Abstract
Rabbits that had been infected intravenously with conidiospores of Aspergillus fumigatus were used as sources of antibody for screening a λ phage cDNA expression library. The cDNA was derived from A. fumigatus mRNA that had been extracted from newly formed, germling hyphae. Thirty-six antigens were identified using antisera from six rabbits. Though many of these antigens were expected to be intracellular proteins because their genes did not encode a signal sequence, the antisera showed consistently a stronger immunoblot reaction with a cell fraction enriched for the fungal cell wall than with a fraction of predominantly intracellular components. Antibodies to eight antigens, including the glycosylhydrolase Asp f 16, were produced by more than one rabbit. In current vaccine studies, Asp f 16 is the only single antigen which has been reported to be capable of inducing protection against invasive aspergillosis in mice (S. Bozza et al., Microb. Infect. 4:1281-1290, 2002). Enolase and Aspergillus HSP90 were detected also; their homologues in Candida albicans have been tested as vaccines and have been reported to provide a partially protective response against invasive candidiasis in mice. The Aspergillus antigens reported here may have value both in diagnostic tests for different forms of aspergillosis and as vaccine candidates for protection against invasive disease.
The widespread use of immunosuppressive drugs for cancer patients and recipients of solid organ and bone marrow transplants has led to a major increase in the incidence of invasive, often lethal, infections caused by opportunistic fungi. Molds of the genus Aspergillus produce many of these infections, nearly half in some instances, and Aspergillus fumigatus is by far the most prominent species especially in leukemia (incidence, 5 to 25%) and in patients transplanted with solid organs, where the incidence is 1 to 10% (15, 16, 28, 46). Compounding the seriousness of the infection, the available antifungal agents are not ideal; indeed, invasive aspergillosis can lead to death in more than half of all patients (30). Therefore, the development of either a protective vaccine for use in high-risk patients or a therapeutic vaccine for those patients already infected would be extremely valuable.
Development of an effective vaccine against A. fumigatus may well be possible because a protective response to Aspergillus has been demonstrated in animals. In the mid-1970s, various researchers demonstrated that mice infected with A. fumigatus developed an increased resistance to later challenge with spores (50). Lehmann and White (29) further reported that when a localized kidney infection had been present for some time, mice developed a systemic resistance to A. fumigatus that was not lost after treatment with cortisone. At the same time, Corbel and Eades (11) found that adult mice had considerably greater resistance to A. fumigatus than did young mice of the same strain. These observations led to several vaccine studies, including one in young turkeys that was conducted with preparations representing different growth stages of the fungus. The inactivated germling vaccine, administered subcutaneously, showed the best protection (38% survival) against an otherwise lethal aerosol challenge with spores (43).
There have been several studies attempting to define the factors responsible for the increased resistance that develops in infected mice. In 1993, de Repentigny et al. reported that the transfer of resistance from sublethally infected to naïve mice could be achieved using splenic macrophages but not serum (18). The role of cellular immunity was further supported when resistance that had been induced by intranasal vaccination with either viable A. fumigatus conidia or culture filtrate was experimentally transferred to naïve recipients using antigen-specific CD4+ T cells (9). In addition, after intranasal vaccinations with the A. fumigatus allergen Asp f 16, mice developed significantly increased resistance to invasive aspergillosis only when the allergen was administered together with a CpG oligodeoxynucleotide adjuvant that promotes a predominantly TH1 immune response (6). The latter result seems to be the first report that resistance to A. fumigatus can be induced by a single antigen.
As aspergilli have genomes that are estimated as containing around 10,000 genes, a variety of other antigens with a potential of inducing a protective immune response are likely to be found. Such antigens may differ in potency, and a vaccine containing mixtures of protective antigens may prove more effective than one containing only a single component. Here, we report on our search for additional immunogenic fungal proteins that are produced following an infection established via the intravenous route and against which antibodies develop in infected rabbits. We have used the antisera to identify proteins expressed by recombinant λ phage containing cDNA derived from the mRNA of young A. fumigatus hyphae (also known as germling hyphae).
MATERIALS AND METHODS
Aspergillus cDNA expression library.
A. fumigatus D141 (NRRL 6585; U.S. Department of Agriculture, Peoria, IL), is a clinical isolate derived from an aspergilloma developing in a 45-year-old human with tuberculosis (51). The fungus was cultured at 37°C on Sabouraud agar (20 g of d-glucose, 10 g of neopeptone, 15 g of agar per liter) for 3 days. Conidia from each petri dish were suspended in 10 ml of 0.9% (wt/vol) NaCl containing 0.1% (vol/vol) Tween 80 (Sigma Chemical Co., St. Louis, MO). Four two-liter flasks containing 250 ml of a glucose-ammonium salts minimal medium (12) were thickly inoculated, each with the conidia from one petri dish, and then were incubated on a rotary shaker (for 16 h at 100 rpm at 37°C) to allow almost all the conidia to germinate and the culture to enter a logarithmic growth phase. The germling hyphae were harvested by filtration through Whatman no. 1 filter paper and then washed twice with cold phosphate-buffered saline. Subsequently, hyphae were frozen in liquid nitrogen and ground to a fine powder using a mortar and pestle. Total RNA was extracted according to standard procedures using guanidinium thiocyanate, followed by centrifugation through a cesium chloride cushion (47). The mRNA fraction was obtained using oligo(dT) cellulose (New England Biolabs, Beverly, MA) to bind poly(A) RNA (47), and the mRNAs were incorporated into a λ phage expression library using the λ phage ZAP Express vector kit (Stratagene, La Jolla, CA) and the protocols of the manufacturer.
Rabbit model.
Invasive aspergillosis in rabbits was established by following the protocol of de Repentigny et al. (17). In order to induce immunosuppression, New Zealand White rabbits weighing 2.5 to 3.5 kg were injected subcutaneously with cortisone acetate (10 mg/kg of body weight/day), beginning 2 days before and ending 3 days after intravenous challenge with conidia (day −2 to day 3). To avoid bacterial superinfections, animals also received ceftazidime (70 mg per kg of body weight; Fortum, GlaxoSmithKline, München) injected subcutaneously in aqueous solution in single daily doses (day −2 to day 3). These same cortisone and ceftazidime treatments were used for both the initial and any subsequent challenges with conidia. Rabbits were infected intravenously on day 0 using conidia obtained from 3-day-old Sabouraud agar cultures. These conidia were prepared as a suspension in 0.9% (wt/vol) NaCl from which almost all large aggregates of conidia, conidiophores, and hyphae had been removed by use of rigorous shaking on a vortex mixer and a brief centrifugation step, followed by passage of the supernatant through woven fabric with a mesh opening of approximately 5 μm (Sefar, Wasserburg, Germany). The concentration of conidia in the filtrate was then determined with a hemacytometer and adjusted to the desired number by the addition of 0.9% NaCl.
Ten rabbits were infected intravenously with a 1.0-ml suspension of conidia. The number of spores per rabbit varied, with the first dose ranging from 5.0 × 103 to 2.5 × 105 conidia (Table 1). Depending on the clinical course and on the detection of an antibody response against Aspergillus antigens by using immunoblotting (see below), injections of conidia were repeated up to two further times using higher doses ranging from 5.0 × 104 to 1.0 × 106 spores per challenge (Table 1). The time interval between challenges was 4 weeks at minimum, but this was extended to a maximum of 10 weeks for some rabbits to allow their recovery from illness. For this purpose, the body weight and temperature of each rabbit were measured daily, with loss in body weight and development of fever (>39.5°C) used as signs of illness. When severely ill rabbits were sacrificed, their inner organs were inspected macroscopically. Suspicious lesions were examined microscopically for fungal elements by employing calcofluor white staining (21). Viable A. fumigatus was detected by streaking tissue samples onto Sabouraud agar plates which were then incubated (at 37°C for 2 days) to allow fungal growth to become obvious.
