the adherence data presented in Fig. 4A. In experiments with whole-cell lysates to detect the Efa-1 protein, the antiserum reacted in Western blots with the Efa-1 protein and no differences in the expression of this protein were observed (data not shown). Overall, these data indicated that the products encoded in the cad operon and produced in these strains by complementation affected the expression of the intimin protein but have no apparent effect on Efa-1.
LDC-negative DEC8 strains are more adhesive to HeLa cells than the LDC-positive strains within the same DEC pathotype.
We have observed that complementation of LDC-negative DEC8 strains with the cad operon had an effect on adherence to HeLa cells. Therefore, we hypothesize that the presence of this operon is detrimental for the adherence abilities of these strains. To explore this hypothesis, we performed comparative analysis of all the DEC8 and DEC9 strains and the Texas outbreak isolate in adherence assays. As shown in 4B, three of the O111 LDC-negative strains (DECs 8A and 8B and the Texas outbreak isolate) adhere better than the LDC-positive counterparts (DECs 8C and 8D). Only the LDC-negative DEC8E strain did not display the same phenotype as the other LDC-negative strains. Such differences could be associated with the inability of the strain to display the surface adhesin responsible for the phenotype (i.e., intimin), an additional regulatory mechanism controlling the expression of the adhesin, and not just strictly associated with its LDC-negative phenotype.
In contrast, the DEC9 strains did not follow the same pattern of adherence observed with the DEC8 strains. Overall, the DEC9 strains were less adhesive than the DEC8 strains, except for DEC9A, and since the LDC-negative phenotype of the DEC9B strain does not strictly correlate with a reduction in adherence, we propose that the cad operon in this particular pathotype is not directly associated with this phenotype. Although further experimentation is required, our result suggests that the absence of a LDC-positive phenotype could represent an advantage during adherence for some of the DEC8 strains.
Effect of cadaverine on adhesion of LDC-negative DEC strains to HeLa cells.
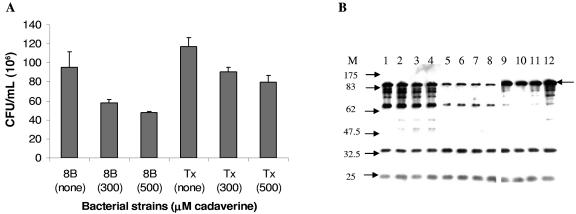
Since adhesion of the LDC-negative strains to tissue culture cells was affected by the presence of the cad operon, and due to previous data indicating that cadaverine has an effect on the expression of Shigella virulence factors (32, 33), we decided to investigate whether cadaverine, a polyamine produced by the breakdown of lysine, has an effect on adhesion of LDC-negative DEC strains to HeLa cells. The data in Fig. 6A reveal that cadaverine at physiological levels (300 and 500 μM) slightly reduced the number of adherent bacteria, the effect being more evident in the DEC8B strain than the Texas outbreak strain. The effect of cadaverine on adhesion was moderate compared with the significant effect produced by the introduction of the cad operon in the LDC-negative strains (Fig. 3G).
FIG. 6.
(A) Quantification of DEC8B and Texas outbreak strains adherent to cultured HeLa cells after 3 h of incubation in the absence or presence of cadaverine (300 and 500 μM). The error bars indicate standard deviations. (B) Effect of different concentrations of cadaverine on expression of intimin in the DEC8B (lanes 1 through 4), DEC8E (lanes 5 through 8), and Texas outbreak (lanes 9 though 12) strains. Whole-cell lysates of bacterial samples in the absence (lanes 1, 5, and 9) or presence of 300 μM (lanes 2, 6, and 10), 500 μM (lanes 3, 7, and 11), or 900 μM (lanes 4, 8, and 12) of cadaverine were fractionated by SDS-10% PAGE and anti-intimin serum was used for the Western blot experiment. M, molecular size standards in kilodaltons. The intimin protein is identified with a black arrow.
