Abstract
A rapid real-time multiplex PCR assay for detecting and differentiating Bordetella pertussis and Bordetella parapertussis in nasopharyngeal swabs was developed. This assay (LC-PCR-IS) targets the insertion sequences IS481 and IS1001 of B. pertussis and B. parapertussis, respectively, and is performed using the LightCycler (Roche Molecular Biochemicals, Indianapolis, Ind.). The analytical sensitivity is less than one organism per reaction. Results for Bordetella culture and/or direct fluorescent antibody testing and a second LightCycler PCR assay (target, pertussis toxin gene) were compared to results of the LC-PCR-IS assay for 111 nasopharyngeal swabs submitted for pertussis testing. Of the specimens, 12 were positive (9 B. pertussis and 3 B. parapertussis) and 68 specimens were negative by all methods. Three other specimens were positive for B. pertussis by at least two of the methods (including the LC-PCR-IS assay), and another 28 specimens were positive for B. pertussis by the LC-PCR-IS assay only. No specimens were negative by the LC-PCR-IS assay and positive by the other methods. A conventional PCR method (target, IS481) was also compared to the LC-PCR-IS assay for a different group of nasopharyngeal swab specimens (n = 96): 44 specimens were positive and 41 specimens were negative for B. pertussis with both PCR methods. Nine specimens were positive for B. pertussis by the LC-PCR-IS assay and negative by the conventional PCR assay, and two specimens were positive for B. pertussis by the conventional PCR assay and negative by the LC-PCR-IS assay. Positivity of the two assays was not significantly different (P = 0.0654). The insertion sequence IS481 is also present in Bordetella holmesii; specimens containing B. holmesii may yield false-positive results. The LC-PCR-IS assay takes approximately 45 min to complete post-nucleic acid extraction, compared to 24 h for the conventional PCR assay previously used in our laboratory. The LC-PCR-IS assay is easier to perform than the conventional PCR assay, and the closed system decreases the chance of contamination. All of these characteristics represent a significant improvement in the detection of B. pertussis and B. parapertussis in nasopharyngeal specimens.
The wide prevalence of pertussis and its changing epidemiology have highlighted the need for rapid and sensitive methods for diagnosing Bordetella pertussis infection. Laboratory criteria for the diagnosis of pertussis include either isolation of B. pertussis from a clinical specimen or a positive PCR assay result for B. pertussis (1). Isolation and identification of B. pertussis by culture may take 3 to 7 days and, though specific, lack sensitivity due to the fastidious nature of the organism. Successful recovery by culture is dependent on the type of swab and transport system used, the time delay between specimen collection and inoculation, prior antimicrobial treatment, the stage and duration of the illness, and the immune status of the patient (4, 6, 16). Direct fluorescent antibody (DFA) testing has been used as a rapid tool for diagnosis but also lacks sensitivity. Such limitations of conventional diagnostic testing have led to the development of PCR assays for detection of B. pertussis. These assays have been developed to identify B. pertussis directly in nasopharyngeal specimens. Different regions of the B. pertussis genome have been used as targets for molecular diagnostic assays, including the insertion sequence IS481, the pertussis toxin gene (PTG), the porin gene, and the adenylate cyclase toxin gene (3, 5, 6, 7, 10, 12, 14). A myriad of studies have shown that PCR is more sensitive than culture for diagnosing B. pertussis infection (11, 12, 18). Bordetella parapertussis may also cause a pertussis-like illness.
We developed a rapid (45-min post-nucleic acid extraction) real-time multiplex LightCycler PCR assay (LC-PCR-IS) for detection of B. pertussis and B. parapertussis in nasopharyngeal swabs. Primers and probes that target the insertion sequence (IS481) of B. pertussis and the insertion sequence (IS1001) of B. parapertussis were used. Detection of B. pertussis and B. parapertussis using the LC-PCR-IS assay was compared to that by conventional DFA testing, culture, and other PCR-based assays.
(This work was presented in part at the 40th Interscience Conference on Antimicrobial Agents and Chemotherapy, Toronto, Ontario, Canada, 17 to 20 September 2000, and at the 101st General Meeting of the American Society for Microbiology, Orlando, Fla., 20 to 24 May 2001.)
MATERIALS AND METHODS
Bacterial strains.
