Abstract
Alternative splicing is widespread in the human genome, and it appears that many genes display different splice forms in cancerous tissue than in normal human tissues. However, since cDNAs for many cancer-associated genes were originally cloned from tumor samples, it is important to ask whether this repertoire of cDNAs provides a complete or representative picture of the transcript isoforms found in normal tissues. To answer this, we used bioinformatics and RT–PCR to identify novel splice forms, focusing on in-frame exonskips, for a panel of 50 cancer-associated genes in normal tissue samples. These data show that in nearly two-thirds of the genes, normal tissues expressed previously unknown splice forms, of which 40% were normally a dominant splice form. Surprisingly, the tumor-associated splice forms were twice as likely to be represented in GenBank than their normal tissue-associated splice forms, most likely because 70% of the mRNAs in GenBank for these genes were cloned from tumor samples. As an example, we describe a novel normal splice form of IKBβ, an important regulator of the NFκB pathway. Our data suggest that systematic re-evaluation of cancer genes' splice forms in normal tissue will yield insights into their distinct functions in normal tissues and in cancer. Our database contains 1308 novel normal splice forms, including many known cancer genes.
INTRODUCTION
Comprehensive and accurate mRNA sequence data are essential for the study of gene function. Researchers rely on sequence data from public databases and large-scale sequencing projects. For example, cancer researchers have sought to identify genes that are differentially expressed in tumors versus normal tissues (1,2), often using techniques such as PCR and microarrays where good probe design relies on existing mRNA sequence information.
Recent studies, however, have indicated that the mRNA sequences in current databases may reflect only a small proportion of all transcripts in the human genome. The unexpectedly low number of predicted genes in the completed human genome has led researchers to new questions about genome complexity. In particular, genomics studies have recently shown that most genes in the human genome are alternatively spliced to produce multiple mRNA products, most of which were previously unknown [reviewed in (3–5)]. A wide variety of experimental approaches including high-throughput expressed sequence tag (EST) sequencing (6), full-length cDNA sequencing (7,8), microarrays (9) and other methods (10) indicate that 40–60% of human genes are alternatively spliced. For example, an analysis of human EST and genome sequence data from January 2002 detected more than 30 000 alternative splices in the human genome, effectively doubling the number of gene products expected from the estimated 32 000 human genes (11).
These new sources of genomic complexity pose a significant challenge and opportunity to biological researchers. On the one hand, these data indicate that public databases may be missing a very large fraction of human gene products. The majority of alternative splice forms identified from EST sequencing were novel (12), i.e. they were not found in any mRNA sequence deposited in GenBank, or in any protein sequence record in SwissProt. Since these isoforms are not represented in public databases, and often cause subtle effects on the protein product (e.g. removal of only a small segment of the protein), they form a worrisome blindspot in biological research. Fortunately, new data from genomics and bioinformatics are rapidly closing this gap by providing detailed sequence information for previously unknown splice forms.
Cancer research is one area where these alternative splicing data may provide a useful new perspective. Recently, there has been growing evidence for the importance of tumor-specific splice variants in cancer, both from bioinformatics studies of expressed sequence databases and from experimental studies (13–18). For example, Bin1, one of the genes identified by our bioinformatics analysis as displaying cancer-specific splicing (14), is an adapter protein with features of a tumor suppressor that binds to and inhibits the oncogenic properties of c-Myc (19). Its expression is lost in many tumors. (20,21). Previous studies by Prendergast and co-workers (22) have shown that Bin1 function is abrogated in melanoma cells by aberrant splicing of a tissue-specific exon. Specifically, most melanoma cells inappropriately expressed exon 12A, which is combined with other alternative exons in Bin1 isoforms in brain, but is not found in isoforms from melanocytes or other non-neuronal cells. Cotransfection experiments in rat fibroblasts showed that exon 12A abolishes the ability of Bin1 to inhibit malignant transformation by c-Myc or adenovirus E1A. Splice variants may also be used as diagnostic markers for cancer (16). For example, a novel splice variant of actinin-4 is specific to SCLC (small cell lung carcinoma) cell lines, suggesting that it may be used as a new diagnostic marker for this disease (23).
Shifting from a paradigm of ‘one gene, one protein’ to recognition of widespread alternative splicing raises an important question for studies of disease genes, such as cancer-associated genes. Under the ‘one gene, one protein’ model, a cancer gene is assumed to produce the same protein product in tumors as in normal tissues, and its role in cancer is attributed to changes in the regulation of its expression, etc. However, if a typical gene can have multiple splice forms, we also need to ask whether there might be changes in splicing between the protein product found in normal tissues versus the form found in tumors. For disease genes, such as genes identified in association with cancer, this raises a specific concern. If such a gene were cloned originally from a tumor cDNA library, how can we be sure that this mRNA sequence is truly the splice form found in normal tissue, and not a tumor-specific splice variant? Since many genes studied by cancer researchers were indeed originally cloned from tumors, this concern warrants further examination.
In this study we have sought to evaluate this question directly, by examining the alternative splicing patterns of 50 genes in normal human tissue samples. We focused on a panel of 50 genes with cancer-associated alternative splicing, i.e. significant shifts in alternative splicing frequencies between tumors versus normal samples. We also focused our study on in-frame exon skips (i.e. alternative splicing events that add or remove an exact multiple of 3 nt, leaving the protein reading frame unchanged), to avoid including alternative splice forms that might cause nonsense-mediated decay (24). Using a combination of bioinformatics analysis and RT–PCR, we identified novel splice forms and evaluated their prevalence in normal tissue samples. These data show that nearly two-thirds of the genes expressed novel, previously unknown splice forms in normal human tissue samples, and indicate that in 40% of the cases the novel splice form was the dominant splice form in normal tissues.
