Abstract
A new fluorescence in situ hybridization (FISH) method with peptide nucleic acid (PNA) probes for identification of Staphylococcus aureus directly from positive blood culture bottles that contain gram-positive cocci in clusters (GPCC) is described. The test (the S. aureus PNA FISH assay) is based on a fluorescein-labeled PNA probe that targets a species-specific sequence of the 16S rRNA of S. aureus. Evaluations with 17 reference strains and 48 clinical isolates, including methicillin-resistant and methicillin-susceptible S. aureus species, coagulase-negative Staphylococcus species, and other clinically relevant and phylogenetically related bacteria and yeast species, showed that the assay had 100% sensitivity and 96% specificity. Clinical trials with 87 blood cultures positive for GPCC correctly identified 36 of 37 (97%) of the S. aureus-positive cultures identified by standard microbiological methods. The positive and negative predictive values were 100 and 98%, respectively. It is concluded that this rapid method (2.5 h) for identification of S. aureus directly from blood culture bottles that contain GPCC offers important information for optimal antibiotic therapy.
The presence of Staphylococcus aureus in blood cultures is first suggested by the detection of gram-positive cocci in clusters (GPCC) in the Gram stain of blood culture bottles that have positive signals. Unfortunately, definitive identification of S. aureus by traditional methods is time-consuming, requiring subculture and biochemical analysis (2). Often, empirical antibiotic therapy is prescribed for patients with blood cultures positive for GPCC, although the majority of cultures are subsequently shown to be coagulase-negative staphylococci (CoNS), such as Staphylococcus epidermidis, a common blood culture contaminant (19). This delayed identification of S. aureus leads to the significant unnecessary use of antibiotics and its sequelae.
Automated blood culture instruments with continuous monitoring are used in clinical microbiology laboratories worldwide and are available with a variety of medium options. Some blood culture media are supplemented either with charcoal (Organon Teknika) or with resins and/or sodium polyanetholesulfonate (SPS; Becton Dickinson) for absorption of antibiotics. These components, along with the blood sample itself and other medium components, may interfere with assays for the direct identification of the organisms in positive blood cultures. Current standard methods therefore rely on subculture followed by a series of biochemical assays for the differentiation of organisms that present as GPCC in positive blood culture bottles. These assays include tests for coagulase and catalase and/or carbon assimilation tests for differentiation between S. aureus and CoNS or other closely related species, such as Micrococcus (2). Standard assays comprising immunological, tube coagulase, and stable endonuclease methods routinely used for the identification of S. aureus isolates have shown variable sensitivities and specificities for use directly on blood culture bottles positive for GPCC (12, 18, 20). Only a relatively few methods for the identification of S. aureus directly from positive blood cultures have been described. Those have mainly been based on molecular biology-based techniques, such as hybridization protection (6), fluorescence in situ hybridization (FISH) (11), and PCR (4).
Peptide nucleic acid (PNA) molecules are pseudopeptides that obey Watson-Crick base-pairing rules for hybridization to complementary nucleic acid targets (RNA and DNA) (9, 13). Due to their uncharged, neutral backbones, PNA probes exhibit favorable hybridization characteristics such as high specificities, strong affinities, and rapid kinetics, resulting in improved hybridization to highly structured targets such as rRNA (21). In addition, the relatively hydrophobic character of PNA compared to that of DNA oligonucleotides enables PNA probes to penetrate the hydrophobic cell wall of bacteria following preparation of a standard smear (22).
rRNA sequence analysis is today a well-established method for phylogenetic analysis of microorganisms (10, 25), and the sequence variations found between the relatively conserved rRNA sequences form the basis for the design of probes specific for most bacteria and yeasts. As a result, molecular biology-based diagnostic methods that use rRNA sequences are rapidly replacing the classic microbiological identification methods based on phenotypic characteristics. Furthermore, the high cellular abundance of rRNA allows individual cells to be identified with labeled probes that target specific rRNA sequences, so-called phylogenetic strains (7).
FISH with PNA probes that target rRNA (the PNA FISH assay) is a novel technique that combines the unique performance characteristics of PNA probes with the advantages of using rRNA as a target, and it has recently been applied for the identification of both bacteria and yeasts in culture (14, 16, 23, 24). In the present study, we applied PNA probes that target the 16S rRNA of S. aureus to the PNA FISH assay for the rapid and specific identification of S. aureus directly in blood cultures positive for GPCC.