TABLE 1.
Antibody production and outcome in rabbits following i.v. infection with A. fumigatus
| Rabbit no. | Challenge no. | Challenge dose (no. of conidia) | Antibody productiona | Serum identifier | Fever and weight lossb | Outcomec |
|---|---|---|---|---|---|---|
| 1 | 1 | 5.0 × 103 | − | − | No clinical illness | |
| 2 | 5.0 × 104 | + | 1 | − | ||
| 2 | 1 | 5.0 × 103 | + | 2a | + | Complete recovery |
| 2 | 5.0 × 104 | ++ | 2b | − | ||
| 3 | 5.0 × 105 | ++ | − | |||
| 3 | 1 | 2.0 × 104 | + | 3a | − | Death 12 weeks after second challenge |
| 2 | 5.0 × 105 | ++ | 3b | + | ||
| 4 | 1 | 2.0 × 104 | + | 4a | − | Complete recovery |
| 2 | 5.0 × 105 | ++ | 4b | + | ||
| 3 | 1.0 × 106 | ++ | − | |||
| 5 | 1 | 5 × 105 | ++ | 5 | + | Death 3 weeks after challenge |
| 6 | 1 | 2.5 × 105 | − | − | Death 2 weeks after second challenge | |
| 2 | 1.0 × 106 | ++ | 6 | + |
Reactive or strongly reactive immunoblot (+ or ++, respectively) or no detectable increase of staining (−) when compared to the preinfection serum. A strongly reactive immunoblot is shown in Fig. 1.
Fever and weight loss present (+) or not detected (−).
Rabbit 3 had a large mycotic focus (4 by 4 mm) in the liver and many lesions in the left kidney. Rabbit 5 had severe fungal infection of kidneys with several isolated foci in liver and lungs. Rabbit 6 had severe fungal infection of the kidneys, liver, and spleen and several foci in the lung.
All experimental work with the rabbits was performed at the University of Göttingen in a facility that adheres to German federal and state laws and guidelines concerning the use of animals in biomedical research. (Protocol 509.42502/01-10.02, Approval date 04/30/2002; District Government [Bezirksregierung] of Braunschweig, Germany). The animal handling standards are consistent with those required for any research using animals that is sponsored by the U.S. Government.
Immunoblots.
One week prior to infection and every week after the first injection of A. fumigatus, a 10-ml blood sample was obtained from each rabbit. Sera were prepared and used for immunoblotting, with the preinfection serum acting as a control. Seroconversion was detected by using two different A. fumigatus preparations, one consisting largely of cell wall proteins and the other enriched for intracellular proteins. For both preparations, A. fumigatus was grown in a rotary shaker (9 to 10 h at 150 rpm at 37°C) in minimal medium (12) to allow germlings to form. Approximately 10 g of germlings was harvested by filtering the culture though a Whatman no. 1 filter paper; then they were washed with 20 mM Na citrate buffer, pH 5.5, and transferred to a 50-ml polystyrene tube. Thirty milliliters of 100 mM Tris-HCl buffer, pH 8.5, and 100 μl of recombinant β(1, 3)-glucanase (Quantazyme ylg, 20 U/μl; Quantum Biotechnologies, Canada) were added, and the cells were incubated with gentle shaking (30 min at 37°C). Then, the suspension was centrifuged (500 × g for 10 min), and the supernatant containing primarily cell wall proteins was separated from the fungal cells. The supernatant was adjusted to pH 7.0 with dilute HCl, and the proteinase inhibitor phenylmethylsulfonyl fluoride was added to a final concentration of 150 μM. This cell wall protein preparation was then concentrated to a volume of 300 to 400 μl and dialyzed against H2O using a Vivaspin concentrator (10,000-molecular-weight exclusion; PES; Vivascience, Germany) according to the instructions of the manufacturer. The protein concentration was determined by the Bradford assay (Bioquant protein reagent solution; Merck, Germany), and aliquots of this “cell wall” preparation were stored at −20°C.
The intracellular protein preparation was made from approximately 2 g of germling cells that had sedimented during the centrifugation step that followed the incubation with β-glucanase (described above). The sediment was suspended in 5 ml of Tris-HCl buffer (50 mM; pH 8.5), and 4 g of glass beads (0.45 to 0.5 mm diameter; Braun, Germany) was added. The germlings were disintegrated by vortexing the sample for 5 min at maximal speed, and the resultant slurry was transferred to a fresh tube and centrifuged (10,000 × g for 10 min). The supernatant was adjusted to pH 7.0 with dilute HCl and supplemented with phenylmethylsulfonyl fluoride (final concentration, 150 μM). The protein concentration was determined using the Bradford assay, and after being placed in aliquots, this preparation, which had been enriched for intracellular proteins, was stored at −20°C.
The two preparations (7.5 μg of protein each), which had been enriched for either cell wall or intracellular antigens, were denatured by heating in Laemmli buffer for 3 min at 95°C (26) and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Following electrophoresis, the proteins were blotted onto a nitrocellulose membrane according to standard protocols (4). After incubation in blocking buffer, consisting of phosphate-buffered saline, pH 7.4, containing both 0.1% (wt/vol) Tween 20 (Merck, Germany) and 10% (wt/vol) skim milk powder, the membranes were incubated with a rabbit serum that had been diluted 1:100 with blocking buffer (at room temperature for 2 h). Unbound immunoglobulins were removed by washing with blocking buffer, and the membranes, together with any attached antibodies, were then incubated with a 1:10,000 dilution of peroxidase-conjugated monoclonal anti-rabbit immunoglobulins (A-2074; Sigma Chemical) prepared in blocking buffer. Following incubation for 2 h, the membranes were washed, and any conjugate that remained was detected via chemiluminescence using the ECL-system (Amersham Bioscience, United Kingdom) and Kodak BIoMax ML imaging films in accordance with the protocols of the suppliers. In all cases, the results for sera taken from an infected rabbit were compared with the result for the preinfection serum taken from the same rabbit, and the relative difference in immunoblot intensity was ranked, by eye, as being insignificant, reactive, or strongly reactive. These rankings were based on the full range of immunoblots used in the current report.
Immunoblotting with recombinant AHP1 (see Table 2, antigen 32) as antigen were performed using the same method.
TABLE 2.