Stock cultures of reference strains and clinical isolates of B. pertussis and B. parapertussis (Table 1) were stored at −70°C on porous beads (Pro-Lab Diagnostics, Austin, Tex.). Dilutions of these organisms were used to optimize PCR conditions, evaluate analytical sensitivity, and perform inhibition studies. B. pertussis and B. parapertussis were grown on charcoal agar containing 10% defibrinated sheep blood and 40 μg of cephalexin/ml. Other Bordetella spp. and a panel of organisms representing commonly isolated pathogenic and nonpathogenic respiratory isolates identified by the Clinical Microbiology Laboratory were used to evaluate analytical specificity (Table 1).
TABLE 1.
Bacterial strains studied
| Bacterial sp. | Straina |
|---|---|
| Bordetella spp. | |
| Bordetella avium | |
| Bordetella bronchiseptica (2 strains) | ATCC 19395, CDC AB9-D19-80b |
| Bordetella holmesii (4 isolates)c | ATCC 51541 |
| Bordetella parapertussis (4 isolates) | ATCC 15311 |
| Bordetella pertussis (5 isolates) | ATCC 9797 |
| Common respiratory and other nonrespiratory isolates | |
| Acinetobacter species | |
| Aeromonas hydrophila | |
| Burkholderia cepacia complex | |
| Campylobacter jejuni | |
| Chlamydia pneumoniae | ATCC VR1310 |
| Escherichia coli | ATCC 25922 |
| Haemophilus influenzae (3 strains) | ATCC 49247, ATCC 49766, ATCC 10211 |
| Klebsiella oxytoca | |
| Klebsiella pneumoniae | |
| Legionella jordanis | ATCC 33623 |
| Legionella pneumophila | ATCC 33152 |
| Listeria monocytogenes | |
| Moraxella catarrhalis | |
| Morganella morganii | |
| Mycoplasma hominis | |
| Mycoplasma pneumoniae | |
| Proteus mirabilis | |
| Proteus vulgaris | |
| Pseudomonas aeruginosa | ATCC 27853 |
| Pseudomonas fluorescens | |
| Staphylococcus epidermidis | CDC AB4-BID-84b |
| Stenotrophomonas maltophilia | CDC AB9-D19-80b |
| Streptococcus pneumoniae | |
| Streptococcus pyogenes | |
| Streptococcus spp. viridans group |
If not otherwise specified, strains represent isolates from the Mayo Clinic Clinical Microbiology Laboratory.
Survey isolates identified by standard microbiologic test methods.
Three isolates were recovered from the blood of Mayo Clinic patients.
Clinical specimens.
Between July and November 1999, the Mayo Clinic Bacteriology Laboratory received 111 nasopharyngeal swabs for culture and/or DFA testing for B. pertussis. The majority of the swabs were rayon tipped with wire shafts and were submitted in Amies charcoal agar transport medium (StarPlex Scientific, Inc., Etobicoke, Ontario, Canada). Time from collection of the swab to receipt in the laboratory was 1 to 7 days. After processing for culture and/or DFA testing, the swabs were replaced in the transport medium and held at 4°C until DNA extraction. Ninety-six nasopharyngeal specimens received in the Mayo Clinic Molecular Microbiology Laboratory for the detection of B. pertussis by conventional PCR were also tested by LC-PCR-IS. Culture and/or DFA testing was not performed on these specimens. Seventy-three of these swabs were received in 1999, and 23 were received from 1996 to 1998. The swabs were placed in 500 μl of IsoQuick sample buffer (Orca Research, Inc., Bothell, Wash.), of which 400 μl was used for DNA extraction for detection of B. pertussis by the conventional PCR assay. The remaining specimen was stored at −70°C until DNA was extracted for this study. Specimen processing was performed in a room separate from reagent preparation. Specimens from Minnesota residents who declined use of their specimens for research were not studied (Minnesota Statute 144.335).
Culture.
The 93 nasopharyngeal swabs submitted for Bordetella culture were inoculated onto selective charcoal agar plates containing cephalexin and sheep blood and trypticase soy agar plates with 5% sheep blood (TSAII; BBL Microbiology Systems, Cockeysville, Md.). The plates were incubated at 35°C and checked daily for up to 7 days for the presence of typical colonies.