MATERIALS AND METHODS
Reference strains and clinical isolates.
Seventeen reference strains representing phylogenetically related and clinically relevant bacterium and yeast species were obtained from the Agricultural Research Service (ARS) Culture Collection (Peoria, Ill.) and the American Type Culture Collection (Manassas, Va.). Forty-eight clinical isolates representing gram-positive cocci in clusters (S. aureus, CoNS including S. epidermidis, Micrococcus, and Stomatococcus) were obtained from the in-house collection at the Clinical Microbiology Laboratory, Cleveland Clinic Foundation, Cleveland, Ohio.
Blood culture media.
The BBL Septi-Chek system with Columbia broth and SPS (Becton Dickinson), the BBL Septi-Chek system with Trypticase soy broth and SPS (Becton Dickinson), the BBL Septi-Chek system with brain heart infusion broth and SPS (Becton Dickinson), the BBL Septi-Chek system with Trypticase soy broth and both resins and SPS (Becton Dickinson), and the FAN BacT/Alert system (Organon Teknika) containing charcoal were all used to evaluate the S. aureus PNA FISH assay. Reference strains were grown in Trypticase soy broth (Difco).
Clinical specimens.
A total of 87 GPCC-positive blood culture bottles (FAN BacT/Alert system; Organon Teknika) from routine tests at the Clinical Microbiology Laboratory, Cleveland Clinic Foundation, were included in the study.
Preparation of smears.
For each smear, 1 drop of phosphate-buffered saline with 1% (vol/vol) Triton X-100 (Aldrich) was placed in a well (diameter, 14 mm) of a Teflon-coated microscope slide (Erie Scientific, Portsmouth, N.H.) and mixed gently with a small drop of resuspended culture. The slide was then placed on a 55°C slide warmer for 20 min. Occasionally, the smears were instead fixed by passing the slide through the blue cone of a Bunsen burner three to four times or by treatment with methanol for 5 min (17). Subsequently, the smears were disinfected by immersion in 80 to 96% (vol/vol) ethanol for 5 to 10 min and air dried.
S. aureus PNA FISH assay.
The S. aureus PNA FISH assay was performed as described previously (23), with minor modifications. Smears were covered with approximately 20 μl of hybridization solution containing 10% (wt/vol) dextran sulfate (Sigma Chemical Co., St. Louis, Mo.), 10 mM NaCl (J. T. Baker), 30% (vol/vol) formamide (Sigma), 0.1% (wt/vol) sodium pyrophosphate (Sigma), 0.2% (wt/vol) polyvinylpyrrolidone (Sigma), 0.2% (wt/vol) Ficoll (Sigma), 5 mM disodium EDTA (Sigma), 1% (vol/vol) Triton X-100 (Aldrich), 50 mM Tris-HCl (pH 7.5), and 500 nM fluorescein-labeled PNA probe (GCTTCTCGTCCGTTC) targeting S. aureus 16S rRNA (Boston Probes, Bedford, Mass.). Coverslips were placed on the smears to ensure even coverage with hybridization solution, and the slides were subsequently placed on a slide warmer with a humidity chamber (Slidemoat; Boeckel Scientific, Feasterville, Pa.) and incubated for 90 min at 55°C. Following hybridization, the coverslips were removed by submerging each slide in approximately 20 ml of prewarmed 5 mM Tris (pH 10), 15 mM NaCl (J. T. Baker), 0.1% (vol/vol) Triton X-100 (Aldrich) in a water bath at 55°C and washed for 30 min. Each smear was finally mounted by using 1 drop of IMAGEN Mounting Fluid (DAKO, Ely, United Kingdom) and covered with a coverslip. Microscopic examination was conducted with a fluorescence microscope (Optiphot [Nikon Corporation, Tokyo, Japan] or BX40 [Olympus, Tokyo, Japan]) equipped with a ×60/1.4 oil objective (Nikon) or a ×60/0.8 objective (Olympus), an HBO 100-W mercury lamp, and both a fluorescein isothiocyanate (FITC)-Texas Red dual-band filter set (Chroma Technology Corp., Brattleboro, Vt.) and a band-pass FITC filter (Omega Optical, Brattleboro, Vt.). S. aureus was identified as multiple clusters of bright fluorescent cocci in multiple fields of view. Images were obtained with a color charge couple device camera (Diagnostic Instruments, Inc., Sterling Heights, Mich.) connected to a computer system.