Putative function, amino acid number, and related proteins in GenBank database for antigens identified with a single rabbit antiserum (antigens 1 to 28) or with more than one antiserum (antigens 29 to 36)
| Antigen | GenBank accession no. and organism | Descriptiona(reference) | No. of amino acidsa | Putative functiona | Serum usedb |
|---|---|---|---|---|---|
| 1 | AAK49451, A. fumigatus | Enolase (27) | 438 | Carbohydrate transport and metabolism | 2a, 2b |
| 2 | EAA65375, A. nidulans | Histone H3 | 141 | Histone | 2a |
| 3 | EAA63190, A. nidulans | Putative Sla2 protein (yeast) | 967 | Cytoskeleton assembly control | 2a |
| 4 | EAA57767, A. nidulans | Putative Acyl CoA binding proteind | 144 | Lipid metabolism | 2b |
| 5 | EAA61425, A. nidulans | Putative cation exchanger | 742 | Sodium/calcium exchanger; integral membrane protein | 2b |
| 6 | CAF32126, A. fumigatus | Putative nuclear segregation protein (40) | 510 | Mitosis and vesicular transport | 2b |
| 7 | EAA60966, A. nidulans | Pyruvate decarboxylase | 585 | Carbohydrate transport and metabolism | 2b |
| 8 | EAA57865, A. nidulans | Formate dehydrogenase | 377 | Energy production and conversion | 2b |
| 9 | EAA63885, A. nidulans | Bromo domain containing protein | 312 | Unknown | 3a |
| 10 | EAA61085, A. nidulans | Protein containing class II aldolase/adducin domain | 312 | Possibly carbohydrate metabolism | 3a |
| 11 | EAA59042, A. nidulans | Hypothetical protein | 106 | Unknown | 3a |
| 12 | EAA62389, A. nidulans | Hypothetical protein with chromosome segregation ATPase-containing region | 779 | Cell division and chromosome partitioning | 3a |
| 13 | EAA61660, A. nidulans | Hypothetical protein with a region of the superfamily I DNA and RNA helicases and helicases subunit | 1,162 | Possibly DNA replication, recombination, and repair | 3a |
| 14 | EAA60227, A. nidulans | Putative pyruvate carboxylase | 1,196 | Carbohydrate transport and metabolism | 3b |
| 15 | EAA65951, A. nidulans | Hypothetical protein with similarities to archain 1 (yeast) | 516 | Intracellular vesicle mediated transport | 3b |
| 16 | EAA63693, A. nidulans | Hypothetical protein with a tetratricopeptide repeat domain | 576 | Unknown | 3b |
| 17 | EAA63343, A. nidulans | Hypothetical protein with DnaJ molecular chaperone homology domain | 466 | Unknown | 3b |
| 18 | EAA62238, A. nidulans | Hypothetical protein with region similar to chromosome segregation ATPases | 941 | Possibly cell division and chromosome partitioning | 4a 4b |
| 19 | EAA59763, A. nidulans | HSP 30 | 180 | Heat shock protein, chaperone | 4a |
| 20 | CAA75754, A. fumigatus | PEP2 (A1) cellular aspartic protease (42) | 398 | Protein turnover (vacuole) | 4a |
| 21 | EAA58700, A. nidulans | Hypothetical protein with regions homologous to DNA mismatch repair enzyme PMS1 (yeast) | 1,228 | Probably DNA replication, recombination, and repair | 4b |
| 22 | EAA66952, A. nidulans | Hypothetical protein; putative choline dehydrogenase | 631 | Amino acid transport and metabolism | 5 |
| 23 | EAA66888, A. nidulans | Hypothetical protein; contains a signal for secretion | 305 | Unknown | 5 |
| 24 | EAA64755, A. nidulans | Hypothetical protein, contains similarity to PRP39 (yeast) | 588 | Possibly pre-mRNA 3′ end processing | 6 |
| 25 | EAA58305, A. nidulans | Hypothetical protein with a WD40 domain and homology to PRP19/PSO4 (yeast) | 475 | Pre-mRNA processing | 6 |
| 26 | EAA64758, A. nidulans | Hypothetical protein with similarity to aminopeptidase N (M1) | 883 | Protein turnover | 6 |
| 27 | EAA59355, A. nidulans | Hypothetical protein with similarity to sterol C14-reductase | 496 | Ergosterol biosynthesis | 6 |
| 28 | EAA58058, A. nidulans | Ribosomal L10 protein | 250 | Ribosome (mRNA-directed protein synthesis) | 6 |
| 29 | CAF32073, A. fumigatus | Putative transketolase (40) | 689 | Carbohydrate transport and metabolism | 1, 6 |
| 30 | EAA58837, A. nidulans | Hypothetical protein | 1,157 | Unknown | 2ac, 4bc |
| 31 | AAB51544, A. fumigatus;EAA59007, A. nidulans | HSP90 (25) | 441 (partial);700 | Heat shock protein, chaperone | 2a, 2b, 5 |
| 32 | O43099, A. fumigatus | AHP1 = allergen Asp f 3 (= PMP20) (23), peroxiredoxin family reductase | 168 | Antioxidant, probable peroxisomal membrane protein | 2a, 2b, 5 |
| 33 | CAF31979, A. fumigatus | Putative HSP88 (40) | 714 | Heat shock protein, chaperone | 3bc, 4a |
| 34 | EAA57903, A. nidulans | Probable translation elongation factor 1 | 411 | translation | 1c, 5, 6 |
| 35 | EAA63438, A. nidulans | Phosphoglucomutase | 556 | Carbohydrate transport and metabolism | 2b, 5, 6 |
| 36 | CAA11266, AAC61261, AAN87849, A. fumigatus | Allergen Asp f 9 (13), allergen Asp f 16 (2), CRH-like protein | 302, 427, 395 | Glycosidase / cell wall metabolism | 2b, 5, 6 |
Descriptions are based on homologues that are described for A. fumigatus, A. nidulans, or yeast (S. cerevisiae) that were found in BLAST searches, performed August 2004.
Serum identifiers are listed in Table 1.
Showed weak reactivity with preinfection serum. All other antigens showed no detectable reactivity with preinfection serum from responding rabbits.
Acyl CoA, acyl coenzyme A.
A. fumigatus cDNA-expression library screening.
Rabbits in which seroconversion had been detected by using immunoblotting (in this case, only 6 of the 10 rabbits) provided the antisera employed for screening the A. fumigatus cDNA expression library. Prior to screening the expression library, antibodies reactive with Escherichia coli and λ phage were removed from the antisera by pseudoscreening (47) using E. coli XL1 Blue MRF′ (Stratagene) which had been transformed with λ ZAP Express phage containing no cDNA insert. Following the manufacturer's (Stratagene's) recommended protocols, approximately 15,000 E. coli plaque lysates per petri dish were generated, and the combined lysate materials were transferred to a Protran BA83 nitrocellulose membrane (Schleicher and Schuell, Germany). Sera diluted in blocking buffer (1:100) were incubated with the membranes in a process similar to that described for the screening (see below). This process was repeated at least five to six times. The absorbed sera were then used for several rounds of screening the cDNA library without detectable loss of activity.
When used for immunoscreening, preparations of E. coli containing the A. fumigatus cDNA expression library in λ phage were cultivated in petri dishes as described above. The library was overlaid with nitrocellulose membranes that had been equilibrated with 0.01 M IPTG (isopropyl-β-d-thiogalactopyranoside) prior to use. This was done to induce the lacZ promoter and thereby allow efficient expression of the recombinant β-galactosidase-Aspergillus fusion proteins produced using the cDNA library inserted into λ ZAP Express. After incubation (for 5 to 6 h at 37°C), the membranes were removed from the agar plates and then blocked with blocking buffer. The membranes were then incubated with the 1:100 dilution of absorbed rabbit serum (for 2 h at room temperature) and, after being washed in blocking buffer, the membrane-bound antibodies were detected using peroxidase-labeled anti-rabbit immunoglobulins by using the procedures described above for immunoblotting. Plaques that bound antibody were excised from the petri dish cultures and transferred to 500 μl of SM-buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM MgSO4, 0.01% [wt/vol] gelatin) mixed with 20 μl of chloroform. After overnight diffusion of phages into the buffer, the phage titer of this stock was determined from the number of plaque-forming units produced on E. coli following standard procedures used for λ phage (47). Then, screening for antibody-reactive plaques was repeated with an agar plate containing only 100 to 200 plaques derived from the stock so that monoclonal phages from a single reactive plaque could be isolated.
Identification of antigens.
The Aspergillus cDNAs in the λ ZAP Express vectors were subcloned into pBK-CMV (where CMV is cytomegalovirus) plasmids following the protocols of the manufacturer (Stratagene). Subsequently, the plasmids were propagated in E. coli XLORL and purified according to standard procedures (47). The cDNAs in the plasmids were partly sequenced either from the N-terminal encoding extremity or from the poly(A) tail or both using the standard sequencing primers for the pBK-CMV plasmid. The sequencing was done at Seqlab Laboratories (Göttingen, Germany).
Sequences were studied using WU-BLAST, version 2.0 (1996), provided by Warren Gish at Washington University in St. Louis via the Washington University BLAST archives. BLAST searches against various databases were performed using sites available via the server of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/).