Since introduction of the cad operon in DEC8 strains caused a reduction in the expression of intimin, we evaluated the role of cadaverine on expression of this protein (Fig. 6B). Addition of different concentrations of cadaverine to the bacterial cultures did not have any effect on the expression of intimin in DECs 8B and 8E and the Texas outbreak strain, not even at higher concentrations of the polyamine (900 μM; Fig. 6B, lanes 4, 8, and 12). Furthermore, the ability of cadaverine to affect the growth of the LDC-negative strains was investigated, and we determined that the growth of DEC8B or the Texas outbreak strain was not affected by the addition of cadaverine (500 μM) (data not shown). Our data indicate that the effect previously observed by the introduction of the cad operon into the LDC-negative strains did not correlate with the phenotype displayed after incubating the bacterial cultures in medium containing cadaverine.
DISCUSSION
In the current study, we have shown that inactivation of the LDC activity, through a deletion of the region containing the cad operon due apparently to natural selection, has an effect on the adherence of DEC8 strains (particularly those from serotype O111:H8) to tissue culture cells. The ability to decarboxylate lysine was restored with the introduction of the cad operon in trans and the LDC-positive phenotype was associated with a decrease in adherence. In contrast, cadaverine added to the experimental medium did not have a significant effect on the adherent phenotype. We also showed by Western blot analysis of the cad operon-carrying strains that LDC-positive activity correlates with a reduction in the expression of intimin, an adhesin required for the formation of attaching and effacing lesions in STEC strains.
Our data strongly suggest that inactivation of the cad operon (considered a pathoadaptive mutation) is an important mechanism to control functions potentially implicated in the pathogenesis of these organisms. The relevance of these findings is supported by previous studies in which the modulation of amino acid metabolism has been linked to the ability of bacterial pathogens to survive and, in some cases, to adhere to the gastrointestinal tract and has been proposed as a property which plays an important role in the colonization of host intestine (3, 18, 20, 31) or the urinary tract (55).
Coordinated regulation of gene expression in response to environmental stimuli is an important requirement for adaptation of pathogenic bacteria to the variety of environments found in the host. For example, pathogenic E. coli possesses glutamate and arginine decarboxylase systems, as part of the acid resistance mechanism, which are used to resist gastric acid shock in the stomach and to establish infection in the appropriate gastrointestinal niche (14, 52). In the case of Salmonella enterica serovar Typhimurium and Vibrio cholerae, the expression of their lysine decarboxylase systems allows the microorganisms to establish an acid tolerance response (37). In E. coli and other indole-producing bacteria, it has been shown that tryptophanase activity (degradation of tryptophan to indole, pyruvate, and ammonia) correlates with the ability of these organisms to form biofilms and it has been proposed that indole acts as a signaling molecule which regulates expression of adhesion and biofilm-promoting factors (31). The results presented in the current manuscript strongly suggest that in the case of STEC O111 strains, the lysine decarboxylase system modulates adhesion and the presence of the cad operon has a detrimental effect on the ability of these organisms to attach to tissue culture cells. These findings could be of significant importance to begin elucidating the regulatory networks controlling the adherence of STEC O111 strains.
E. coli O111 strains are the second most common non-O157 STEC reported in the United States (8) and among the most common reported in Europe (10). Outbreaks caused by this organism are becoming more frequent and have been reported in Canada (28), Australia (40), Japan (56), and Europe (5, 9). In the United States, two clusters of STEC O111 infections have been identified (1, 11) and recently, an outbreak caused by STEC of serotype O111:H8 was reported (7, 41, 42). The identification of these organisms as causative agents of outbreaks seems to be of importance because E. coli O111 appears to be a non-O157 STEC associated particularly with hemolytic uremic syndrome cases (51, 59), with an apparently underestimated pathogenic potential (4). Therefore, it is necessary to elucidate the pathogenic mechanisms of STEC O111 strains, particularly those of serotype O111:H8.