Identification of GPCC.
The contents of positive blood culture bottles that demonstrated GPCC on Gram staining were subcultured onto 5% sheep blood agar plates, and the plates were incubated overnight at 35°C in an environment with 5 to 10% CO2. Gamma- and beta-hemolytic colonies were considered possible staphylococci. These were differentiated from gamma- and beta-hemolytic streptococci by testing for catalase. Slide and tube coagulase tests were used to differentiate S. aureus (positive for both slide and tube coagulase tests) from CoNS. Some CoNS were further characterized as S. epidermidis by carbohydrate utilization. Micrococcus and Stomatococcus were suspected by the typical colony morphologies for these organisms and were identified by biochemical and susceptibility assays.
RESULTS
Optimization of S. aureus PNA FISH assay.
Initially, the S. aureus PNA FISH assay method was optimized for performance on various standard blood culture media by standard smear preparation techniques. In particular, for the charcoal-containing medium of the FAN BacT/Alert system from Organon Teknika, modification of the previously published PNA FISH assay methods was required as the charcoal unspecifically bound to the PNA probe, and, thus, depending on the actual amount of charcoal in the smear, provided variable results and occasionally led to false-negative results. Centrifugation to remove the charcoal was not an option, as S. aureus clusters have a tendency to adhere to the charcoal particles. Detergents, such as Triton X-100, were found to be effective blocking reagents, and 1% Triton X-100 was added to both the phosphate-buffered saline used for smear preparation and the hybridization buffer. The S. aureus PNA FISH assay was found to perform well on all blood culture media listed above.
A variety of standard smear preparation methods such as heat fixation (20 min at 55°C), flame fixation, and methanol fixation were all compatible with the S. aureus PNA FISH assay. As a precautionary step, all smears were disinfected by immersion in 80 to 96% ethanol for 5 to 10 min prior to performance of the S. aureus PNA FISH assay.
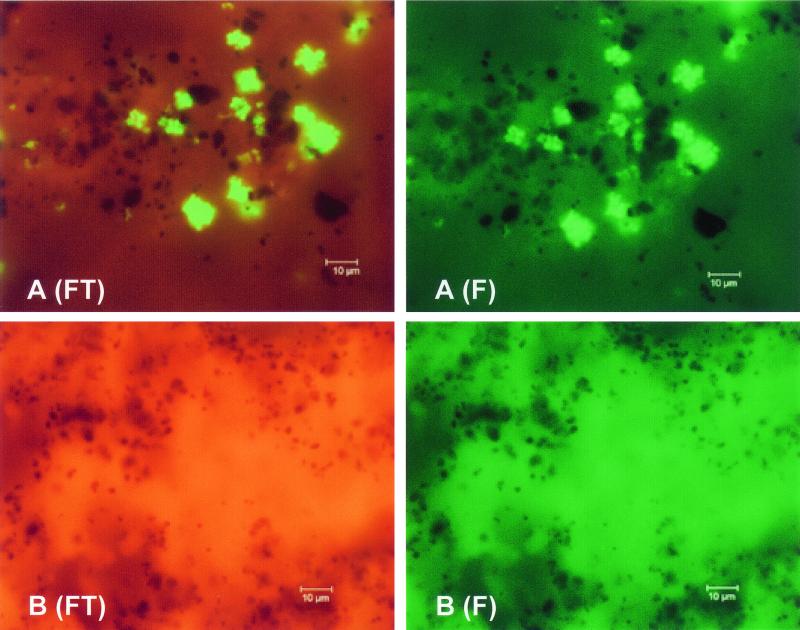
In agreement with other PNA FISH assays, the use of an FITC-Texas Red double filter provided an excellent means of visual discrimination between autofluorescence and a specific signal, such that clusters of S. aureus were observed as bright green fluorescent clusters on a reddish autofluorescent smear background (Fig. 1). In contrast, autofluorescence from the smear itself was greenish when a standard FITC filter was used (Fig. 1) and thus would interfere with the visual interpretation, increasing the risk for false identification.
FIG. 1.
Images of a blood culture positive for S. aureus and GPCC (A) and a blood culture negative for S. aureus but positive for GPCC (B). S. aureus is identified as bright-green fluorescent clusters of cocci on a reddish smear background. Images were obtained with both the recommended FITC-Texas Red double filter (FT) and a band-pass FITC filter (F). The dark areas are charcoal particles in the medium provided with the FAN BacT/Alert system.