Genomic data for A. fumigatus were provided by The Institute for Genomic Research (www.tigr.org/tdb/e2k1/afu1) and The Wellcome Trust Sanger Institute (www.sanger.ac.uk/Projects/A_fumigatus); genomic data for Aspergillus nidulans were provided by The Broad Institute (http://www.broad.mit.edu/annotation/fungi/aspergillus/); and genomic data for Aspergillus oryzae was provided by The National Institute of Advanced Industrial Science and Technology (http://oryzae.cbrc.jp/ and www.bio.nite.go.jp/dogan/Top). The analysis of these data was enabled by an international collaboration involving more than 50 institutions from 10 countries and coordinated from Manchester, United Kingdom (www.cadre.man.ac.uk and www.aspergillus.man.ac.uk).
Preparation of recombinant AHP1 and its use in immunoblots.
A 168-amino-acid peptide corresponding to the entire AHP1 protein (GenBank accession no. O43099 (Table 2, antigen 32) was produced using the pET expression system from Novagen (Darmstadt, Germany). Plasmid pET-11a was first modified by incorporating 5′-CATGCACCATATGCACCATATGCACCATGGTAAGGATC-3′ between the unique NheI and BamHI cloning sites. This insert encoded a Met-His6 amino acid sequence, and the sixth His residue of the newly generated plasmid pET-11aH6 was encoded by a unique NcoI cloning site. The primers 5′-GTTCCATGGGATGTCTGGACTCAAGGCCGGTG-3′ and 5′-CTTGGATCCTTACAGGTGCTTGAGGACGG-3′ were used in a PCR to amplify the cDNA for APH1 that had been subcloned into the pBK-CMV plasmid when originally sequencing the gene (described above). The PCR products were then digested with NcoI and BamHI and cloned into the NcoI and BamHI sites of pET-11aH6. The generated plasmid was named pAHP1.
E. coli BL21 cells that had been transformed with pAHP1 were grown at 37°C until they reached a concentration of approximately 0.6 optical density at 600 nm. Expression of recombinant 6-His-tagged AHP1 was induced by adding IPTG (0.1 mM final concentration) and continuing the incubation for 4 h. Bacteria were then collected by centrifugation, and the 6-His tagged APH1 was extracted using guanidine hydrochloride buffer and Ni-nitrilotriacetic acid resin columns using the method described by the manufacturer (QIAGEN, Germany). When used as the antigen in immunoblotting, approximately 0.5 μg of purified AHP1 per lane was applied to a 15% SDS-polyacrylamide gel, subjected to electrophoresis, then blotted onto nitrocellulose.
RESULTS
Infection of rabbits.
Of the 10 rabbits infected, 4 succumbed without developing a strongly positive immunoblot. Death was quite soon after either the initial infection or a later challenge, and the sera from these rabbits were not employed for expression library screening. The remaining six rabbits, which are listed in Table 1, developed strongly positive immunoblots but had variable clinical responses: one rabbit had no clinical illness, two became ill but completely recovered, and the three others developed severe illness and weight loss. These last three were sacrificed, and postmortem inspection of their organs showed evidence of macroscopic lesions which contained fungal hyphae (Table 1).
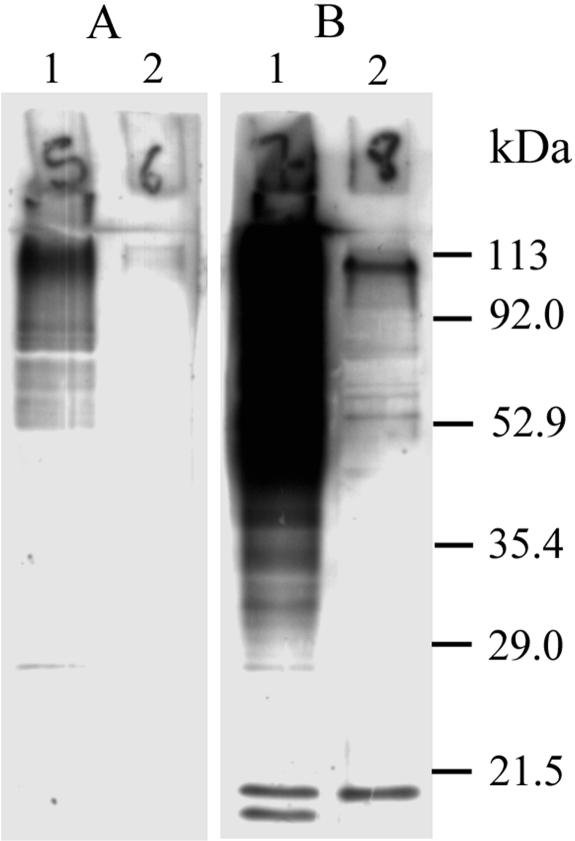
Antibodies reactive with A. fumigatus proteins were produced in 6 of the 10 rabbits following their infection with a range of doses of conidia (summarized in Table 1). Seroconversion, which was detected by immunoblotting, usually was found 14 days after an infection, and Fig. 1 shows findings that were typical for the sera from the six responders that were used in screening the gene expression library. In particular, it was notable that postinfection the sera became strongly reactive with the fraction that was enriched for A. fumigatus cell wall components (Fig. 1, lane 1). On the other hand, the differences between the preinfection and postinfection sera (Fig. 1, lane 2) were less striking for antigens present in the A. fumigatus fraction that was enriched for intracellular components. Indeed, for the postinfection sera that showed only a weak immunoblot, seroconversion was only obvious for immunoblots made with the fraction that was enriched for cell wall components.
FIG. 1.
Immunoblot from a 12.5% SDS-PAGE showing A. fumigatus proteins reacting with rabbit serum obtained either prior to infection (A) or 14 days after infection (B). Lane 1, cell wall extract; lane 2, intracellular extract. The infectious serum tested corresponds to the second infectious serum of rabbit no. 2 (Table 1, serum identifier 2b) which was gained 2 weeks after a second challenge with 50,000 conidia (compare with Fig. 2).
For three rabbits, the sera obtained 14 days after different challenges were tested separately (Table 1). The four rabbits that were not used to provide antisera either succumbed rapidly or failed to seroconvert. It was noticed that the severity of the infection was not strictly dependent on the inoculum dose.
Detection of antigens expressed by the A. fumigatus cDNA library.
Antisera used for screening the A. fumigatus cDNA expression library were always derived from blood drawn 14 days after intravenous challenge with conidia (Table 1 and Fig. 2). Approximately 2.3 million plaques were screened using nine different sera obtained from the infected rabbits, and 245 reactive plaques (∼1:10,000 total plaques) were detected. As expression of the Aspergillus-E. coli β-galactosidase fusion protein requires the codons of the cDNA insert and the gene for β-galactosidase to be in frame, we would expect that two-thirds of the inserts would be unproductive. Therefore, our results suggest that only one out of about 3,500 mRNAs used to make the cDNA represents a protein with antigenic reactivity.
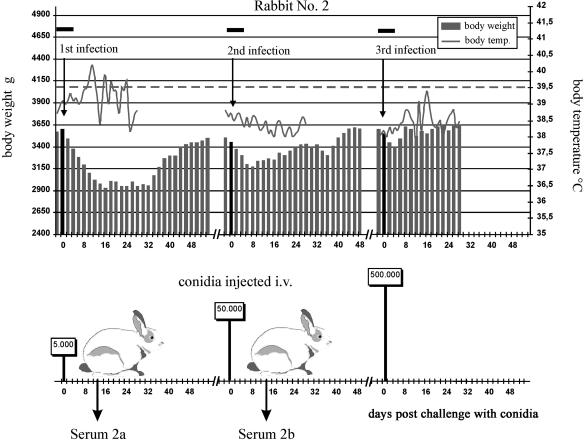
FIG. 2.