The inability of STEC O111 to decarboxylate lysine is a property previously observed in isolates collected in Brazil, and it was proposed as a presumptive marker to identify STEC strains of this particular serogroup (21, 22, 60). The authors of these investigations made the initial observation that STEC O111:NM strains fail to decarboxylate lysine and this observation was confirmed in a large number of STEC O111 strains. Our results are in accordance with their observations because we found that STEC O111 strains, particularly those of serotype O111:H8, were unable to decarboxylate lysine. Moreover, in the current study we have confirmed that the LDC-negative phenotype is caused by a large deletion of the region containing the cad operon and showed that restoration of LDC activity had an effect on the adherence of these strains.
Our results also supported a previous observation made by Paton et al. (45) indicating that an enhanced capacity to adhere to intestinal cells is one of the factors which distinguishes human-virulent STEC strains from those of lesser clinical significance. In that study, it was showed that an STEC O111:H− strains isolated from patients suffering hemolytic uremic syndrome adhere significantly better than STEC strains found in the contaminated food source and not in the patients (44). In our current study, we demonstrated that the cad operon had an effect on intimin expression, resulting in a decrease in adherence to tissue culture cells. It is not known whether their O111:H− isolate lacked the LDC system.
The role of the cad operon in virulence expression and as an inducible system that responds to environmental factors was first elucidated by Maurelli et al., who found that introduction of cadA in Shigella flexneri 2a causes attenuation of its virulence and inhibition of enterotoxin production (32) and showed that cadaverine had an effect on the migration of polymorphonuclear leukocytes across the intestinal epithelial monolayers as well as inducing compartmentalization of Shigella species to the phagolysosome (18, 33). Furthermore, they revealed that the lack of LDC activity in Shigella strains is due to a large chromosomal deletion up to 90 kb in the vicinity of the cadA gene (16) and proposed that the presence of “black holes” is a mechanism which provides an evolutionary advantage enabling S. flexneri to increase its pathogenic potential in host tissues. Our results indicate that the presence of the region containing the cad operon is responsible for the phenotype observed and the modest effect observed when cadaverine is added exogenously could be the result of a decrease in outer membrane permeability because it has been reported that polyamines, such as cadaverine, can act as endogenous modulators of outer membrane permeability and that this effect is part of an adaptive response to acidic conditions (50).
Our data mainly suggest that a protein(s) encoded in the cad operon is playing a regulatory role in STEC adhesion. The cad operon encodes three proteins, CadA, B, and C, and al least two of them has been assigned a regulatory role. Our group recently found that cadA is involved in the hyperadherent phenotype of enterohemorrhagic E. coli O157:H7 and proposed that the cad system acts as a regulatory system controlling functions associated with the virulence of that microorganism (58). We recently constructed an EHEC O157:H7 cadA isogenic mutant and confirmed that disruption of the cad system in this strain enhanced its adherence to tissue culture cells, a phenotype that could be eliminated by complementation with a cad+ plasmid (A. G. Torres, unpublished data). In contrast, we were unable to decrease adherence of the LDC-negative strains by introduction of a plasmid containing the cadA gene (data not shown), suggesting that these strains might require more than one of the protein products encoded in the cad operon to have an effect on the adherence phenotype.
In the case of CadC, the positive regulator controlling the cadAB operon in response to acid stress, it has been reported that the expression of this protein in the absence of cadaverine leads to a reduction of porin expression and it has been suggested that CadC is part of a novel mechanism for the environmental control of porin expression (50). Furthermore, it has been proposed that the main strategy adopted by Shigella and enteroinvasive E. coli strains for silencing the cad operon relies on mutations in cadC (13). Recent data indicate that Shigella and enteroinvasive E. coli strains have silenced the cad locus through several strategies, i.e., lack of the entire cad locus in S. flexneri or the presence of the cad locus with only one point mutation in the cadC promoter in one enteroinvasive E. coli strain (12). These investigators (12) have proposed that the first step in silencing the cad locus is the inactivation of the cadC gene followed by a spread of insertion sequences in the cadB and cadA genes, inducing deletions within the cad locus or extending to the flanking genes (12).
