Abstract
Natural antisense transcripts are reported from all kingdoms of life and several recent reports of genomewide screens indicate that they are widely distributed. These transcripts seem to be involved in various biological functions and may govern the expression of their respective sense partner. Very little, however, is known about the degree of evolutionary conservation of antisense transcripts. Furthermore, none of the earlier analyses has studied whether antisense relationships are solely dual or involved in more complex relationships. Here we present a systematic screen for cis- and trans-located antisense transcripts based on open reading frames (ORFs) from five fungal species. The relative number of ORFs involved in antisense relationships varies greatly between the five species. In addition, other significant differences are found between the species, such as the mean length of the antisense region. The majority of trans-located antisense transcripts is found to be involved in complex relationships, resulting in highly connected networks. The analysis of the degree of evolutionary conservation of antisense transcripts shows that most antisense transcripts have no ortholog in any other species. An annotation of antisense transcripts based on Gene Ontology directs to common terms and shows that proteins of genes involved in antisense relationships preferentially localize to the nucleus with common functions in the regulation or maintenance of nucleic acids.
INTRODUCTION
Antisense transcripts are RNA sequences that are complementary to known (sense) transcripts. Experimental as well as in silico investigations have revealed substantial evidence that antisense transcripts are much more widespread than thought previously. However, it is still unclear to what extent antisense transcripts are generally functional. There are numerous examples of the detailed analysis of antisense transcripts functioning at the transcriptional and posttranscriptional level [for review see (1,2)] ranging from transcriptional control, splicing control, and degradative control of mRNA, up to higher order functions, such as gene silencing and imprinting (3,4). This implies that there is no single molecular mechanism of antisense function.
Several published investigations using different detection strategies report the occurrence of antisense transcripts in the human and mouse genomes. For example Lehner et al. (5) used pairwise BLAST analysis of curated sets of two mRNA libraries, while Shendure et al. (6) and Yelin et al. (7) used mRNA and Expressed sequence tag (EST)-libraries combined with information like exon–intron splicing structures and poly(A) signals to detect overlapping antisense transcripts from the same genomic locus.
Most analyses of antisense transcripts have focused on how widespread they are within one organism. The conservation across different species has been less studied, although many fundamental principles on the nature of antisense transcripts could be deduced from these studies. For instance, a comparison of antisense genes from human and mouse showed that less than half of all antisense genes between humans and mice have an ortholog in either species and that for only one-fifth of all pairs of antisense genes the antisense relationship is conserved (8).
To gain more insights into the abundance of antisense transcripts across eukaryotes, we decided to perform an analysis in four ascomycetes and one microsporidian fungi. A genomewide screening was performed in the genomes of the budding yeast Saccharomyces cerevisiae (9), its close relative Ashbya gossypii (10), the fission yeast Schizosaccharomyces pombe (11), the multicellular fungi Neurospora crassa (12) and the distant parasitic microsporidian Encephalitozoon cuniculi (13). At least some of these species are very diverged: orthologous genes between S.cerevisiae and S.pombe have on average the same distance between them as they have to their respective human orthologs (11). The species also show dramatic differences in organization of their genome: e.g. the number of genes with introns is greatly varying [S.cerevisiae (4%) (9) and S.pombe (43%) (11)]. They differ also in the capability to perform RNAi. There exist well known examples of RNAi in S.pombe, with important functions in the organization of heterochromatin (14,15) and cell divison (16) and in N.crassa there are even two antisense systems (quelling and the classical RNAi) (17,18). However, there are no known examples of RNAi in S.cerevisiae, which lacks the main components of the RNAi machinery (19). Thus, it is of interest if these differences could influence the individual equipment with antisense transcripts in each species.
It has been shown both experimentally and computationally that antisense transcripts can also arise from trans-genomic loci (5,20). However, we are not aware of any systematic investigation of this phenomenon and, therefore, decided to perform a systematic search for both cis- and trans-located antisense transcripts based on nucleotide sequences of open reading frames (ORFs). We analyze the relationships of antisense transcripts leading to complex network structures, describe the degree of conservation across the species and functionally annotate the antisense transcripts based on the Gene Ontology (GO) vocabulary.