Fluorescent antibody testing.
Fluorescent antibody testing was used to confirm colonies on culture plates as either B. pertussis or B. parapertussis and for direct detection of B. pertussis and B. parapertussis in nasopharyngeal swabs (DFA testing). Seventy-two nasopharyngeal swabs submitted for DFA testing were placed in 1% Casamino Acids for 1 h prior to smearing two spots marked with a china pencil on a microscope slide. For a culture isolate, the organism was suspended in phosphate-buffered saline before spreading two spots marked with a china pencil on a microscope slide. One spot was stained with fluorescein isothiocyanate-conjugated B. pertussis antiserum, and the other spot was stained with B. parapertussis antiserum (Difco Laboratories, Detroit, Mich.).
Conventional PCR.
The 96 nasopharyngeal swabs submitted for B. pertussis DNA detection by the conventional PCR assay were swished in 500 μl of IsoQuick sample buffer. DNA was extracted from two separate 200-μl aliquots of this specimen with the IsoQuick nucleic acid extraction kit. Each of the two extracts was subjected to conventional PCR. A 262-bp target sequence from IS481 of B. pertussis was amplified (primers: B. pertussis 642, 5′ CTT CAC CGA CAT CCA CCC CGA CGA G 3′; B. pertussis 879, 5′ GTG AGC GTA AGC CCA CTC ACG CAA G 3′ ), electrophoresed in a 2% agarose gel, and Southern blotted using a B. pertussis IS481-specific probe (174 bp, positions 671 to 844, IS481) (A. Algeciras, C. P. Kolbert, and D. H. Persing, Abstr. 94th Gen. Meet. Am. Soc. Microbiol. 1994, abstr. C-26, p. 495, 1994). Concordant positive or negative results between the two conventional PCR assays run on each patient specimen were recorded as positive or negative, respectively. Discordant results were resolved by repeat testing of the original specimen. Conventional PCR was performed in a room separate from the reagent preparation room and the specimen processing room.
Specimen preparation for LightCycler PCR.
The swabs obtained after culture and/or DFA testing were swished in 500 μl of IsoQuick sample buffer; 200 μl of IsoQuick sample buffer was added to the specimens originally processed for conventional PCR testing. DNA was extracted from the nasopharyngeal swab, using the IsoQuick nucleic acid extraction kit according to the manufacturer’s instructions, and reconstituted in 100 μl of sterile water. Extracted DNA was either tested immediately or stored at −20°C until testing was performed.
Primers and probes.
The LC-PCR-IS primers and probes used in this study, LightCycler—Bordetella pertussis/parapertussis, Primers/Hybridization Probes, are available from Roche Molecular Biochemicals (Indianapolis, Ind.). Primers and probes were designed from GenBank sequences M28220 for B. pertussis and X66858 for B. parapertussis to target the insertion sequence IS481 of B. pertussis and the insertion sequence IS1001 of B. parapertussis, respectively. Probes were synthesized by IT Biochem (Salt Lake City, Utah).
Controls.
Pure culture isolates of B. pertussis ATCC 9797 and B. parapertussis ATCC 15311 were used as PCR controls. Bacterial colonies were suspended in sterile water to approximately an 0.5 McFarland standard before lysis at 100°C for 10 min. Tenfold dilutions of the supernatant were analyzed, and the control extracts were used at a final dilution of <100 organisms/reaction. The sterile water used to suspend the extracted DNA pellet was used as the negative control and comprised 10% of specimens included in each run.
LC-PCR-IS assay.