Based on these and our observations, we can speculate that the cad locus in STEC O111 LDC-negative strains is located in a region undergoing active rearrangements in which the cadC gene has been already inactivated and the cadA gene is disrupted and only portions of the gene remained in the chromosome of these strains. Although the cadB gene was amplified by PCR, we cannot rule out the possibility that this gene has accumulated point mutations. Since CadA is responsible for the synthesis of cadaverine from lysine and CadB is the lysine-cadaverine antiporter, we hypothesize that the STEC O111 strains initially acquire the LDC-negative phenotype by inactivating the positive regulator CadC. Our data also suggest that the cad system in STEC O111 strains serves to fine tune the virulence of this organism by regulating the expression of putative adhesion molecules under specific environmental circumstances. Currently, we are testing whether one or more of these genes is having a direct regulatory effect on the expression of STEC O111 intimin and therefore on the reduction in the adherence phenotype.
Acknowledgments
We thank Susana Oaxaca-Torres for technical assistance and Vanessa Sperandio for critical reading of the manuscript. We also thank Gad Frankel (Imperial College) for the anti-intimin serum, Elizabeth Hartland (Monash University) for the anti-Efa-1 serum, and Tony Maurelli (USUHS) and George Bennett (Rice University) for plasmids pKER65 and pSM10.
R.C.V.-J. received a summer fellowship from the American Chemical Society and the Academia Mexicana de Ciencias. This work was supported by institutional funds from the University of Texas Medical Branch John Sealy Memorial Endowment Fund for Biomedical Research and the Gastro Intestinal Research Interdisciplinary Program to A.G.T.
Editor: J. T. Barbieri
REFERENCES
- 1.Banatvala, N., M. M. Debeukelaer, P. M. Griffin, T. J. Barrett, K. D. Greene, J. H. Green, and J. G. Wells. 1996. Shiga-like toxin-producing Escherichia coli O111 and associated hemolytic-uremic syndrome: a family outbreak. Pediatr. Infect. Dis. J. 15:1008-1011. [DOI] [PubMed] [Google Scholar]
- 2.Banatvala, N., P. M. Griffin, K. D. Greene, T. J. Barrett, W. F. Bibb, J. H. Green, J. G. Wells, and H.U.S.S. Collaborators. 2001. The United States National Prospective Hemolytic Uremic Syndrome Study: microbiologic, serologic, clinical, and epidemiologic findings. J. Infect. Dis. 183:1063-1070. [DOI] [PubMed] [Google Scholar]
- 3.Bearson, S., B. Bearson, and J. W. Foster. 1997. Acid stress responses in enterobacteria. FEMS Microbiol. Lett. 147:173-180. [DOI] [PubMed] [Google Scholar]
- 4.Bettelheim, K. A., and P. N. Goldwater. 2004. Outbreak of Shiga toxin-producing Escherichia coli O111:H8 infection. Clin. Infect. Dis. 39:148. [DOI] [PubMed] [Google Scholar]
- 5.Blanco, J. E., M. Blanco, M. E. Molinero, E. Peiro, A. Mora, and J. Blanco. 1996. Brote de gastroenteritis asociado con un Escherichia coli verotoxigenico O111:H- VT1+ eae+. Alimentaria 275:109-113. [Google Scholar]
- 6.Blattner, F. R., G. r. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Brooks, J. T., D. Bergmire-Sweat, M. Kennedy, K. Hendricks, M. Garcia, L. Marengo, J. Wells, M. Ying, W. Bibb, P. M. Griffin, R. M. Hoekstra, and C. R. Friedman. 2004. Outbreak of Shiga toxin-producing Escherichia coli O111:H8 infections among attendees of a high school cheerleading camp. Clin. Infect. Dis. 38:190-198. [DOI] [PubMed] [Google Scholar]
- 8.Brooks, J. T., E. G. Sowers, J. G. Wells, K. D. Greene, P. M. Griffin, and N. A. Strockbine. 2001. Presented at the 39th Annual Meeting of the Infectious Diseases Society of America, San Francisco.