METHODS
Data sources
We used the annotated genomic sequences (NC_*), which are provided by the different fungal genome projects and which are redistributed by the RefSeq project (21) at NCBI. We downloaded the respective RefSeq releases of all five complete fungal genomes: S.cerevisiae (dated June 18, 2003), A.gossypii (dated March 10, 2004), S.pombe (dated January 9, 2004), N.crassa (dated January 22, 2004) and E.cuniculi (dated December 17, 2003). We extracted all sequence information for the ORFs as well as their annotation directly from the respective RefSeq files.
Antisense database
We used a self developed SQL-based database-schema (‘antisense-db’, unpublished data) suited to store and process antisense-related information. It is flexible and serves as a prototype implementation for the analysis of large sets of antisense transcripts. Virtually all analyses were performed by means of the ‘antisense-db’.
Identification of cis natural antisense transcripts (NATs)
The overlaps of cis NATs were determined directly from the genomic coordinates of ORF sequences that are specified in the respective RefSeq genome annotations. We report every antisense overlap occuring between ORFs with a minimal genomic overlap size of at least 1 bp. Since typical untranslated region (UTR) sizes are larger than 50 bp, the true overlap lengths are always larger than the size measured by the ORF overlap length.
Identification of trans NATs
To identify all ORFs that are antisense to each other, a pairwise BLAST-search using WU-BLASTN (W. Gish, personal communication) with the nucleotide sequences of all predicted ORFs was performed (E-value <10−9). Besides the classical wobble-base pairs we also allowed the non-canonical basepairing of U–G. For that, we altered the nucleotide substitution matrices: the setting was M = 1, N = −2, Q = 3, R = 2, W = 9, wordmask = seg, lcmask, V = 10000, B = 10000, E = 0.000001, −altscore = ‘C T 1’ and −altscore =‘A G 1’. When more than one high-scoring pair (HSP) was reported, we combined their coordinates to one spanning region along the transcripts. For a given query sequence all HSPs from one subject sequence as reported by WU-BLAST were analyzed as follows: assume that there are n (overlapping) HSPs, such that ik and jk denote the first and last position, respectively, of the k-th HSP of the query, and ik′ and jk′ denote the first and last position, respectively, of the k-th HSP of the subject. Then take the HSP with the highest score, labeled m, and order all the remaining n−1 HSPs consistently with respect to their position, such that jk < m < ik+1 and jk′ > m > i′k+1 for all k ∈ {1,2,n−1}. The longest consistent ordering is reported as the antisense overlap region of putative NAT partners. All cis NATs identified during the BLAST search are removed.
Identification of longest continuous stretch (LCS)-regions
The LCS is reported as the maximum length of matching base pairs (canonical and non-canonical basepairing U–G) in the antisense overlap regions of trans NATs.
Classification of overlap regions
The classification of overlap regions follows a simple schematic rule: we distinguish 5′-overlaps, 3′-overlaps and inside-overlaps. Any sequence was always considered from 5′ to 3′. If an overlap occurs within the first quarter of the sequence and ends before the third quarter, we classify it as 5′-overlap. Vice versa, if an overlap occurs within the third quarter of the sequence and starts after the first quarter, we classify it as 3′-overlap. The remainders show no bias to one end of a sequence and are classified as inside-overlaps.