Protocol used for infection of rabbit 2, which became severely ill on primary challenge with 5 × 103 A. fumigatus conidia but which, after recovery, tolerated doses 10-fold and 100-fold higher when given 9 weeks after the first or 7 weeks after the second challenge. Data from monitoring body weight and body temperature have been plotted, with fever being reported when the body temperature rose above 39.5°C (dashed line), a temperature that is normal in healthy rabbits (3). The black horizontal bars at the top of the figure show times rabbits received cortisone and ceftazidime as described in Materials and Methods. The darker vertical bar for rabbit weight shows the weight at the time of challenge. Sera 2a and 2b are listed in Table 1.
The plaques that bound antibody from infected rabbits were considered to be infection specific when the plaques showed either no reaction or a reaction that was significantly weaker with serum taken from the same rabbit prior to infection. In spite of the preabsorption of anti-E. coli antibodies from the rabbit sera, plaques expressing no recombinant Aspergillus protein were always detectable; thus, the detection of plaques having significant reactivity required the use of negative-control plaques in which no recombinant A. fumigatus protein was expressed. The negative control was provided by selecting a λ phage plaque derived from the A. fumigatus cDNA library which expressed β-galactosidase after induction with IPTG. This phage did not have a cDNA insertion in lacZ, and its plaque developed a blue color when incubated with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (47).
Phage without cDNA inserts and each monoclonal phage that formed an infection-specific plaque were mixed 20:1 and used to develop plaques on agar plates. Each plate exhibited approximately 800 plaques that included an expected average of 40 antigen-expressing plaques per plate. Nitrocellulose membranes containing IPTG were laid on the plates in the manner described above for inducing β-galactosidase expression, but after removal from plates the membranes were divided into two halves. One half was then incubated with dilute rabbit serum obtained prior to infection, and the other half was incubated with antiserum of the same rabbit obtained after infection and previously used for the screening and isolation of the Aspergillus cDNA-containing phage. In both cases, these sera had been preabsorbed with E. coli and prepared as a 1:100 dilution as described in Materials and Methods. After development, both halves were compared. Only those plaques that clearly bound more antibodies from the sera of infected rabbits than from preinfection sera were used for further analysis.
Of the 245 plaques that were detected using E. coli-absorbed sera from infected rabbits, 193 were infection specific. Of the latter, 156 did not react with preinfection serum from the same rabbit, and the remaining 37 showed a weaker reaction. Examples are shown in Fig. 3.
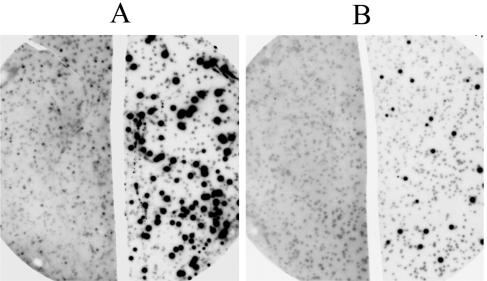
FIG. 3.
Detection of recombinant λ phage-generated plaques expressing an antigen in a background of control plaques produced by phages lacking an insert of cDNA derived from A. fumigatus mRNA. The left side of the figure shows reactivity of membrane blots with preinfection serum; the right side shows reactivity with antiserum obtained from the same rabbit 14 days after challenge with A. fumigatus conidia. Panel A shows an example of plaques expressing an antigen which was weakly reactive with serum obtained prior to infection but was strongly reactive with antiserum generated following infection. Panel B shows an example of an antigen having no detectable reactivity with the preinfection serum.
Identification of antigens.
The cDNAs from infection-specific plaques were subcloned and partially sequenced. The sequences represented 36 different antigens (Table 2). The redundancy in sequences likely reflects the various numbers of different mRNA molecules in the mRNA population used for library construction. Five antigens were detected by sera from two rabbits, and three antigens, including Asp f 16 (2), were detected with sera obtained from three different rabbits. Table 2 includes putative functions and accession numbers for antigens or their homologues that were derived from the annotated A. nidulans genome: in the case of a match to a known A. fumigatus protein, the accession number and original article are listed.
Immunoblotting on recombinant AHP1.
We examined whether the immune response to an antigen, which was detected for only some rabbits when screening the phage library, might be detected in other infected rabbits by using a standard serological detection method. The antigen AHP1, which had been detected by gene screening with antisera from only rabbits 2 and 5, was produced as a recombinant protein and used as the antigen in immunoblotting. While the immunoblot was negative using all preinfection sera, in contrast to the gene screening approach, immunoblots were positive to different extents using postinfection sera obtained from each of the six rabbits, though in the case of rabbit 1, the activity was quite weak (Fig. 4).
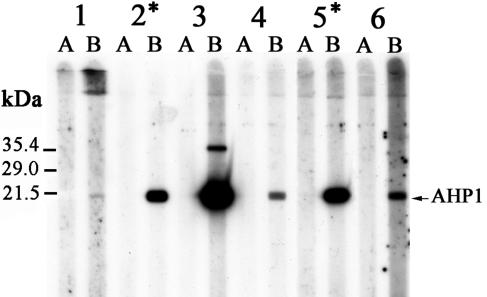
FIG. 4.
Recombinant AHP1 immunoblot using preinfection (A) and postinfection sera (B) of rabbits (1 to 6) obtained 2 weeks after the challenge (Table 1, sera 1, 2b, 3b, 4b, 5, and 6). The arrow points to the position calculated for AHP1 (molecular mass of ∼18.5 kDa). The additional reactive upper band (∼35 kDa) that was detected with antiserum from rabbit 3 may reflect dimerized AHP1. An asterisk indicates rabbits in which postinfection sera led to the isolation of the AHP1 antigen by phage library screening.
DISCUSSION
Though a gene screening approach has been used to characterize A. fumigatus antigens expressed in human allergic aspergillosis (13), the application of this procedure to the detection of antigens expressed during invasive aspergillosis appears novel. For these antigens, previous studies have largely relied on Western blotting where, typically, the antigens are described on the basis of their molecular mass (28). The dominant immunogens produced early in a primary infection of the rabbits appear most likely to be those associated with young, invasive mycelium. It seems unlikely that earlier stages, i.e., germ tubes and young hyphae, would cause a specific immune response directly since their biomass is minute and likely to be well below the amount needed for antibody induction. Furthermore, at the time of the conidial challenges, the rabbits are also receiving cortisone, which is immunosuppressive. In contrast to the situation with the earliest stages of fungal growth that follow infection by conidia, the invasive mycelium expands rapidly, and the antigens associated with this stage are likely to induce antibody production within 1 or 2 weeks. Several of these antibodies likely bind antigens present on both earlier growth forms and the invasive mycelium, and we believe that this serological response to the early invasive mycelium provides the basis for much of the spectrum of antibody specificities that we have exploited in screening the A. fumigatus gene expression library.
Although the most common form of invasive aspergillosis in humans follows the germination of inhaled conidia within the lung, here we have employed an intravenous route for administering conidia because, in previous experiments, infection by this route had induced resistance to A. fumigatus in mice (29). Taking advantage of the ongoing Aspergillus genome projects, we were able to identify 36 proteins that were bound by rabbit antibodies produced very early during the course of an Aspergillus infection. Three of these antigens showed weak reactivity with preinfection serum, while the other 33 antigens had no reactivity. Other antigens were detected that were equally reactive with sera obtained both before and during infection: these antigens were not further analyzed and may be cross-reactive with antigens of a number of environmental fungi that do not produce an invasive infection (44).