MATERIALS AND METHODS
Genome-wide detection of cancer-specific alternative splicing
We based our analysis on our previously validated identification of alternative splicing from human ESTs aligned to genomic sequence (11,12). Cancer-specific splicing was detected as described previously in (14). Briefly, to identify changes in splicing that are characteristic of the transformed state, we pooled 4067 EST libraries from tumor samples and compared against a separate pool of 1737 EST libraries from normal tissue. By pooling many different tumors, we sought specific characteristics that are shared by many cancer types, and which are present far more frequently in tumors than in normal samples. We used histological information provided by ORESTES (Ludwig Institute for Cancer Research, http://www.ludwig.org.br/ORESTES), NCI-CGAP (Cancer Genome Anatomy Project, http://cgap.nci.nih.gov/) and NIH-MGC (Mammalian Gene Collection, http://mgc.nci.nih.gov/). We also performed text searches to classify other EST libraries. All tumor types were combined into a single pool, as were all normal tissue libraries. A total of 1160 EST libraries were excluded because they could not be clearly assigned to either cancer or normal (for example, if its histology was unclear or pre-cancerous). To assess the statistical significance based on EST coverage, we assigned an LOD score to each pair of splices, giving the log-odds ratio for a statistically significant change in the ratio of the two splice forms, as described in detail in (14). By this LOD score measure, we detected cancer-specific alternative splicing in 1284 genes at low confidence (P-value < 0.05) and 316 genes at high confidence (LOD score 2 or greater, i.e. P < 0.01), with 89 genes above LOD 3 (P < 0.001). A splice was considered novel, if there were no complete mRNAs deposited in GenBank matching this splice event. For this study, we focused on novel splices that were detected in normal tissues, referred to as ‘novel normals’. For experimental validation, we selected a random sample of ‘novel normal’ splices that are in-frame exon skips, i.e. which add or remove an exact multiple of 3 nt, causing no change to the protein reading frame. We used this criterion to avoid alternative splicing events that might cause frame shifts in protein coding regions and thus introduce premature stop codons, possibly resulting in nonsense-mediated decay of the transcript (24).
To measure the representation of our normal-specific versus cancer-specific splice forms in human mRNAs from GenBank, we downloaded the human UniGene dataset for February 2005 and selected all sequences of type mRNA. For each splice being tested, we concatenated the nucleotide sequences of the two exons joined by that splice (including up to 100 nt of sequence adjoining the splice from each exon) and searched the mRNA database using BLAST and an expectation value cutoff of 0.001. Any hit of at least 99% identity to the probe sequence was treated as a match to that specific splice form.
Cell lines
Cell lines used were SKNSH, SKNMC and U87 from ATCC (www.atcc.org); F-508 and W-98 (25,26) were kindly provided by Dr Linda Liau and Dr Stan Nelson (UCLA, Los Angeles, CA). SKNSH and SKNMC are neuroblastoma cell lines. U87, F-508 and W-98 are all glioblastoma cell lines. All cell lines were grown in RPMI 1640 medium supplemented with l-glutamine, 10% fetal calf serum, 100 U/ml penicillin and 100 U/ml streptomycin at 5% CO2.
RNA preparation
Total RNA was isolated from cell lines by using the Absolutely RNA microprep kit (Stratagene, La Jolla, CA). To remove genomic DNA contamination, DNase treatment was performed as recommended by the kit manufacturers. Total RNA from normal human bone marrow, brain, breast, skeletal muscle, lung, placenta and testis were purchased from BD-Biosciences Clontech (Palo Alto, CA).
cDNA synthesis and RT–PCR
cDNA was synthesized using either oligo(dT)12–18 or random hexamers and Stratascript reverse transcriptase using the StrataScript First-Strand Synthesis System (Stratagene). cDNAs from both reactions were pooled before performing PCR. This was performed to increase coverage of the entire gene.
Gene-specific primers were designed using MIT Primer3 software and synthesized by MWG Biotech (High Point, NC). All primers flanked at least one exon–intron junction (to rule out artefacts from genomic DNA contamination) and all had Tm between 55 and 60°C. Primer sequences are available online. GAPDH levels were monitored as a control. Touchdown PCR was performed on the MJR PTC-0200 thermal cycler (MJ Research, South San Francisco, CA) using Taq polymerase (Qiagen, Valencia, CA). Touchdown PCR conditions were as follows: 95°C for 2 min; 10× (95°C for 1 min; 65°C for 1 min with a decrease of 1°C per cycle; 72°C for 1 min), 30× (95°C for 1 min; 55°C for 1 min; 72°C for 1 min); 72°C for 10 min; hold at 4°C. Reaction products were run on a 2–2.5% agarose gel and visualized by staining with Ethidium Bromide (Sigma–Aldrich, St Louis, MO). As an internal control for successful PCR, we required that at least one band, corresponding to the known splice form was observed. PCR products were gel purified using a Qiaquick gel purification kit (Qiagen). Gel purified products were sequenced in both directions using gene-specific primers and Amersham MegaBACE 1000 sequencers (Amersham Pharmacia Biotech, Piscataway, NJ). The results confirmed the expected DNA sequences in all cases.