The reaction mixture was prepared in a controlled access reagent preparation room and consisted of 15 μl of PCR master mix plus 5 μl of DNA extract per cuvette. The PCR master mix consisted of 4 mM MgCl2, 50 mM KCl, 20 mM Tris-HCl (pH 8.4), 0.2 mM PCR Nucleotide MixPLUS (1× dATP, dCTP, and dGTP and 3× dUTP [Roche Molecular Biochemicals]), 0.75 μM (each) primers, 0.025% bovine serum albumin (Sigma, St. Louis, Mo.), 0.2% HK-UNG thermolabile uracil-N-glycosylase (Epicentre Technologies, Madison, Wis.), 0.2 μM (each) fluorescein probe, 0.3 μM (each) LC Red-640 probe, 0.5% dimethyl sulfoxide (Sigma), and 0.03 U of Platinum Taq DNA polymerase (Life Technologies, Rockville, Md.)/μl. The carousel containing the sealed cuvettes was centrifuged and placed in the LightCycler. The cycling protocol consisted of 1 cycle of 37°C for 5 min and 95°C for 3 min (activation and inactivation, respectively, of uracil-N-glycosylase), followed by 40 cycles of 95°C for 0 s, 60°C for 20 s, and 72°C for 14 s. A melting curve was generated by measuring the fluorescent signal generated while raising the temperature slowly from 55 to 85°C. Since the same fluorophores were used to detect both insertion sequences, melting curve analysis was used to differentiate the two insertion sequences, hence the two species of Bordetella. The LightCycler software was used to analyze the amplification and melting curve generated in the assay.
Cross-reactivity and inhibition studies.
Boiled lysates of different isolates of common respiratory organisms, other nonrespiratory organisms, and other Bordetella species (Table 1) were tested for cross-reactivity in the LC-PCR-IS assay. The presence of amplifiable DNA was confirmed using broad-range 16S rRNA gene amplification by standard methods (8). A specific PCR assay was used to verify the presence of nucleic acid in the Chlamydia pneumoniae extracted (data not shown). Negative DNA extracts from patient specimens spiked with low concentrations (approximately 10 organisms/μl) of B. pertussis and B. parapertussis were used to assess PCR inhibition. Inhibition was indicated by a loss of amplification signal or appearance of a signal at a later cycle number than noted with similar concentrations of organisms, diluted in sterile water.
LC-PCR-PTG assay.
The LightCycler PCR PTG (LC-PCR-PTG) assay, targeting the PTG, was performed as previously described (L. M. Sloan, M. K. Hopkins, P. S. Mitchell, E. A. Vetter, and F. R. Cockerill, Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., abstr. D-885, p. 147, 2000) using the 111 nasopharyngeal swabs submitted for culture and/or DFA testing. Briefly, a 320-bp target sequence was amplified using primers PTG 2314 (5′ CAA GGA TCT CAA GCG TCC 3′) and PTG 2615 (5′ TGA TTC TG CCC TTC AGTT 3′). The amplified product was detected with a 46-bp (positions 2387 to 2433 of PTG) B. pertussis PTG-specific probe. The probe is positioned over a region containing a 1-bp change in sequence from B. pertussis to B. parapertussis. This results in a melting temperature shift allowing differentiation of the two Bordetella species.
Statistical method.
The positivity rates of the assays were compared using a McNemar test for the samples. The alpha level was set at 0.05 for statistical significance. Agreement of the positivity result by the assays was assessed with a kappa statistic, and a 95% confidence interval was calculated. Interpretations of the relative meaning of the kappas calculated refer to a paper by Cohen (2).
RESULTS
Analytical sensitivity, analytical specificity, and inhibition rates of LC-PCR-IS assay.
The analytical sensitivity of the LC-PCR-IS assay was evaluated using a 10-fold dilution series (105 to 10−1 organism/μl) of B. pertussis and B. parapertussis in water. A sensitivity of approximately 0.75 B. pertussis organisms (or 60 IS481 copies) per reaction and approximately 0.75 B. parapertussis organisms (or 15 IS1001 copies) per reaction was demonstrated. The LC-PCR-IS assay was able to detect both B. pertussis and B. parapertussis in the same tube at less than one organism per reaction.
A panel of 25 common respiratory isolates and other nonrespiratory isolates (Table 1) showed no cross-reactivity in the LC-PCR-IS assay. Seven isolates of three species of Bordetella (Bordetella bronchiseptica, Bordetella avium, and Bordetella holmesii) were also tested. The primer and probe set detecting the insertion sequence IS481 of B. pertussis cross-reacted with all four B. holmesii isolates tested. No cross-reactivity with the other Bordetella species was detected.
Fifty-two nucleic acid extracts of the original sample, from patients negative for B. pertussis and B. parapertussis, were spiked with a low concentration of B. pertussis and B. parapertussis organisms and assayed by the LC-PCR-IS to test for the presence of inhibitors. Complete inhibition of LC-PCR-IS amplification was demonstrated for 5 of the 52 (9.6%) specimens. Repeat testing of the 52 specimens generated the same results.