The detection limit for the S. aureus PNA FISH assay was determined to be approximately 105 CFU per ml with serial dilutions of an S. aureus-positive culture.
S. aureus PNA FISH assay performance.
The PNA probe targets the same species-specific region of the 16S rRNA of S. aureus that previously published DNA probes target (5, 11); however, the PNA probe is shorter than those DNA probes due to the higher affinity of the PNA probe. Initially, the specificity of the PNA probe was tested by the PNA FISH assay with a panel of reference strains representing phylogenetic representatives of the Staphylococcus genus as well as clinically relevant bacterium and yeast species (Table 1). These results indicate that the assay has a high degree of specificity, with the only limitation being weak cross-hybridization to Staphylococcus schleiferi. This species was included in the evaluation only because a BLAST search (1) revealed that the sequence of the PNA probe has only a single mismatch to some S. schleiferi 16S rRNA sequences. Fortunately, this species is not of significant clinical relevance. A search with the BLAST algorithm also confirmed that the probe sequence has two or more mismatches to the rRNA sequences of all other bacterial species and included sequences from clinically relevant Staphylococcus species, such as S. epidermidis, Staphylococcus haemolyticus, Staphylococcus warneri, Staphylococcus cohnii, Staphylococcus intermedius, and Staphylococcus lugdunensis.
TABLE 1.
Reaction of S. aureus PNA FISH assay with a panel of reference strains representing clinically relevant and phylogenetically related bacterium and yeast species
| Organism | Straina | S. aureus PNA FISH assay result |
|---|---|---|
| Staphylococcus aureus | ATCC 6538 | + |
| Staphylococcus epidermidis | ATCC 14990 | − |
| Staphylococcus haemolyticus | ATCC 29970 | − |
| Staphylococcus schleiferi | ATCC 43808 | +b |
| Acinetobacter calcoaceticus | ATCC 23065 | − |
| Bacillus subtilis | ATCC 6633 | − |
| Burkholderia cepacia | ATCC 25416 | − |
| Candida albicans | NRRL Y-12983 | − |
| Candida dubliniensis | NRRL Y-17512 | − |
| Citrobacter freundii | ATCC 8090 | − |
| Escherichia coli | ATCC 8739 | − |
| Micrococcus luteus | ATCC 9341 | − |
| Proteus mirabilis | ATCC 12453 | − |
| Pseudomonas aeruginosa | ATCC 27853 | − |
| Pseudomonas putida | ATCC 12633 | − |
| Salmonella choleraesuis | ATCC 29946 | − |
| Yersinia enterocolitica | DSCC 8P25 | − |
ATCC, American Type Culture Collection; NRRL, ARS Culture Collection; DSCC, Stock Culture Collection, Department of Bacteriology, University of Wisconsin—Madison.
Weak positive reaction.
The sensitivity and specificity of the S. aureus PNA FISH assay were further examined with 48 clinical isolates representing methicillin-resistant and methicillin-susceptible S. aureus strains, species of CoNS, and other gram-positive cocci in clusters (Micrococcus and Stomatococcus) (Table 2). The assay correctly identified all S. aureus isolates and gave negative results for all other isolates except one Stomatococcus isolate.
TABLE 2.
Reaction of S. aureus PNA FISH assay with a panel of clinical isolates representing clinically relevant GPCC
| Organism | No. of samples with the following S. aureus PNA FISH assay result:
|
|
|---|---|---|
| Positive | Negative | |
| S. aureus | 16 | 0 |
| CoNS | 0 | 14 |
| Micrococcus | 0 | 13 |
| Stomatococcus | 1 | 4 |
Finally, the diagnostic applicability of the S. aureus PNA FISH assay was evaluated directly with 87 blood culture specimens positive for GPCC, and the results were compared to those obtained by standard methods. Of the 37 blood culture specimens that were positive for S. aureus by standard methods, 36 (97%) were positive by the S. aureus PNA FISH assay, whereas 1 specimen that was S. aureus positive by standard methods was negative by the S. aureus PNA FISH assay. Interestingly, the S. aureus-positive blood culture that was negative by the S. aureus PNA FISH assay was from an aerobic bottle for which the corresponding anaerobic bottle that was positive for GPCC was negative for S. aureus by both the S. aureus PNA FISH assay and standard methods. Of the remaining 50 blood culture specimens positive for GPCC, all specimens were correctly identified as negative by the S. aureus PNA FISH assay. From these data, the performance specifications for the S. aureus PNA FISH assay were calculated and are as follows: the diagnostic sensitivity was 97% (36 of 37 specimens), the diagnostic specificity was 100% (50 of 50 specimens), the positive predictive value was 100% (36 of 36 specimens), and the negative predictive value was 98% (50 of 51 specimens).