RESULTS
Screening of ‘novel normal’ splice forms of 50 genes in normal tissue samples
Using a set of genes with cancer-associated shifts in alternative splicing (14), we searched for novel splice forms via a bioinformatics analysis of human ESTs. A splice form was defined as novel if it was not observed in any complete mRNA deposited in GenBank (see Materials and Methods). In particular, we focused on novel splices that were detected in ESTs derived from normal tissues, which we will refer to as ‘novel normal’ splice forms, since they appeared to be novel splice forms expressed in normal tissues. We detected a total of 1308 ‘novel normal’ splices (with LOD scores of >1.0) in 804 genes, of which 472 were exon skip events.
To assess the validity of these novel normal splices, we selected a random sample of 50 single-exon skip events and used RT–PCR to detect both the proposed novel splice form and the known splice form, i.e. that reported in GenBank. We designed primer pairs directed to the exons flanking the alternatively spliced exon, so that a single primer pair would amplify both the known and novel splice forms, yielding different PCR product sizes. Thus, all our primer pairs spanned at least two introns. This probe design rules out artifacts due to genomic contamination or incomplete mRNA processing, since the transcript must be correctly spliced to yield the expected product sizes for the known and novel splice forms. We selected one or more normal tissues to test for each gene, based on the tissue origin of the ESTs in which the novel splice form was reported by our bioinformatics study.
A total of 32 of the 50 novel splice forms (64%) were detected by RT–PCR at the expected molecular weight (Table 1). In all but two of these cases (DCTN1 and LEF1, detailed in Table 1), the known splice form was also detected at its expected molecular weight. In about one-third of the genes (10/32), the novel splice form was detected as the dominant isoform in the tissues sampled. Finally, in 18 cases we did not detect the novel splice form, but since we only screened two tissue samples on average in these cases, it is possible that the novel splice form would be detected in other normal tissues.
Table 1.
Validation of novel splice forms
| Gene symbol | Gene title | Effect of novel splice | Novel form detected in |
|---|---|---|---|
| ACAA1 | Acetyl-CoA acyl transferase 1 | N/A | Not detected (BRN, LNG and TST) |
| BAT3§ | HLA-B associated transcript 3/human Scythe | Deletion of 48 amino acids | LNG and TST |
| BCAS1§ | Breast carcinoma amplified sequence 1 | Insertion of 45 amino acids | BRN |
| CACNA2D4 | Calcium channel, voltage-dependent, α 2/δ subunit 4 | Deletion of 15 amino acids from the N-terminus | BMR and SPL |
| CAPNS1 | Calpain, small subunit 1 | Deletion of 112 amino acids from the N-terminus | BRN and TST |
| CCT6A | Chaperonin containing TCP1, subunit 6A | N/A | Not detected (BRN, TST) |
| CCND3 | Cyclin D3 | N/A | Not detected (BRN, LNG, TST) |
| CHFR§ | Checkpoint with forkhead and ring finger domains | Deletion of 92 amino acids | TST |
| COCH | Coagulation factor C, cochlin | Deletion of 16 amino acids | TST |
| DARS | Aspartyl-tRNA synthetase | N/A | Not detected (BRN) |
| DCTN1 | Dynactin 1, p150 | Insertion of seven amino acids | BRN, LNG and SPL |
| DCTN3 | Dynactin 3 (p22) | N/A | Not detected (PLC and SPL) |
| EIF4G2/p97/DAP5§ | Eukaryotic translation initiation factor 4-gamma, 2 | Deletion of 38 amino acids | PLC and BRST |
| ELN | Elastin | N/A | Not detected (PLC, LNG and SPL) |
| EPB49 | Erythrocyte membrane protein band 4.9 | N/A | Not detected (BRN) |
| ESRRA | Estrogen-related receptor alpha | N/A | Not detected (BMR and SPL) |
| FXR1 | Fragile X, mental retardation, autosomal homolog 1 | N/A | Not detected (SPL) |
| GOLGA2 | Golgi autoantigen, golgin subfamily a, 2 | Insertion of 27 amino acids | KDN |
| GOSR2 | Golgi SNAP receptor complex member 2 | Deletion of 47 amino acids | BRN, TST and PLC |
| HLA-DMB§ | Major histocompatibility complex, class II, DM beta | Deletion of 39 amino acids | BMR and PLC |
| ITGAE | Integrin, alpha E (antigen CD103, human mucosal lymphoyte antigen 1; alpha polypeptide) | Deletion of 32 amino acids | BMR, SPL and TST |
| KHK | Ketohexokinase | Deletion of 84 amino acids | BRN |
| LCK | Lymphocyte-specific protein tyrosine kinase | Deletion of 51 amino acids | BMR and SPL |
| LEF1 | Lymphoid enhancer-binding factor 1, TCF1-alpha | Deletion of 28 amino acids | BRN and TST |
| LMAN2L/VIPL | Lectin, mannose-binding 2-like | Insertion of 11 amino acids | BRN |
| MAPT/TAU | Microtubule associated protein tau | Deletion of 29 amino acids with reference to var 1 (insertion with reference to var 2) | BRN |
| MEF2B | MADS box transcription enhancer factor 2 | N/A | Not detected (LNG, TST, PLC) |
| NFKBIB/IKBβ | I-kappa-B-beta | Deletion of 85 amino acids from the N-terminus | BRN |
| NOS2A§ | Nitric oxide synthase 2A | Deletion of 65 amino acids | TST |
| PCBP2§ | Heterogeneous nuclear ribonucleoprotein E2/poly(rC) binding protein 2 | Deletion of 31 amino acids | BMR, BRN, BRST, PLC |
| PDHA | Pyruvate dehydrogenase complex, E1-alpha polypeptide 1 precursor | Insertion of 38 amino acids | LNG, BRN |
| PPT1 | Palmitoyl-protein thioesterase 1 | N/A | Not detected (BRST, LNG) |
| PRP18 | Pre-mRNA processing factor 18 | Insertion of nine amino acids | SkM |