Detection and discrimination of B. pertussis and B. parapertussis.
The LC-PCR-IS assay was designed to detect and differentiate B. pertussis from B. parapertussis in a single tube using primer and probe sets. This multiplexing did not affect the analytical sensitivity of the assay compared to performing the B. pertussis and B. parapertussis assays in separate tubes. The difference in the melting temperature between B. pertussis (75 ± 2°C) and B. parapertussis (65 ± 2°C) in the multiplex assay was 10°C.
Clinical specimen studies.
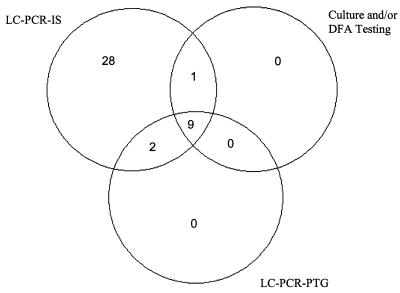
Of the 111 nasopharyngeal specimens tested by culture and/or DFA testing and the LC-PCR-IS and LC-PCR-PTG assays (hereafter referred to as the “combined test panel”), 12 (11%) were positive for B. pertussis by at least one culture and/or DFA testing or the LC-PCR-PTG assay as well as the LC-PCR-IS assay (Fig. 1). Of the 111, 71 (64%) were negative for B. pertussis by the combined test panel. Two of the 12 nasopharyngeal specimens referred to above were positive for B. pertussis by the LC-PCR-IS and LC-PCR-PTG assays but not by culture and/or DFA. One of the 12 specimens was positive for B. pertussis by the LC-PCR-IS and culture but not by the LC-PCR-PTG assay. Twenty-eight of the 111 specimens were positive for B. pertussis by the LC-PCR-IS assay but negative by culture and/or DFA testing and the LC-PCR-PTG assay. A significantly higher positivity for the LC-PCR-IS assay was seen (P < 0.0001). There was fair agreement between the assays as measured with the kappa statistic, κ = 0.35 (95% confidence interval: 0.20, 0.51). Three specimens were positive and 108 specimens were negative for B. parapertussis by the combined test panel.
FIG. 1.
B. pertussis detection using culture and/or DFA testing and the LC-PCR-IS and LC-PCR-PTG assays (test panel). Seventy-one specimens yielded negative results by the test panel.
Ninety-six nasopharyngeal specimens were used to compare B. pertussis detection by the conventional PCR assay with that by the LC-PCR-IS assay. Neither culture nor DFA testing was performed on these specimens. Forty-four specimens were positive and 41 specimens were negative for B. pertussis by both methods. Nine specimens were positive for B. pertussis by the LC-PCR-IS assay and negative by the conventional PCR assay, and two specimens were negative by the LC-PCR-IS assay and positive by the conventional PCR assay. The positivity rates of the two assays were not significantly different (P = 0.0654). There was substantial agreement between the two assays as measured by the kappa statistic, κ = 0.77 (95% confidence interval: 0.65, 0.90). Two specimens negative for B. pertussis by the conventional PCR assay were positive for B. parapertussis by the LC-PCR-IS assay. The conventional assay was not designed to detect B. parapertussis.
DISCUSSION
PCR has consistently been demonstrated to be the most sensitive means of detecting B. pertussis infection (11, 12, 18). The LightCycler PCR assay described herein represents an advance beyond conventional PCR because it takes 45 min (post-nucleic acid extraction) to complete, is easier to perform than a conventional assay, and is performed in a closed system, decreasing the chance of contamination. A further strength of this assay is that it detects and differentiates B. pertussis and B. parapertussis. As a result of these advantages, this assay has become the primary diagnostic test for pertussis at our institution.