- 9.Caprioli, A., I. Luzzi, F. Rosmini, C. Resti, A. Edefonti, F. Perfumo, C. Farina, A. Goglio, A. Gianviti, and G. Rizzoni. 1994. Community-wide outbreak of hemolytic-uremic syndrome associated with non O157 verocytotoxin-producing Escherichia coli. J. Infect. Dis. 169:208-211. [DOI] [PubMed] [Google Scholar]
- 10.Caprioli, A., and A. E. Tozzi. 1998. Epidemiology of Shiga toxin producing Escherichia coli infections in continental Europe, p. 38-48. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. American Society for Microbiology Press, Washington, D.C.
- 11.Carlson, C. 2002. Presented at the International Conference on Emerging Infectious Diseases, Atlanta, Ga.
- 12.Casalino, M., M. C. Latella, G. Prosseda, P. Ceccarini, F. Grimont, and B. Colonna. 2005. Molecular evolution of the lysine decarboxylase-defective phenotype in Shigella sonnei. Int. J. Med. Microbiol. 294:503-512. [DOI] [PubMed] [Google Scholar]
- 13.Casalino, M., M. C. Latella, G. Prosseda, and B. Colonna. 2003. CadC is the preferential target of a convergent evolution driving enteroinvasive Escherichia coli toward a lysine decarboxylase-defective phenotype. Infect. Immun. 71:5472-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castanie-Cornet, M. P., T. A. Penfound, D. Smith, J. F. Elliott, and J. W. Foster. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention. 2004. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food-selected sites, United States, 2003. Morb. Mortal Wkly. Rep. 53:338-343. [PubMed] [Google Scholar]
- 16.Day, W. A. J., R. E. Fernandez, and A. T. Maurelli. 2001. Pathoadaptive mutations that enhance virulence: genetic organization of the cadA regions of Shigella spp. Infect. Immun. 69:7471-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandez, I. M., M. Silva, R. Schuch, W. A. Walker, A. M. Siber, A. T. Maurelli, and B. A. McCormick. 2001. Cadaverine prevents the escape of Shigella flexneri from the phagolysosome: a connection between bacterial dissemination and neutrophil transepithelial signaling. J. Infect. Dis. 184:743-753. [DOI] [PubMed] [Google Scholar]
- 19.Fitzhenry, R. J., D. J. Pickard, E. L. Hartland, S. Reece, G. Dougan, A. D. Phillips, and G. Frankel. 2002. Intimin type influences the site of human intestinal mucosal colonisation by enterohaemorrhagic Escherichia coli O157:H7. Gut 50:180-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorden, J., and P. L. Small. 1993. Acid resistance in enteric bacteria. Infect. Immun. 61:364-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guth, B. E., T. A. Gomes, T. M. Vaz, and K. Irino. 2003. Inability to decarboxylate lysine as a presumptive marker to identify Shiga toxin-producing Escherichia coli strains of serogroup O111. J. Clin. Microbiol. 41:3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guth, B. E., S. R. Ramos, A. M. Cerqueira, J. R. Andrade, and T. A. Gomes. 2002. Phenotypic and genotypic characteristics of shiga toxin-producing Escherichia coli strains isolated from children in Sao Paulo, Brazil. Mem. Inst. Oswaldo Cruz 97:1085-1089. [DOI] [PubMed] [Google Scholar]
- 23.Jaeger, J. L., and D. W. Acheson. 2000. Shiga toxin-producing Escherichia coli. Curr. Infect. Dis. Rep. 2:61-67. [DOI] [PubMed] [Google Scholar]
- 24.Jerse, A. E., K. G. Gicquelais, and J. B. Kaper. 1991. Plasmid and chromosomal elements involved in the pathogenesis of attaching and effacing Escherichia coli. Infect. Immun. 59:3869-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaper, J. B., J. P. Nataro, and H. L Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123-140. [DOI] [PubMed] [Google Scholar]
- 26.Karch, H., M. Bielaszewska, M. Bitzan, and H. Schmidt. 1999. Epidemiology and diagnosis of Shiga toxin-producing Escherichia coli infections. Diagn. Microbiol. Infect. Dis. 34:229-243. [DOI] [PubMed] [Google Scholar]