| RPN2 | Ribophorin II | N/A | Not detected (TST) |
| SCAMP2 | Secretory carrier membrane protein 2 | N/A | Not detected (BRN) |
| SCML1§ | Sex comb on midleg-like 1 | Insertion of 28 amino acids | TST |
| SH3BGR | SH3-binding domain and glutamic acid-rich protein | N/A | Not detected (LNG and TST) |
| SLC3A2 | Solute carrier family 3, member 2 | N/A | Not detected (BRN, SPL and PLC) |
| SUPT5H | Suppressor of TY 5, Saccharomyces cerevesiae, homolog of | Deletion of four amino acids | BRN= |
| SRRM1/SRM160§ | Serine/arginine repetitive matrix protein 1 | Insertion of 14 amino acids | LNG, SPL and PLC |
| TAF2 | TATA box-binding protein-associated factor, 2B | N/A | Not detected (BMR, BRN and SPL) |
| TEX27/Hs.6120 | Testis expressed gene, 27 | Deletion of 33 amino acids | BRN |
| TIE1 | Tyrosine kinase with immunoglobulin and EGF factor homology domains | Deletion of 43 amino acids | LNG and BRN |
| TIM17b | Translocase of inner mitochondrial membrane 17 | N/A | Not detected (BRN and LNG) |
| TPD52L2§ | Tumor protein D52-like 2 (D54) | Insertion of 23 amino acids | BRN |
| UBE1C | Ubiquitin-activating enzyme E1C | Deletion of 27 amino acids | Not detected (PLC and SPL) |
| WARS | Trytophanyl-tRNA synthetase | Truncation of the C-terminus | TST and LNG |
| WBP2 | WW domain-binding protein 2 | Deletion of 45 amino acids | BRN and TST |
| Z391G | Immunoglobulin superfamily protein Z391G | Deletion of 94 amino acids | LNG and PLC |
| TBC1D7 | TBC1 domain family, member 7 | Deletion of 27 amino acids | BM, BRST and LNG |
We validated novel splice forms using RT–PCR, in one or more of the following tissues: BMR-bone marrow; BRN-brain; LNG-lung; KDN-kidney; SPL-spleen; SKM-skeletal muscle; TST-testis; PLC-placenta; BRST-breast. To indicate the relative abundance of the novel splice form in each tissue, we write the tissue's name in boldface if the novel form was more abundant than the known form; in plain font if the known form was more abundant than the novel form; in italics if only the novel form (and not the known form) was observed; =denotes that both the known and the novel forms were equally abundant. §indicates that these genes were selected for further expression analysis (see Supplementary Data).
To ascertain the exact identity of the bands matching the expected molecular weight of the novel splice form, we selected a random sample of 15 genes. For each gene, we gel purified and sequenced the PCR product for the novel normal splice form. In 15 of 15 cases tested, the independent sequencing of these PCR products exactly matched the novel splice form reported by our earlier bioinformatics study (12). Thus, these novel splice forms are easily detected even in a small sample of human tissues and correspond precisely to the alternative splice forms reported by bioinformatics analysis of ESTs.
Evaluation of literature and GenBank mRNA data
Given the ease with which these splice forms can be detected in human tissue, it is reasonable to ask why they were not reported by the original studies that cloned these genes, or by subsequent entries in GenBank. To answer this question, we first examined the original literature to identify the tissue sources from which these genes were originally cloned. For a random sample of 20 genes, represented in GenBank by 44 mRNAs, 31 mRNAs (70%) were cloned from tumor samples versus 13 from normal tissue samples (see Supplementary Data for a complete list). Thus, it is perhaps not surprising that the GenBank records for these genes often lack the normal splice form.
It is well known that public mRNA databases are not complete in their coverage of alternative splice forms. But are they nearing the goal of truly complete coverage? During the last 3 years, over 90 000 human mRNA sequences have been added to GenBank. We checked the latest GenBank data (February 2005) to assess what fraction of novel normal splice forms are now included in GenBank mRNA sequences. Of the novel normal splice forms identified from the January 2002 dataset, only 11% were successfully found in the February 2005 GenBank mRNA data. The remainder were not represented by any February 2005 GenBank human mRNA, indicating that current sequence databases are still far from complete in their representation of human alternative splice forms. Moreover, for this set of genes the tumor-associated splice form was much more likely than the normal tissue-associated splice form (by about 2-fold) to be represented by an mRNA sequence in the February 2005 data.
Screening of a panel of normal tissues and tumor-derived cell lines
To assess the expression patterns of novel normal splice forms, we selected a random sample of 10 genes and screened each in a panel of 8 normal tissues (bone marrow, brain, breast, lung, skeletal muscle, placenta, spleen and testes), and 5 cell lines derived from glioblastomas and neuroblastomas. In 4 of the 10 genes the novel splice form was the dominant isoform in most normal tissues, or had approximately equal representation in normal tissues to that of the known splice form (see Supplementary Data). In two other genes, the novel splice form was strongly restricted to specific tissues, and in three other genes, the novel splice form was ubiquitously expressed, but at a much lower level than the known splice form (Supplementary Data).