The screening method enabled the repeated detection of eight proteins (Table 2, no. 29 to 36). Five were detected with antisera obtained from two rabbits, and, for three antigens, specific antibodies were produced by three rabbits. One of these infection-related antigens, Asp f 16 (Table 2, no. 36), is reported to induce a protective cellular immune response against invasive aspergillosis in mice (6). The repeated detection of the same protein and the finding of a known immunoprotective antigen show that the gene screening method both has some consistency when used to detect antigens produced during infection in the rabbit model for invasive aspergillosis and could be useful for isolating antigens that are suitable for use in vaccines. In contrast to the eight antigens detected by more than one rabbit, the majority of antigens were detected by only a single antiserum during gene screening. The failure of these 28 antigens to be detected using antisera from the remaining rabbits is most easily explained by recognizing that the gene screening method is not equivalent to traditional antigen detection methods which employ antibodies and extracts of fungi. Gene screening can be insensitive to potent protein antigens which may not be expressed effectively as fusion proteins in λ phage or for which mRNA is not present in a fairly high concentration relative to the other mRNAs used for generating the cDNA library. When mRNA is rare, we would expect to detect an antigen only occasionally within the gene expression library, and so the antigen may fail to be represented in each set of plaques employed for screening. In this situation, a failure to detect the protein by gene screening will not necessarily mean that a rabbit lacks antibody; for instance, while the AHP1 antigen was detected by the gene screen approach with sera from only two of the six rabbits, all infected rabbits showed an anti-AHP1 response in an immunoblot with the purified recombinant antigen (Fig. 4). Furthermore, a strong AHP1 immunoblot reaction did not necessarily predict that AHP1 would be detected by the expression library screening method; for example, for rabbit 3 there was a strong AHP1 immunoblot but the same antiserum did not detect the AHP1 gene among the ∼250,000 phage plaques that were screened. Similar investigations with other antigens are under way.
As the antibodies that we studied represented the specificities associated with the development of an early immune response and primary protection (29), it seems likely that they will be directed predominantly against surface-associated or secreted antigens produced by intact invasive hyphae rather than detecting mainly intracellular proteins. These intracellular proteins are likely to become more prominent antigens during later stages of infection when the mycelial hyphae begin to die or are killed by neutrophils. But, in contrast to Asp f 16, which has a signal for secretion (2), the amino acid sequences of most antigens we have detected lacked the signal sequence needed for protein secretion via the classical secretory pathway. Were these antigens representing strictly intracellular proteins, largely hidden from the cellular and humoral mediators of host defenses, we would not expect them to be promising vaccine candidates, though they could play a significant role in enhancing a protective response by inducing both increased inflammation and a more effective clearance of dead or moribund fungal cells later in infection. However, in spite of their lack of a signal peptide, we believe that many of the antigens we describe can act as targets for a protective response because they are likely to have an additional surface location in Aspergillus. This assumption is supported by the consistently strong immunoblot reaction of the rabbits' postinfection sera against the Aspergillus antigens present in the cellular fraction enriched for cell wall-associated proteins. Furthermore, several of the antigens have homologues in other fungi that also lack a signal peptide but have been shown to have cell wall-associated forms. This finding may be explained by invoking the activity of nonclassical secretory pathways that do not require the presence of a signal peptide for protein secretion. These pathways have been best described for yeast and mammalian cells (10, 24, 36), and we consider it likely that similar pathways are present in A. fumigatus.
Asp f 16, enolase, transketolase and others antigens involved in carbohydrate metabolism.
The Asp f 16 antigen has been reported to induce a protective cellular immune response against invasive aspergillosis in mice (6). It shows strong homology to another A. fumigatus protein, Asp f 9, that was detected by Crameri et al. using sera obtained from patients with a history of Aspergillus-induced allergic disease (13). It has been suggested that both Asp f 16 and Asp f 9 are encoded by the same gene and that the differences in nucleotide sequences and molecular sizes are attributable to the splicing of mRNA that occurs during transcription (2). Probably, the very same gene also encodes a third highly homologous protein (GenBank accession no. AAN87849), which is described as Congo red hypersensitive (CRH)-like due to sequence homology. These proteins belong to a group of cell wall O-glycosylhydrolases which include several β-endoglucanases (glycosylhydrolase family 16 in the Pfam database at http://www.sanger.ac.uk/). Their nomenclature is derived from “CRH” and “CRH-like” which were the identifiers given to homologous proteins in Saccharomyces cerevisiae (45). In contrast to Asp f 16 and Asp f 9, the putative CRH-like protein of A. fumigatus has a hydrophobic tail and may be anchored by glycosylphosphatidylinositol, a feature also reported for CRH and CRH-like proteins in S. cerevisiae and Candida albicans (14, 45). Indeed, glycosylphosphatidylinositol anchorage of CRH-like proteins and possibly even of the Asp f 9 has also been shown in A. fumigatus by proteome analysis of membrane preparations (7), and our own BLAST search on the A. fumigatus genome has revealed at least three additional, related genes, all of which likely encode other surface-expressed glycosylhydrolases of the CRH-family.
The A. fumigatus enolase (Table 2, no. 1) has been described previously (27) and has a homologue in C. albicans that has been reported to induce a partially protective immune response against systemic candidiasis in mice (38, 54). Enolase can be detected intracellularly but, in this site, it is likely to be inaccessible to mediators of an anti-enolase response produced during infection. However, though the enolase sequence lacks a signal for secretion, enolase activity can be detected within or on the cell walls of both fungi and bacteria (1, 37, 39, 48), indicating, as considered in the first part of the Discussion, that a signal sequence may not be needed for a cell wall location. When exposed on or within the cell wall, enolase could act as a specific target for the protective immune response.
Among other proteins associated with carbohydrate metabolism, we have identified pyruvate decarboxylase, a putative aldolase, a putative pyruvate carboxylase, a putative transketolase, and a phosphoglucomutase (Table 2, no. 7, 10, 14, 29, and 35). Expression of mRNA encoding these enzymes is not totally unexpected as the fungal cell must produce very large amounts of sugar derivatives which are needed not only for its energy supply but also for the synthesis of the carbohydrate-rich cell wall (for a review, see reference 20). As with enolase, the C. albicans transketolase, pyruvate carboxylase, and pyruvate decarboxylase each lack a signal sequence for secretion but can be found both intracellularly and in cell wall locations (41, 53).
Antigens of the HSP family.
While antibodies to heat shock protein 30 (HSP30) were produced by only one rabbit, HSP88 and HSP90 were detected using antisera from three and two rabbits, respectively. Antibodies against HSP90 and HSP88 have been found previously in various types of infections caused by A. fumigatus in humans (8), and proteins showing homology to these HSPs have also been characterized in C. albicans where, in addition to an intracellular location, they are located in the cell wall (31, 35, 53). HSPs are immunodominant self-presenting protein antigens which can act as adjuvants (22, 52), and for this reason, they could be a useful component of a vaccine. HSPs may also act directly as protective antigens, an example being Histoplasma capsulatum HSP60 (19). A protective activity may also be afforded by the response to C. albicans HSP90. Here, the development of antibody against HSP90 has been associated with recovery from systemic candidiasis in humans, and both human and murine monoclonal antibodies generated against the HSP90 epitope, LKVIRK, showed some protective activity against invasive candidiasis in mice, even when administered 2 h after injection of a lethal dose of C. albicans (34).
Additional antigens and cell wall location.
Several of the proteins listed in Table 2 represent homologues of proteins known to be expressed intracellularly but may have an additional location in the fungal cell wall. Some of these proteins appear to be involved in nucleic acid replication, mRNA processing, and cell division. λ-phage-generated plaques expressing AHP1 and translation elongation factor 1 (Table 2, no. 32 and 34, respectively) were isolated with screening sera of two and three rabbits, respectively. AHP1 (also known as Asp f 3) is an allergen produced by A. fumigatus (23) and is thought to be a peroxisomal membrane protein. Like Asp f 9, it was detected previously using sera obtained from patients with allergic aspergillosis (13). Proteins showing homology to the A. fumigatus translation elongation factor 1 have been found in the cell wall of C. albicans (41, 53) and in various bacteria, for example, Mycobacterium leprae or Listeria monocytogenes (33, 48). Although a protective activity of elongation factor 1 has not been studied, the leishmanial elongation factor 4a (LeIF) protected mice from uncontrolled infection by Leishmania spp. (49). The response to LeIF uses both innate and adaptive mechanisms for it included increased gamma interferon production by natural killer cells and an increased production of interleukin-12 and interleukin-18 by dendritic cells (5). These cytokines favor the expansion of LeIF-specific Th1 cells (49), and a similar activity would be expected for several other cytokines produced by activated cells of the innate immune system (32).