- 27.Karmali, M. A. 1989. Infection by verocytotoxin-producing Escherichia coli. Clin. Microbiol. Rev. 2:15-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karmali, M. A., M. Petric, M. Winkler, M. Bielaszewska, J. Brunton, N. van de Kar, T. Morooka, G. B. Nair, S. E. Richardson, and G. S. Arbus. 1994. Enzyme-linked immunosorbent assay for detection of immunoglobulin G antibodies to Escherichia coli Vero cytotoxin 1. J. Clin. Microbiol. 32:1457-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 30.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Martino, P. D., R. Fursy, L. Bret, B. Sundararaju, and R. S. Phillips. 2003. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can. J. Microbiol. 49:443-449. [DOI] [PubMed] [Google Scholar]
- 32.Maurelli, A. T., R. E. Fernandez, C. A. Bloch, C. K. Rode, and A. Fasano. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. USA 95:3943-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCormick, B. A., M. I. Fernandez, A. M. Siber, and A. T. Maurelli. 1999. Inhibition of Shigella flexneri-induced transepithelial migration of polymorphonuclear leucocytes by cadaverine. Cell. Microbiol. 1:143-155. [DOI] [PubMed] [Google Scholar]
- 34.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meng, S. Y., and G. N. Bennett. 1992. Nucleotide sequence of the Escherichia coli cad operon: a system for neutralization of low extracellular pH. J. Bacteriol. 174:2659-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meng, S. Y., and G. N. Bennett. 1992. Regulation of the Escherichia coli cad operon: location of a site required for acid induction. J. Bacteriol. 174:2670-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merrell, D. S., and A. Camilli. 2002. Acid tolerance of gastrointestinal pathogens. Curr. Opin. Microbiol. 5:51-55. [DOI] [PubMed] [Google Scholar]
- 38.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicholls, L., T. H. Grant, and R. M. Robins-Browne. 2000. Identification of a novel genetic locus that is required for in vitro adhesion of a clinical isolate of enterohaemorrhagic Escherichia coli to epithelial cells. Mol. Microbiol. 35:275-288. [DOI] [PubMed] [Google Scholar]
- 40.No authors listed. 1995. Community outbreak of hemolytic uremic syndrome attributable to Escherichia coli O111:NM-South Australia 1995. Morb. Mortal. Wkly. Rep. 44:550-551. [PubMed] [Google Scholar]
- 41.No authors listed. 2000. Escherichia coli O111:H8 outbreak among teenage campers-Texas, 1999. Morb. Mortal. Wkly. Rep. 49:321-324. [PubMed] [Google Scholar]
- 42.No authors listed. 2000. Centers for Disease Control and Prevention Escherichia coli O111:H8 outbreak among teenage campers-Texas, 1999. JAMA 283:2517-2518. [PubMed] [Google Scholar]
- 43.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 44.Paton, A. W., R. M. Ratcliff, R. M. Doyle, J. Seymour-Murray, D. Davos, J. A. Lanser, and J. C. Paton. 1996. Molecular microbiological investigation of an outbreak of hemolytic-uremic syndrome caused by dry fermented sausage contaminated with Shiga-like toxin-producing Escherichia coli. J. Clin. Microbiol. 34:1622-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paton, A. W., E. Voss, P. A. Manning, and J. C. Paton. 1997. Shiga toxin-producing Escherichia coli isolates from cases of human disease show enhanced adherence to intestinal epithelial (Henle 407) cells. Infect Immun. 65:3799-3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perna, N. T., G. R. Plunkett, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-553. [DOI] [PubMed] [Google Scholar]
- 48.Phillips, A. D., and G. Frankel. 2000. Intimin-mediated tissue specificity in enteropathogenic Escherichia coli interaction with human intestinal organ cultures. J. Infect. Dis. 181:1496-1500. [DOI] [PubMed] [Google Scholar]
- 49.Phillips, A. D., S. Navabpour, S. Hicks, G. Dougan, T. Wallis, and G. Frankel. 2000. Enterohaemorrhagic Escherichia coli O157:H7 target Peyer's patches in humans and cause attaching/effacing lesions in both human and bovine intestine. Gut. 47:377-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Samartzidou, H., and A. H. Delcour. 1999. Excretion of endogenous cadaverine leads to a decrease in porin-mediated outer membrane permeability. J. Bacteriol. 181:791-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scheutz, F., L. Beutin, and H. R. Smith. 2000. Presented at the 4th International Symposium and Workshop on Shiga Toxin (Verocytotoxin) Producing Escherichia coli Infections, Kyoto, Japan.