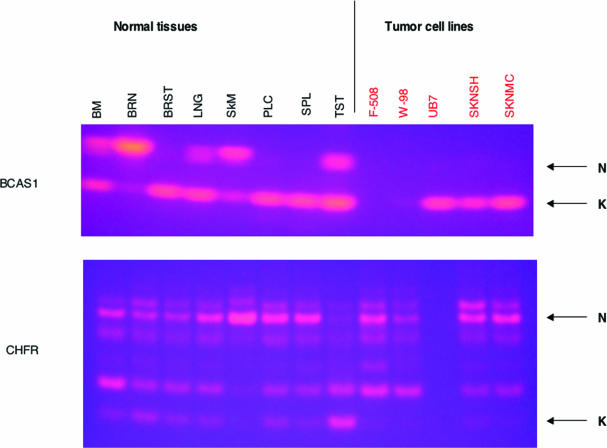
In two genes (BCAS1 and CHFR), the novel splice form appeared to be lost in glioblastoma and neuroblastoma cell lines (Figure 1). BCAS1 (Breast carcinoma amplified sequence 1) was originally cloned from breast carcinomas and is overexpressed in breast cancers (27). Though little is known about its function, BCAS1 is a candidate oncogene. BCAS1 was expressed only as the novel splice form in brain, but only the known splice form was detected in the brain tumor-derived cell lines. In CHFR, the picture was somewhat more complex. The novel splice form was expressed at a low level in most normal tissues, but was apparently absent from all the tumor cell lines tested. CHFR encodes a protein with forkhead-associated and RING finger domains and defines a checkpoint that delays entry into metaphase (28). Methylation-dependent silencing of CHFR is seen in many tumors (29). EST-genomic alignments indicated that the known form is observed in both tumor and normal tissues (four ESTs from tumors and three ESTs from normal tissues), whereas a novel splice form is observed in two ESTs both from normal testis. Our RT–PCR results showed the novel splice form to be the dominant form in testis (in agreement with the EST results), and to be present in most normal tissues (including brain), but usually at a lower level than the known form. In contrast, the novel splice form was missing from the five tumor cell lines. Sequencing of the band corresponding to the novel splice form matched the novel splice form sequence predicted by the ESTs and results in an exon skip removing 92 amino acids (Table 1). However, CHFR displays additional alternative splice forms and requires further study.
Figure 1.
Screening of a panel of normal tissues and tumor-derived cell lines using RT–PCR. RT–PCR was performed on cDNAs from normal, non-cancerous tissues (lettered in black) and from tumor-derived cell lines (lettered in red). 2% agarose gels were run, and bands visualized by ethidium bromide staining. N denotes the band corresponding to the novel splice form, and K denotes the band corresponding to the known splice form. The genes shown are BCAS1 (upper gel) and CHFR (lower gel).
Bioinformatics analysis of a novel normal splice form of IKBβ
As an example of our results that illustrates their potential interest to cancer researchers, we have identified a novel splice variant of IKBβ (NFKBIB), a key regulator of NFκB activation (Supplementary Data). The complete mRNA sequence for IKBβ that is deposited in GenBank is from a pancreas epithelioid carcinoma library [MGC 70; (30)]. A total of 17 ESTs (primarily from tumors or tumor cell lines) support this mRNA. However, three ESTs from a normal brain library (MGC 119) support a novel alternative splice variant. We validated this novel splice variant from normal brain cDNA by both RT–PCR and sequencing (Supplementary Data). This alternative splice variant (Supplementary Figure 2A) replaces the first exon with a shorter exon, resulting in a truncation of the N-terminus, removing 86 amino acids (Supplementary Data). The region removed by the novel splice encodes important functional motifs, including two phosphorylation sites and the first ankyrin domain. In an inactive state, NFκB exists as homo- or heterodimers associated with IKB proteins (both IKBα and IKBβ). An inducing signal results in the phosphorylation, polyubiquitination and subsequent degradation of IKB, leading to dissociation of NFκB, localization to the nucleus and activation [reviewed in (31,32)]. The shorter splice form of IKBβ lacks the phosphorylation sites at S19 and S23, and this seems very likely to affect NFκB signaling. Experimental studies of its impact on NFκB function and regulation may require development of new IKBβ antibodies, since most existing anti-IKBβ antibodies were raised using N-terminal peptide fragments and may not be able to detect the novel isoform.
DISCUSSION
Our experimental data clearly show that the novel normal splice forms predicted by our previous bioinformatics work are a real feature of normal human tissues. Even examining only a small sample of human tissues (typically only 1–3 different tissues), we were able to find ∼60% of the novel normal splice forms predicted by bioinformatics, and sequencing of these forms exactly matched the form predicted by bioinformatics in every case tested. It should be emphasized that our study focused on in-frame exon skips (i.e. alternative splicing events that add or remove an exact multiple of 3 nt, leaving the protein reading frame unchanged), to avoid including alternative splice forms that might cause nonsense-mediated decay (24). Thus, the absence of database records for the novel normal splice forms cannot be attributed to nonsense-mediated decay of ‘aberrant’ splice forms that do not give rise to a protein product. Our analysis of GenBank (both from 2002 data and 2005 data) shows both that public databases lack many alternative splice forms, and intriguingly, that this effect is much more pronounced for splice forms that are associated with normal tissues, than for the splice forms associated with tumors. At least for the set of ∼300 genes in this study, the tumor-associated splice forms appear to be ‘over-represented’ relative to the normal-tissue-associated splice forms. This highlights both a problem and an opportunity for cancer researchers.