While our study did not specifically resolve the status of specimens that were positive by the LC-PCR-IS assay and not by culture, DFA testing, or the LC-PCR-PTG assay, several points support the specificity of the LC-PCR-IS assay. First, a previous study where such discrepant results were resolved by review of patient histories concluded that PCR is a highly specific method for the detection of B. pertussis (M. J. Loeffelholz, C. J. Thompson, K. S. Long, and M. J. R. Gilchrist, Letter, J. Clin. Microbiol. 38:467, 2000). Second, the disparity in sensitivities between the two LightCycler assays, the LC-PCR-IS and the LC-PCR-PTG assays, may reflect the copy number of IS481 in the B. pertussis chromosome (50 to 100 copies) and the copy number of IS1001 in the B. parapertussis chromosome (35 to 50 copies) versus the single copy of the PTG in B. pertussis and B. parapertussis. The sensitivity of the LC-PCR-PTG assay is <10 organisms per reaction for B. pertussis and <100 organisms per reaction for B. parapertussis (Sloan et al., 40th ICAAC). Third, the comparability of results generated by the LC-PCR-IS assay to those generated using a conventional PCR assay supports the specificity of the LC-PCR-IS assay.
One limitation of the insertion sequence IS481 as a target for B. pertussis detection is that this insertion sequence is also present in B. holmesii. However, in our opinion, the sensitivity for detecting B. pertussis provided by IS481 outweighs this limit in specificity. Specimens positive for B. pertussis IS481 DNA by PCR could be subjected to a second PCR assay which uses another target such as PTG; however, given that the PTG target is a less sensitive target, this approach is of limited use. B. holmesii was first described in 1995 (19) and has been associated with septicemia, endocarditis, and respiratory symptoms (15). Cephalexin, an antimicrobial currently recommended for the isolation of Bordetella spp., is widely used in various culture media and has an inhibitory effect on B. holmesii, which likely explains why most laboratories have not isolated B. holmesii from nasopharyngeal specimens submitted for pertussis culture (13). In a recently published study, B. holmesii was isolated from 0.3% of nasopharyngeal specimens (versus B. pertussis, which was isolated from 7% of nasopharyngeal specimens) from patients suspected of having pertussis (20). Further studies are needed to determine the role of B. holmesii in causing cough and the prevalence of asymptomatic nasopharyngeal carriage of B. holmesii.
The change from culture-based techniques to PCR-based techniques for detection of B. pertussis and B. parapertussis requires a reevaluation of what represents a suitable specimen for testing. Collection of nasopharyngeal specimens for culture and DFA testing is uncomfortable for the patient; although throat swabs are not ideal for B. pertussis culture, a reassessment of throat swabs for B. pertussis detection using PCR should be considered (4). The ideal swab and transport medium for PCR-based assays remains to be defined; calcium alginate fibers and aluminum shaft components have been shown to inhibit some PCR assays used to detect B. pertussis (17). This issue needs to be reevaluated in the context of the specific nucleic acid extraction method used. The optimal nucleic acid extraction method for pertussis PCR also needs to be defined. All of these issues are being evaluated in our laboratory.
A final point must be considered if PCR-based assays are used as stand-alone diagnostic tests for detection of B. pertussis. Isolates of B. pertussis will not be available for conventional antimicrobial susceptibility testing and nucleic acid fingerprinting for epidemiological purposes. Despite recent isolated reports of erythromycin resistance in B. pertussis, routine antimicrobial susceptibility testing of B. pertussis isolates is not currently recommended (9). Culture and erythromycin susceptibility testing could be performed from specimens that are positive by PCR or in cases of therapeutic failure. Molecular biology-based assays could be designed to detect erythromycin resistance-associated rRNA gene mutations.
Acknowledgments
This work was supportd by a grant from the National Insitutes of Health (R03 AR47009 to R.P.) and by the Mayo Foundation.
We acknowledge the secretarial assistance of Gretchen A. Thomason.