- 52.Shin, S., M. P. Castanie-Cornet, J. W. Foster, J. A. Crawford, C. Brinkley, and J. B. Kaper. 2001. An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol. Microbiol. 41:1133-1150. [DOI] [PubMed] [Google Scholar]
- 53.Sokurenko, E. V., D. L. Hasty, and D. E. Dykhuizen. 1999. Pathoadaptive mutations: gene loss and variation in bacterial pathogens. Trends Microbiol. 7:191-195. [DOI] [PubMed] [Google Scholar]
- 54.Stevens, M. P., P. M. van Diemen, G. Frankel, A. D. Phillips, and T. S. Wallis. 2002. Efa-1 influences colonization of the bovine intestine by shiga toxin-producing Escherichia coli serotypes O5 and O111. Infect. Immun. 70:5158-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sturgill, G., and P. N. Rather. 2004. Evidence that putrescine acts as an extracellular signal required for swarming in Proteus mirabilis. Mol. Microbiol. 51:437-446. [DOI] [PubMed] [Google Scholar]
- 56.Tanaka, H., M. Ohseto, Y. Yamashita, N. Shinohara, H. Inoue, Y. Sasaki, Y. Kakihara, T. Tsukamoto, T. Yutsudo, Y. Oku, et al. 1989. Bacteriological investigation on an outbreak of acute enteritis associated with verotoxin-producing Escherichia coli O111:H- [in Japanese]. Kansenshogaku Zasshi 63:1187-1194. [DOI] [PubMed] [Google Scholar]
- 57.Torres, A. G., J. A. Giron, N. T. Perna, V. Burland, F. R. Blattner, F. Avelino-Flores, and J. B. Kaper. 2002. Identification and characterization of lpfABCC′DE, a fimbrial operon of enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 70:5416-5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Torres, A. G., and J. B. Kaper. 2003. Multiple elements controlling adherence of enterohemorrhagic Escherichia coli O157:H7 to HeLa cells. Infect. Immun. 71:4985-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tozzi, A. E., A. Caprioli, F. Minelli, A. Gianviti, L. De Petris, A. Edefonti, G. Montini, A. Ferretti, T. De Palo, M. Gaido, G. Rizzoni, and H.U.S.S. Group. 2003. Shiga toxin producing Escherichia coli infections associated with hemolytic uremic syndrome, Italy, 1988-2000. Emerg. Infect. Dis. 9:106-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vaz, T. M., K. Irino, M. A. Kato, A. M. Dias, T. A. Gomes, M. I. Medeiros, M. M. Rocha, and B. E. Guth. 2004. Virulence properties and characteristics of Shiga toxin-producing Escherichia coli in Sao Paulo, Brazil, from 1976 through 1999. J. Clin. Microbiol. 42:903-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whittam, T. S., M. L. Wolfe, I. K. Wachsmuth, F. Orskov, I. Orskov, and R. A. Wilson. 1993. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect. Immun. 61:1619-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]