Our results suggest several reasons why normal splice forms for many cancer genes may have been missed. Many such genes were originally cloned from tumor samples, introducing a potential bias for tumor-specific splice forms instead of the normal splice form. This bias could be particularly strong for genes such as CHFR and BCAS1, where the novel splice form appears to be lost in tumor cell lines. Indeed, when we scrutinized the tissue origins of the mRNAs for the genes in this study, we found that 70% were cloned from tumor samples (Supplementary Data). Thus, it is not surprising that the tumor splice form is known, and that many normal splice forms have been missed. In about half of the genes we have studied, the novel splice form was restricted to certain tissues or expressed at a much lower level than the known splice form. For such genes, detecting the novel splice form probably would have required sequencing multiple cDNAs from different normal tissue samples. Unfortunately, it has been common practice, even in many sophisticated sequencing projects (e.g. the Mammalian Gene Collection), to halt further sequencing of a gene once a single full-length mRNA is deposited. It should also be noted that these alternative splice forms often produce protein variants whose molecular weight is very similar to the known form, differing by only a few kilodaltons, and thus may be hard to distinguish by electrophoresis and related methods.
Our database of novel normal splice forms can be of value for cancer researchers for several reasons. First, the sequence of such a splice form may immediately suggest important implications for the function or regulation of a gene involved in cancer. For example, in the case of IKBβ, the novel normal splice form that we identified removes phosphorylation sites that are important for regulation of the protein's stability, and thus its regulation of NFκB signaling (see Results and Supplementary Data). Such implications deserve further experimental study, and could lead to new insights into the gene's function (and thus regulation of the NFκB pathway) in normal tissue. Predicted functional effects for some of our novel splice variants are shown in Table 2. Second, our data show that public databases are far from complete in their cataloging of alternative splice forms, even for well-studied genes. About 60% of the splice forms detected by both our bioinformatics and experimental analyses are not represented in GenBank. In a recent experimental study of 162 well-studied genes that are the targets of existing drug therapies, Jin et al. (10) identified novel splice forms (not reported in GenBank or RefSeq) in 70% of the genes by performing RT–PCR on a pooled panel of human tissue samples. Thus, even for genes that have been intensively studied because of their role in human disease processes, the existence of novel splice forms is the rule, not the exception. Third, our data indicate that in a large fraction of cases (30–40%), the novel normal splice forms in our database actually constitute the dominant splice form in normal tissues (see Results and Supplementary Data). Thus, the absence of these splice forms from public databases means that researchers are not just missing a splice form, but are actually missing the main splice form of the gene. Fourth, the GenBank sequence for a given gene may actually reflect a tumor splice form. While this is a valid concern for any gene, it is of particular concern for genes involved in cancer, since many such genes were originally cloned and sequenced from tumor libraries.
Table 2.
Predicted functional effects of selected novel splice forms
| Gene symbol | Gene title | Effect of novel splice | Domain affected |
|---|---|---|---|
| CAPNS1 | Calpain small subunit 1/CAPN4 | Deletion of 112 amino acids from N-terminus | Removal of the N-terminal glycine-rich domain (domain V) implicated in interaction with lipids |
| GOSR2 | Golgi SNAP receptor complex member 2 | Deletion of 47 amino acids | Removes part of the potential cytoplasmic domain |
| HLA-DMB | MHC II DM beta | Removes 39 amino acids | Removes the transmembrane domain |
| ITGAE | Integrin, alpha E (antigen CD103) | Deletion of 32 amino acids | Removes part of the integrin alpha E heavy chain domain |
| GOLGA2 | Golgi matrix protein GM130 | Insertion of 27 amino acids | Disrupts the p115 binding site |
| LEF1 | Lymphoid enhancer-binding factor 1, TCF1-alpha | Deletion of 28 amino acids | Removes part of the proline rich domain |
| NFKBIB/IKBβ | I-kappa-B-beta | N-terminus truncated | Removal of two phosphorylation sites and an ankyrin domain |
| NOS2A | Nitric oxide synthase 2A | Deletion of 65 amino acids | Truncation of C-terminus, removing 2 NAD-binding domains |
| PCBP2 | Heterogeneous nuclear ribonucleoprotein E2/poly(rC) binding protein 2 | Deletion of 45 amino acids | Removes the region between KH2 and KH3 domains |
| PDHA | Pyruvate dehydrogenase complex, E1-alpha polypeptide 1 precursor | Insertion of 38 amino acids | Disrupts the region responsible for mitochondrial import of precursor protein |
| TIE1 | Tyrosine kinase with immunoglobulin and EGF factor homology domains | Deletion of 43 amino acids | Removes EGF-like domain 3 |
| WBP2 | WW domain-binding protein 2 | Deletion of 45 amino acids | Removes part of a proline rich domain |
Discovery of the normal tissue forms of these genes, as provided by our data, and comparison with their tumor forms, could shed a useful new light on the action of many cancer genes. Our novel-normal database currently contains 1308 novel normal splice forms for 804 human genes, including exon skip events, alternative 5′ and alternative 3′ splicing, and alternative initiation and alternative termination. This database, available at http://www.bioinformatics.ucla.edu/ASAP (33), can provide cancer researchers with a shortcut for identifying whether there are novel normal splice forms for their gene of interest, and whether these forms reveal likely functional changes in their sequences. Our database also tabulates evidence for tumor-specific shifts in alternative splicing, when a splice form is observed much more frequently in tumors than in normal tissue samples (14).