DISCUSSION
We have shown that the S. aureus PNA FISH assay with PNA probes that target the rRNA of S. aureus can be used for the direct identification of S. aureus in blood culture bottles positive for GPCC. The test is performed with smears made from the blood culture bottles, and interpretation of results is conducted by microscopy, such that the PNA FISH assay procedure simply adds the high specificity of PNA probes to standard microbiological procedures (i.e., smear preparation and microscopy) to provide the definitive identification of S. aureus in an amount of time not possible by conventional methods. These attributes make this method adaptable to typical clinical microbiology settings.
The S. aureus PNA FISH assay procedure resembles previously published PNA FISH methods for culture identification of Mycobacterium species (8, 15) and Candida species (14). The assay may therefore easily be supplemented with a palette of other specific PNA probes for establishment of a diagnostic concept that will permit the rapid and definitive identification of organisms in blood cultures in routine clinical microbiology laboratories.
Rapid identification of S. aureus may potentially eliminate empirical treatment based on Gram staining results and thus lead to an overall reduction in the level of use of antibiotics, such as vancomycin, which may be used empirically to “cover” patients with blood cultures with GPCC (3). This may help to reduce the prevalence of nosocomial infections with drug-resistant bacteria, in particular, vancomycin-resistant enterococci. We are evaluating the S. aureus PNA FISH assay in this respect.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- 2.Baron, E. J. 1998. Processing and interpretation of blood cultures, p.58–62. In H. D. Isenberg (ed.), Essential procedures for clinical microbiology. ASM Press, Washington, D.C.
- 3.Bartlett, J. G., and J. W. Froggatt III. 1995. Antibiotic resistance. Arch. Otolaryngol. Head Neck Surg. 121:392–396. [DOI] [PubMed] [Google Scholar]
- 4.Benito, M. J., M. M. Rodriguez, M. G. Cordoba, E. Aranda, and J. J. Cordoba. 2000. Rapid differentiation of Staphylococcus aureus from staphylococcal species by arbitrarily primed-polymerase chain reaction. Lett. Appl. Microbiol. 31:368–373. [DOI] [PubMed] [Google Scholar]
- 5.Bentley, R. W., N. M. Harland, J. A. Leigh, and M. D. Collins. 1993. A Staphylococcus aureus-specific oligonucleotide probe derived from 16S rRNA gene sequences. Lett. Appl. Microbiol. 16:203–206. [DOI] [PubMed] [Google Scholar]
- 6.Davis, T. E., and D. D. Fuller. 1991. Direct identification of bacterial isolates in blood cultures by using a DNA probe. J. Clin. Microbiol. 29:2193–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243:1360–1363. [DOI] [PubMed] [Google Scholar]
- 8.Drobniewski, F. A., P. G. More, and G. S. Harris. 2000. Differentiation of Mycobacterium tuberculosis complex and nontuberculous mycobacterial liquid cultures by using peptide nucleic acid-fluorescence in situ hybridization probes. J. Clin. Microbiol. 38:444–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egholm, M., O. Buchard, L. Christensen, C. Behrens, S. M. Freier, D. A. Driver, R. H. Berg, S. K. Kim, B. Norden, and P. E. Nielsen. 1993. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen bonding rules. Nature 365:556–568. [DOI] [PubMed] [Google Scholar]
- 10.Fox, G. E., E. Stackebrandt, R. B. Hespell, J. Gibson, J. Maniloff, T. A. Dyer, R. S. Wolfe, W. E. Black, R. S. Tanner, L. J. Magrum, L. B. Zablen, R. Blakemore, R. Gupta, L. Bonen, B. J. Lewis, D. A. Stahl, K. R. Luehrsen, K. N. Chen, and C. R. Woese. 1980. The phylogeny of prokaryotes. Science 209:457–463. [DOI] [PubMed] [Google Scholar]