The antigens we have described here may prove to have some value in serodiagnostic testing for patients who remain immunocompetent. For example, different profiles of antigen reactivity might characterize different types of aspergillosis. However, because the fungal components are detected by antibodies present 14 days postinfection, a time that is similar to that associated with the development of immunity by sublethally infected mice, we believe that their major value will be as candidates for testing in experimental vaccines against invasive aspergillosis.
Acknowledgments
We are indebted to Birgit Riemenschneider for technical assistance at various stages of this investigation and to Barbara Léchenne for technical expertise in preparing recombinant AHP1 antigen.
The project was supported by the German Research Foundation grant RE 953/3 and by the German José Carreras Leukemia Foundation. Sequencing of A. fumigatus was funded by the National Institutes of Allergy and Infectious Disease grant U01 AI 48830 to David Denning (University of Manchester, United Kingdom) and William Nierman (TIGR, Rockville, MD), the Wellcome Trust, and Fondo de Investicagiones Sanitarias.
Editor: T. R. Kozel
REFERENCES
- 1.Angiolella, L., M. Facchin, A. Stringaro, B. Maras, N. Simonetti, and A. Cassone. 1996. Identification of a glucan-associated enolase as a main cell wall protein of Candida albicans and an indirect target of lipopeptide antimycotics. J. Infect. Dis. 173:684-690. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee, B., V. P. Kurup, P. A. Greenberger, B. D. Johnson, and J. N. Fink. 2001. Cloning and expression of Aspergillus fumigatus allergen Asp f 16 mediating both humoral and cell-mediated immunity in allergic bronchopulmonary aspergillosis (ABPA). Clin. Exp. Allergy 31:761-770. [DOI] [PubMed] [Google Scholar]
- 3.Berghoff, P. C. 1989. Kleine Heimtiere und ihre Erkrankungen. In P. C. Berghoff (ed.), Tierärztliche Heimtierpraxis I. Parey Verlag, Berlin, Germany.
- 4.Bollag, D. M., and S. J. Edelstein. 1991. Protein methods. Wiley-Liss, Inc., New York, N.Y.
- 5.Borges, M. M., A. Campos-Neto, P. Sleath, K. H. Grabstein, P. J. Morrissey, Y. A. Skeiky, and S. G. Reed. 2001. Potent stimulation of the innate immune system by a Leishmania brasiliensis recombinant protein. Infect. Immun. 69:5270-5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bozza, S., R. Gaziano, G. B. Lipford, C. Montagnoli, A. Bacci, P. Di Francesco, V. P. Kurup, H. Wagner, and L. Romani. 2002. Vaccination of mice against invasive aspergillosis with recombinant Aspergillus proteins and CpG oligodeoxynucleotides as adjuvants. Microbes Infect. 4:1281-1290. [DOI] [PubMed] [Google Scholar]
- 7.Bruneau, J. M., T. Magnin, E. Tagat, R. Legrand, M. Bernard, M. Diaquin, C. Fudali, and J. P. Latge. 2001. Proteome analysis of Aspergillus fumigatus identifies glycosylphosphatidylinositol-anchored proteins associated to the cell wall biosynthesis. Electrophoresis 22:2812-2823. [DOI] [PubMed] [Google Scholar]
- 8.Burnie, J. P., and R. C. Matthews. 1991. Heat shock protein 88 and Aspergillus infection. J. Clin. Microbiol. 29:2099-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cenci, E., A. Mencacci, A. Bacci, F. Bistoni, V. P. Kurup, and L. Romani. 2000. T cell vaccination in mice with invasive pulmonary aspergillosis. J. Immunol. 165:381-388. [DOI] [PubMed] [Google Scholar]
- 10.Cleves, A. E., D. N. Cooper, S. H. Barondes, and R. B. Kelly. 1996. A new pathway for protein export in Saccharomyces cerevisiae. J. Cell Biol. 133:1017-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corbel, M. J., and S. M. Eades. 1977. Examination of the effect of age and acquired immunity on the susceptibility of mice to infection with Aspergillus fumigatus. Mycopathologia 60:79-85. [DOI] [PubMed] [Google Scholar]
- 12.Cove, D. J. 1966. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta 113:51-56. [DOI] [PubMed] [Google Scholar]
- 13.Crameri, R. 1998. Recombinant Aspergillus fumigatus allergens: from the nucleotide sequences to clinical applications. Int. Arch. Allergy Immunol. 115:99-114. [DOI] [PubMed] [Google Scholar]
- 14.De Groot, P. W. J., K. J. Hellingwerf, and F. M. Klis. 2003. Genome-wide identification of fungal GPI proteins. Yeast 20:781-796. [DOI] [PubMed] [Google Scholar]
- 15.Denning, D. W. 2000. Aspergillus species, p. 2674-2685. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases, 5 ed., vol. 2. Churchill Livingstone, Philadelphia, Pa.
- 16.Denning, D. W. 1998. Invasive aspergillosis. Clin. Infect. Dis. 26:781-805. [DOI] [PubMed] [Google Scholar]
- 17.de Repentigny, L., E. Kilanowski, L. Pedneault, and M. Boushira. 1991. Immunoblot analyses of the serologic response to Aspergillus fumigatus antigens in experimental invasive aspergillosis. J. Infect. Dis. 163:1305-1311. [DOI] [PubMed] [Google Scholar]
- 18.de Repentigny, L., S. Petitbois, M. Boushira, E. Michaliszyn, S. Senechal, N. Gendron, and S. Montplaisir. 1993. Acquired immunity in experimental murine aspergillosis is mediated by macrophages. Infect. Immun. 61:3791-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez, F. J., R. Allendoerfer, and G. S. Deepe, Jr. 1995. Vaccination with recombinant heat shock protein 60 from Histoplasma capsulatum protects mice against pulmonary histoplasmosis. Infect. Immun. 63:2587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gooday, G. W. 1995. Cell walls, p. 43-62. In N. A. R. Gow and G. M. Gadd (ed.), The growing fungus. Chapman and Hall, London, United Kingdom.