We wish to emphasize that care is required in interpreting comparisons between splice forms observed in normal versus tumor samples. First, such differences need not be absolutely black and white. While in some cases we have observed a strong switch from one splice form (S) in normal tissues to a different splice form (S′) in tumors, other mechanisms are also common, such as gain of S′ (in which the S′ form, rare in normal tissue, becomes common in tumors, while the S form remains constant), and loss of S (in which the S form, common in normal tissue, becomes significantly depressed in tumors, while the S′ form remains constant) (14). Thus, there is no contradiction in noting that the normal tissue splice form (S) may also be observed in tumors; this is what is expected under the gain of S′ mechanism. Second, tumors are highly heterogeneous, so one cannot conclude that a splice form is ‘cancer-specific’ from examination of just a few samples. To address this more challenging question, would require a large panel of tumor samples, each with a tissue-matched normal control sample. Moreover, since the results would likely vary significantly from tumor to tumor (due to tumor heterogeneity), careful statistical analysis would be required to demonstrate a significant association with a specific splice form. In this paper, we have deliberately not addressed this challenging question, since in our view it is first necessary to prove the existence of our putative ‘novel normal’ forms in normal tissues, before attempting to show that these genes shift to a different splice form in tumors. However, our current results, combined with our ‘novel normal’ isoforms database, should make it possible for many researchers to begin the studies necessary for assessing whether these genes indeed show cancer-specific changes in splicing in different tumor types.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
Supplementary Material
Acknowledgments
The authors thank Drs O. Bogler, K. Miura, S. Nelson, I. Newsham and Y. Xing for helpful discussion and comments on this work. C.L. was supported by the Dreyfus Foundation Teacher-Scholar Award, and NIH Grant U54-RR21813, and M.R. by DOE grant DEFG0387ER60615. Funding to pay the Open Access publication charges for this article was provided by National Institutes of Health, Bethesda, Maryland, USA.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lal A., Lash A.E., Altschul S.F., Velculescu V., Zhang L., McLendon R.E., Marra M.A., Prange C., Morin P.J., Polyak K., et al. A public database for gene expression in human cancers. Cancer Res. 1999;59:5403–5407. [PubMed] [Google Scholar]
- 2.Strausberg R.L. The Cancer Genome Anatomy Project: new resources for reading the molecular signatures of cancer. J. Pathol. 2001;195:31–40. doi: 10.1002/1096-9896(200109)195:1<31::AID-PATH920>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 3.Modrek B., Lee C. A genomic view of alternative splicing. Nature Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 4.Boue S., Letunic I., Bork P. Alternative splicing and evolution. Bioessays. 2003;25:1031–1034. doi: 10.1002/bies.10371. [DOI] [PubMed] [Google Scholar]
- 5.Lareau L.F., Green R.E., Bhatnagar R.S., Brenner S.E. The evolving roles of alternative splicing. Curr. Opin. Struct. Biol. 2004;14:273–282. doi: 10.1016/j.sbi.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Strausberg R.L., Buetow K.H., Emmert-Buck M.R., Klausner R.D. The cancer genome anatomy project: building an annotated gene index. Trends Genet. 2000;16:103–106. doi: 10.1016/s0168-9525(99)01937-x. [DOI] [PubMed] [Google Scholar]
- 7.Strausberg R.L., Feingold E.A., Klausner R.D., Collins F.S. The mammalian gene collection. Science. 1999;286:455–457. doi: 10.1126/science.286.5439.455. [DOI] [PubMed] [Google Scholar]
- 8.Wiemann S., Weil B., Wellenreuther R., Gassenhuber J., Glassl S., Ansorge W., Bocher M., Blocker H., Bauersachs S., Blum H., et al. Toward a catalog of human genes and proteins: sequencing and analysis of 500 novel complete protein coding human cDNAs. Genome Res. 2001;11:422–435. doi: 10.1101/gr.154701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson J.M., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 10.Jin P., Fu G.K., Wilson A.D., Yang J., Chien D., Hawkins P.R., Au-Young J., Stuve L.L. PCR isolation and cloning of novel splice variant mRNAs from known drug target genes. Genomics. 2004;83:566–571. doi: 10.1016/j.ygeno.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 11.Xu Q., Modrek B., Lee C. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res. 2002;30:3754–3766. doi: 10.1093/nar/gkf492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Modrek B., Resch A., Grasso C., Lee C. Genome-wide analysis of alternative splicing using human expressed sequence data. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z., Lo H.S., Yang H., Gere S., Hu Y., Buetow K.H., Lee M.P. Computational analysis and experimental validation of tumor-associated alternative RNA splicing in human cancer. Cancer Res. 2003;63:655–657. [PubMed] [Google Scholar]