REFERENCES
- 1.Centers for Disease Control and Prevention. 1997. Case definitions for infectious conditions under public health surveillance. Morb. Mortal. Wkly. Rep. 46(RR-10):25. [PubMed] [Google Scholar]
- 2.Cohen, J. 1960. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 20:37–46. [Google Scholar]
- 3.Douglas, E., J. Coote, R. Parton, and W. McPheat. 1993. Identification of Bordetella pertussis in nasopharyngeal swabs by PCR amplification of a region of the adenylate cyclase gene. J. Med. Microbiol. 38:140–144. [DOI] [PubMed] [Google Scholar]
- 4.Farrell, D., G. Daggard, and T. Mukkur. 1999. Nested duplex PCR to detect Bordetella pertussis and Bordetella parapertussis and its application in diagnosis of pertussis in nonmetropolitan southeast Queensland, Australia. J. Clin. Microbiol. 37:606–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glare, E., J. Paton, R. Premier, A. Lawrence, and I. Nisbet. 1990. Analysis of a repetitive DNA sequence from Bordetella pertussis and its application to the diagnosis of pertussis using the polymerase chain reaction. J. Clin. Microbiol. 28:1982–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hallander, H. 1999. Microbiological and serological diagnosis of pertussis. Clin. Infect. Dis. 28(Suppl. 2):S99–S106. [DOI] [PubMed] [Google Scholar]
- 7.Houard, S., C. Hackel, A. Herzog, and A. Bollen. 1989. Specific identification of Bordetella pertussis by polymerase chain reaction. Res. Microbiol. 140:477–487. [DOI] [PubMed] [Google Scholar]
- 8.Johnson, J. 1994. Similarity analysis of rRNAs, p. 683–700. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 9.Korgenski, E., and J. Daly. 1997. Surveillance and detection of erythromycin resistance in Bordetella pertussis isolates recovered from a pediatric population in the intermountain west region of the United States. J. Clin. Microbiol. 35:2989–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li, Z., D. Jansen, T. Finn, S. Halperin, A. Kasina, A. O’Connor, T. Aoyama, C. Manclark, and M. Brennan. 1994. Identification of Bordetella pertussis infection by shared-primer PCR. J. Clin. Microbiol. 32:783–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loeffelholz, M., C. Thompson, K. Long, and M. Gilchrist. 1999. Comparison of PCR, culture and direct fluorescent-antibody testing for detection of Bordetella pertussis. J. Clin. Microbiol. 37:2872–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mastrantonio, P., P. Stefanelli, and M. Giuliano. 1996. Polymerase chain reaction for the detection of Bordetella pertussis in clinical nasopharyngeal aspirates. J. Med. Microbiol. 44:261–266. [DOI] [PubMed] [Google Scholar]
- 13.Mazengia, E., E. Silva, J. Peppe, R. Timperi, and H. George. 2000. Recovery of Bordetella holmesii from patients with pertussis-like symptoms: use of pulsed-field gel electrophoresis to characterize circulating strains. J. Clin. Microbiol. 38:2330–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller, F., J. Hoppe, and C. Wirsing von Konig. 1997. Laboratory diagnosis of pertussis: state of the art in 1997. J. Clin. Microbiol. 35:2435–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang, Y., M. Hopkins, C. Kolbert, P. Hartley, P. Severance, and D. Persing. 1998. Bordetella holmesii-like organisms associated with septicemia, endocarditis, and respiratory failure. Clin. Infect. Dis. 26:389–392. [DOI] [PubMed] [Google Scholar]
- 16.Van der Zee, A., C. Agterberg, M. Peeters, J. Schellekens, and F. Mooi. 1993. Polymerase chain reaction assay for pertussis: simultaneous detection and discrimination of Bordetella pertussis and Bordetella parapertussis. J. Clin. Microbiol. 31:2134–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wadowsky, R., S. Laus, T. Libert, S. States, and G. Ehrlich. 1994. Inhibition of PCR-based assay for Bordetella pertussis by using calcium alginate fiber and aluminum shaft components of a nasopharyngeal swab. J. Clin. Microbiol. 32:1054–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wadowsky, R., T. Libert, and G. Ehrlich. 1994. Detection of Bordetella pertussis using PCR, p. 621–632. In G. D. Ehrlich and S. J. Greenberg (ed.), PCR-based diagnostics in infectious disease. Blackwell Scientific Publications, Boston, Mass.
- 19.Weyant, R., D. Hollis, R. Weaver, M. Amin, A. Steigerwalt, S. O’Connor, A. Whitney, M. Daneshvar, C. Moss, and D. Brenner. 1995. Bordetella holmesii sp. nov., a new gram-negative species associated with septicemia. J. Clin. Microbiol. 33:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yih, W., E. Silva, J. Ida, N. Harrington, S. Lett, and H. George. 1999. Bordetella holmesii-like organisms isolated from Massachusetts patients with pertussis-like symptoms. Emerg. Infect. Dis. 5:441–443. [DOI] [PMC free article] [PubMed] [Google Scholar]