- 21.Hageage, G. H., Jr., and B. J. Harrington. 1984. Use of calcofluor white in clinical mycology. Lab. Med. 15:109-112. [Google Scholar]
- 22.Heike, M., B. Noll, and K. H. Meyer zum Buschenfelde. 1996. Heat shock protein-peptide complexes for use in vaccines. J. Leukoc. Biol. 60:153-158. [DOI] [PubMed] [Google Scholar]
- 23.Hemmann, S., K. Blaser, and R. Crameri. 1997. Allergens of Aspergillus fumigatus and Candida boidinii share IgE-binding epitopes. Am. J. Respir. Crit. Care Med. 156:1956-1962. [DOI] [PubMed] [Google Scholar]
- 24.Kuchler, K., and J. Thorner. 1992. Secretion of peptides and proteins lacking hydrophobic signal sequences: the role of adenosine triphosphate-driven membrane translocators. Endocr. Rev. 13:499-514. [DOI] [PubMed] [Google Scholar]
- 25.Kumar, A., L. V. Reddy, A. Sochanik, and V. P. Kurup. 1993. Isolation and characterization of a recombinant heat shock protein of Aspergillus fumigatus. J. Allergy Clin. Immunol. 91:1024-1030. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Lai, H. Y., M. F. Tam, R. B. Tang, H. Chou, C. Y. Chang, J. J. Tsai, and H. D. Shen. 2002. cDNA cloning and immunological characterization of a newly identified enolase allergen from Penicillium citrinum and Aspergillus fumigatus. Int. Arch. Allergy Immunol. 127:181-190. [DOI] [PubMed] [Google Scholar]
- 28.Latgé, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehmann, P. F., and L. O. White. 1976. Acquired immunity to Aspergillus fumigatus. Infect. Immun. 13:1296-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, S. J., J. Schranz, and S. M. Teutsch. 2001. Aspergillosis case-fatality rate: systematic review of the literature. Clin. Infect. Dis. 32:358-366. [DOI] [PubMed] [Google Scholar]
- 31.López-Ribot, J. L., H. M. Alloush, B. J. Masten, and W. L. Chaffin. 1996. Evidence for presence in the cell wall of Candida albicans of a protein related to the hsp70 family. Infect. Immun. 64:3333-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lutz, M. B., and G. Schuler. 2002. Immature, semi-mature and fully mature dendritic cells: which signals induce tolerance or immunity? Trends Immunol. 23:445-449. [DOI] [PubMed] [Google Scholar]
- 33.Marques, M. A., S. Chitale, P. J. Brennan, and M. C. V. Pessolani. 1998. Mapping and identification of the major cell wall-associated components of Mycobacterium leprae. Infect. Immun. 66:2625-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matthews, R., S. Hodgetts, and J. Burnie. 1995. Preliminary assessment of a human recombinant antibody fragment to hsp90 in murine invasive candidiasis. J. Infect. Dis. 171:1668-1671. [DOI] [PubMed] [Google Scholar]
- 35.Matthews, R., C. Wells, and J. P. Burnie. 1988. Characterisation and cellular localisation of the immunodominant 47-Kda antigen of Candida albicans. J. Med. Microbiol. 27:227-232. [DOI] [PubMed] [Google Scholar]
- 36.Michaelis, S. 1993. STE6, the yeast a-factor transporter. Semin. Cell Biol. 4:17-27. [DOI] [PubMed] [Google Scholar]
- 37.Mölkänen, T., J. Tyynelä, J. Helin, N. Kalkkinen, and P. Kuusela. 2002. Enhanced activation of bound plasminogen on Staphylococcus aureus by staphylokinase. FEBS Lett. 517:72-78. [DOI] [PubMed] [Google Scholar]
- 38.Montagnoli, C., S. Sandini, A. Bacci, L. Romani, and R. La Valle. 2004. Immunogenicity and protective effect of recombinant enolase of Candida albicans in a murine model of systemic candidiasis. Med. Mycol. 42:319-324. [DOI] [PubMed] [Google Scholar]
- 39.Ngondi-Ekome, J., F. Thiebault, J.-M. Strub, A. Van Dorsselaer, R. Bonaly, C. Contino-Pepin, M. Wathier, B. Pucci, and J. Coulon. 2003. Study on agglutinating factors from flocculent Saccharomyces cerevisiae strains. Biochimie 85:133-143. [DOI] [PubMed] [Google Scholar]
- 40.Pain, A., J. Woodward, M. A. Quail, M. J. Anderson, R. Clark, M. Collins, N. Fosker, A. Fraser, D. Harris, N. Larke, L. Murphy, S. Humphray, S. O'Neil, M. Pertea, C. Price, E. Rabbinowitsch, M.-A. Rajandream, S. Salzberg, D. Saunders, K. Seeger, S. Sharp, T. Warren, D. W. Denning, B. Barrell, and N. Hall. 2004. Insight into the genome of Aspergillus fumigatus: analysis of a 922-kb region encompassing the nitrate assimilation gene cluster. Fungal Genet. Biol. 41:443-453. [DOI] [PubMed] [Google Scholar]
- 41.Pitarch, A., M. Sánchez, C. Nombela, and C. Gil. 2002. Sequential fractionation and two-dimensional gel analysis unravels the complexity of the dimorphic fungus Candida albicans cell wall proteome. Mol. Cell Proteomics 1:967-982. [DOI] [PubMed] [Google Scholar]
- 42.Reichard, U., G. T. Cole, R. Rüchel, and M. Monod. 2000. Molecular cloning and targeted deletion of PEP2 which encodes a novel aspartic proteinase from Aspergillus fumigatus. Int. J. Med. Microbiol. 290:85-96. [DOI] [PubMed] [Google Scholar]
- 43.Richard, J. L., J. R. Thurston, R. C. Cutlip, and A. C. Pier. 1982. Vaccination studies of aspergillosis in turkeys: subcutaneous inoculation with several vaccine preparations followed by aerosol challenge exposure. Am. J. Vet. Res. 43:488-492. [PubMed] [Google Scholar]
- 44.Ripatti, T., P. Koskela, M. Kotimaa, E. Koskinen, and P. H. Mäenpää. 1990. Serum IgG antibody concentrations against environmental microbes in mares and foals during different seasons and effect of stabling practices. Am. J. Vet. Res. 51:550-555. [PubMed] [Google Scholar]
- 45.Rodríguez-Peña, J. M., V. J. Cid, J. Arroyo, and C. Nombela. 2000. A novel family of cell wall-related proteins regulated differently during the yeast life cycle. Mol. Cell. Biol. 20:3245-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rüchel, R., and U. Reichard. 1999. Pathogenesis and clinical presentation of aspergillosis, p. 21-43. In A. Brakhage, B. Jahn, and A. Schmidt (ed.), Aspergillus fumigatus. Biology, clinical aspects and molecular approaches to pathogenicity, vol. 2. Karger AG, Basel, Switzerland. [DOI] [PubMed]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2 ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Schaumburg, J., O. Diekmann, P. Hagendorff, S. Bergmann, M. Rohde, S. Hammerschmidt, L. J.änsch, J. Wehland, and U. Kärst. 2004. The cell wall subproteome of Listeria monocytogenes. Proteomics 4:2991-3006. [DOI] [PubMed] [Google Scholar]
- 49.Skeiky, Y. A. W., M. Kennedy, D. Kaufman, M. M. Borges, J. A. Guderian, J. K. Scholler, P. J. Ovendale, K. S. Picha, P. J. Morrissey, K. H. Grabstein, A. Campos-Neto, and S. G. Reed. 1998. LeIF: a recombinant Leishmania protein that induces an IL-12-mediated Th1 cytokine profile. J. Immunol. 161:6171-6179. [PubMed] [Google Scholar]
- 50.Smith, G. R. 1972. Experimental aspergillosis in mice: aspects of resistance. J. Hyg. 70:741-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Staib, F., S. K. Mishra, C. Rajendran, R. Voigt, J. Steffen, K. H. Neumann, C. A. Hartmann, and G. Heins. 1980. A notable Aspergillus from a mortal aspergilloma of the lung. New aspects of the epidemiology, serodiagnosis and taxonomy of Aspergillus fumigatus. Zentbl. Bakteriol. A 247:530-536. [PubMed] [Google Scholar]
- 52.Suzue, K., and R. A. Young. 1996. Heat shock proteins as immunological carriers and vaccines. EXS 77:451-465. [DOI] [PubMed] [Google Scholar]
- 53.Urban, C., K. Sohn, F. Lottspeich, H. Brunner, and S. Rupp. 2003. Identification of cell surface determinants in Candida albicans reveals Tsa1p, a protein differentially localized in the cell. FEBS Lett. 544:228-235. [DOI] [PubMed] [Google Scholar]
- 54.van Deventer, H. J., W. H. Goessens, A. J. van Vliet, and H. A. Verbrugh. 1996. Anti-enolase antibodies partially protective against systemic candidiasis in mice. Clin. Microbiol. Infect. 2:36-43. [DOI] [PubMed] [Google Scholar]