- 14.Xu Q., Lee C. Discovery of novel splice forms and functional analysis of cancer-specific alternative splicing in human expressed sequences. Nucleic Acids Res. 2003;31:5635–5643. doi: 10.1093/nar/gkg786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hui L., Zhang X., Wu X., Lin Z., Wang Q., Li Y., Hu G. Identification of alternatively spliced mRNA variants related to cancers by genome-wide ESTs alignment. Oncogene. 2004;23:3013–3023. doi: 10.1038/sj.onc.1207362. [DOI] [PubMed] [Google Scholar]
- 16.Brinkman B.M. Splice variants as cancer biomarkers. Clin. Biochem. 2004;37:584–594. doi: 10.1016/j.clinbiochem.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 17.Venables J.P. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- 18.Reis E.M., Ojopi E.P., Alberto F.L., Rahal P., Tsukumo F., Mancini U.M., Guimaraes G.S., Thompson G.M., Camacho C., Miracca E., et al. Large-scale transcriptome analyses reveal new genetic marker candidates of head, neck, and thyroid cancer. Cancer Res. 2005;65:1693–1699. doi: 10.1158/0008-5472.CAN-04-3506. [DOI] [PubMed] [Google Scholar]
- 19.Sakamuro D., Elliott K.J., Wechsler-Reya R., Prendergast G.C. BIN1 is a novel MYC-interacting protein with features of a tumour suppressor. Nature Genet. 1996;14:69–77. doi: 10.1038/ng0996-69. [DOI] [PubMed] [Google Scholar]
- 20.Ge K., Duhadaway J., Sakamuro D., Wechsler-Reya R., Reynolds C., Prendergast G.C. Losses of the tumor suppressor BIN1 in breast carcinoma are frequent and reflect deficits in programmed cell death capacity. Int. J. Cancer. 2000;85:376–383. [PubMed] [Google Scholar]
- 21.Ge K., Minhas F., Duhadaway J., Mao N.C., Wilson D., Buccafusca R., Sakamuro D., Nelson P., Malkowicz S.B., Tomaszewski J., et al. Loss of heterozygosity and tumor suppressor activity of Bin1 in prostate carcinoma. Int. J. Cancer. 2000;86:155–161. doi: 10.1002/(sici)1097-0215(20000415)86:2<155::aid-ijc2>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 22.Ge K., DuHadaway J., Du W., Herlyn M., Rodeck U., Prendergast G.C. Mechanism for elimination of a tumor suppressor: aberrant splicing of a brain-specific exon causes loss of function of Bin1 in melanoma. Proc. Natl Acad. Sci. USA. 1999;96:9689–9694. doi: 10.1073/pnas.96.17.9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Honda K., Yamada T., Seike M., Hayashida Y., Idogawa M., Kondo T., Ino Y., Hirohashi S. Alternative splice variant of actinin-4 in small cell lung cancer. Oncogene. 2004;23:5257–5262. doi: 10.1038/sj.onc.1207652. [DOI] [PubMed] [Google Scholar]
- 24.Lewis B.P., Green R.E., Brenner S.E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl Acad. Sci. USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liau L.M., Lallone R.L., Seitz R.S., Buznikov A., Gregg J.P., Kornblum H.I., Nelson S.F., Bronstein J.M. Identification of a human glioma-associated growth factor gene, granulin, using differential immuno-absorption. Cancer Res. 2000;60:1353–1360. [PubMed] [Google Scholar]
- 26.Yang I., Kremen T.J., Giovannone A.J., Paik E., Odesa S.K., Prins R.M., Liau L.M. Modulation of major histocompatibility complex Class I molecules and major histocompatibility complex-bound immunogenic peptides induced by interferon-alpha and interferon-gamma treatment of human glioblastoma multiforme. J. Neurosurg. 2004;100:310–319. doi: 10.3171/jns.2004.100.2.0310. [DOI] [PubMed] [Google Scholar]
- 27.Collins C., Rommens J.M., Kowbel D., Godfrey T., Tanner M., Hwang S.I., Polikoff D., Nonet G., Cochran J., Myambo K., et al. Positional cloning of ZNF217 and NABC1: genes amplified at 20q13.2 and overexpressed in breast carcinoma. Proc. Natl Acad. Sci. USA. 1998;95:8703–8708. doi: 10.1073/pnas.95.15.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scolnick D.M., Halazonetis T.D. Chfr defines a mitotic stress checkpoint that delays entry into metaphase. Nature. 2000;406:430–435. doi: 10.1038/35019108. [DOI] [PubMed] [Google Scholar]
- 29.Toyota M., Sasaki Y., Satoh A., Ogi K., Kikuchi T., Suzuki H., Mita H., Tanaka N., Itoh F., Issa J.P., et al. Epigenetic inactivation of CHFR in human tumors. Proc. Natl Acad. Sci. USA. 2003;100:7818–7823. doi: 10.1073/pnas.1337066100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strausberg R.L., Feingold E.A., Grouse L.H., Derge J.G., Klausner R.D., Collins F.S., Wagner L., Shenmen C.M., Schuler G.D., Altschul S.F., et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl Acad. Sci. USA. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh S., Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109(Suppl):S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 32.Ting A.Y., Endy D. Signal transduction. Decoding NF-kappaB signaling. Science. 2002;298:1189–1190. doi: 10.1126/science.1079331. [DOI] [PubMed] [Google Scholar]
- 33.Lee C., Atanelov L., Modrek B., Xing Y. ASAP: The Alternative Splicing Annotation Project. Nucleic Acids Res. 2003;31:101–105. doi: 10.1093/nar/gkg029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.