- 11.Kempf, V. A. J., K. Trebesius, and I. B. Autenrieth. 2000. Fluorescent in situ hybridization allows rapid identification of microorganisms in blood cultures. J. Clin. Microbiol. 38:830–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDonald, C. L., and K. Chapin. 1995. Rapid identification of Staphylococcus aureus from blood culture bottles by a classic 2-hour tube coagulase test. J. Clin. Microbiol. 33:50–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nielsen, P. E., M. Egholm, and O. Buchard. 1994. Peptide nucleic acids (PNA). A DNA mimic with a peptide backbone. Bioconjugate Chem. 5:3–7. [DOI] [PubMed] [Google Scholar]
- 14.Oliveira, K., G. Haase, C. Kurtzman, J. J. Hyldig-Nielsen, and H. Stender. 2001. Differentiation between Candida albicans and Candida dubliniensis by fluorescence in situ hybridization using peptide nucleic acid probes. J. Clin. Microbiol. 39:4138–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Padilla, A., J. M. Manterola, O. F. Rasmussen, J. Lonca, J. Doninguez, L. Matas, A. Hernandez, and V. Ausina. 2000. Evaluation of a fluorescence hybridization assay using peptide nucleic acid probes for identification and differentiation of tuberculous and non-tuberculous mycobacteria in liquid cultures. Eur. J. Microbiol. Infect. Dis. 19:140–145. [DOI] [PubMed] [Google Scholar]
- 16.Perry-O’Keefe, H., S. Rigby, K. Oliveira, D. S. Sorensen, H. Stender, J. Coull, and J. J. Hyldig-Nielsen. 2001. Identification of indicator microorganisms using a standardized PNA FISH method. J. Microbiol. Methods 47:281–292. [DOI] [PubMed] [Google Scholar]
- 17.Pezzlo, M. 1998. Interpretation of aerobic bacterial growth on primary culture media, p.51–57. In H. D. Isenberg (ed.), Essential procedures for clinical microbiology. ASM Press, Washington, D.C.
- 18.Rappaport, T., K. P. Sawyer, and I. Nachamkin. 1988. Evaluation of several commercial biochemical and immunological methods for rapid identification of gram-positive cocci directly from blood cultures. J. Clin. Microbiol. 26:1335–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Souvenir, D., D. E. Anderson, Jr., S. Palpant, H. Mroch, S. Askin, J. Anderson, J. Claridge, J. Eiland, C. Malone, M. W. Garrison, P. Watson, and M. D. Campbell. 1998. Blood cultures positive for coagulase-negative staphylococci: antisepsis, pseudobacterimia, and therapy of patients. J. Clin. Microbiol. 36:1923–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Speers, D. J., T. R. Olma, and G. L. Gilbert. 1998. Evaluation of four methods for rapid identification of Staphylococcus aureus from blood cultures. J. Clin. Microbiol. 36:1032–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stefano, K., and J. J. Hyldig-Nielsen. 1997. Diagnostic applications of PNA oligomers, p.19–37. In S. A. Minden and L. M. Savage (ed.), Diagnostic gene detection & quantification technologies. IBC Library Series, Southborough, Mass.
- 22.Stender, H., T. A. Mollerup, K. Lund, K. H. Petersen, P. Hongmanee, and S. E. Godtfredsen. 1999. Direct detection and identification of Mycobacterium tuberculosis in smear-positive sputum samples by fluorescence in situ hybridization (FISH) using peptide nucleic acid (PNA) probes. Int. J. Tuberc. Lung Dis. 3:830–837. [PubMed] [Google Scholar]
- 23.Stender, H., K. Lund, K. H. Petersen, O. F. Rasmussen, P. Hongmanee, H. Miorner, and S. E. Godtfredsen. 1999. Fluorescence in situ hybridization assay using peptide nucleic acid probes for differentiation between tuberculous and nontuberculous Mycobacterium species in smears of Mycobacterium cultures. J. Clin. Microbiol. 37:2760–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stender, H., C. Kurtzman, J. J. Hyldig-Nielsen, D. Sørensen, A. Broomer, K. Oliveira, H. Perry-O’Keefe, A. Sage, B. Young, and J. Coull. 2001. Identification of Brettanomyces (Dekkera bruxellensis) from wine by fluorescence in situ hybridization using peptide nucleic acid probes. Appl. Environ. Microbiol. 67:938–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221–271. [DOI] [PMC free article] [PubMed] [Google Scholar]