Detection of orthologous/paralogous sets
The determination of unique orthologs is not always possible due to the unequal expansion of protein family sizes (22) or the impact of genome duplication, e.g. as seen in S.cerevisiae (10). Therefore we decided to calculate sets of orthologs for an individual ORF. Thus we use a more relaxed definition of orthology, allowing for a specific ORF more than one orthologous partner in the other species [hence, we include also paralogs in our orthologous sets, similar to the definition of orthologous sets of paralogs given by Tatusov et al. (23) as used for the COG database]. The sets were identified first by using FastA (24) to compute pairwise alignments between the deduced protein sequences of the ORFs of each of the five species. We used a cutoff E-value of 10−9 in all similarity searches. A given ORF was classified to have an ortholog in another species if it is ranked within the top three positions according to the E-value, and if its reciprocally reported ORF was also ranked within its best three hits. Thus, for a given ORF we allow maximally three orthologous ORFs in another species, and two ORFs from one species can share the same ortholog. Let Oij, j ≠ i be the relative number of ORFs in organism i that have an orthologous ORF in organism j. Then we denote by Oi* the mean of all Oij, j = 1, …, 5, j ≠ i. When we refer to the individual species, we use the letters C for S.cerevisiae, A for A.gossypii, P for S.pombe, N for N.crassa and E for E.cuniculi.
For the detection of paralogous (multigene) families within one species a similar approach was performed. Here, sets of ORFs were classified to be paralogous if every possible pair of ORFs is reported with an E-value <10−9. In addition, to discriminate between members of multigene families and remote homologs a bidirectional overlap of at least 60% was required.
Visualization of networks
To visualize the network of trans NATs we used yEd, a graphical editor included in the yFiles library (25). In the resulting graph a vertex represents an ORF involved in at least one antisense relationship and an edge is drawn between two vertices if the two corresponding ORFs form an antisense relationship.
Annotation of NATs
The annotation of antisense transcripts is based on an ontology maintained by the GO consortium (26). The mapping from ontology terms to individual ORFs is performed by means of associaton files downloaded from the GO-website, which are available only for S.cerevisiae and S.pombe (http://www.geneontology.org). Since there are no association files provided for the other three species, orthology from S.cerevisiae ORFs to A.gossypii, N.crassa and E.cuniculi ORFs was used to create the respective gene association files. A further refinement to gene associations was carried out by mapping the full ontology to GO-slim terms, a cut-down version of the full ontology containing a subset of the terms in the whole ontology. Then all common terms shared by NATs were detected using the GO-TermFinder perl modules (http://search.cpan.org/dist/GO-TermFinder/). This provides an object oriented set of libraries for dealing with files produced by the Gene Ontology project. From this analysis all significant common GO-terms with a P-value <0.1 are reported. The P-values of a set of GO annotated genes are determined for a set of genes against the background of all genes in the genome sharing the same GO annotation. The P-value is calculated using the hypergeometric distribution as the probability of x or more out of n NAT ORFs having a given annotation, given that X of N (equal the total number of ORFs) have that annotation in the genome in general.
RESULTS
Genomewide detection and classification of NATs
Our genomewide prediction of NATs is based on predicted ORFs. Analysis of the coding sequences of the five fungal genomes revealed a high number of ORFs exhibiting regions of base complementarity. Though the total number of antisense ORFs is varying to a great extent, we detected antisense ORFs in all five species (Table 1). The number of ORFs involved in antisense relationships ranges from 14.8% in S.cerevisiae, 12.8% in E.cuniculi, 6.4% in N.crassa to 2.5% in A.gossypii and 1.0% in S.pombe (compared with the total number of ORFs annotated in each genome).
Table 1.
Number of coding sequences (ORFs) involved in antisense relationships of five fungal organisms
| S.cerevisiae | A.gossypii | S.pombe | N.crassa | E.cuniculi | |
|---|---|---|---|---|---|
| Total number of ORFs | 6304 | 4718 | 5041 | 10079 | 1996 |
| cis | 708 (11.2%) | 91 (1.9%) | 40 (0.8%) | 87 (0.9%) | 182 (9.1%) |
| trans | 295 (4.7%) | 29 (0.6%) | 9 (0.2%) | 561 (5.6%) | 100 (5.0%) |
| cis/trans | 73 (1.2%) | 1 (0.02%) | 1 (0.02%) | 2 (0.02%) | 26 (1.3%) |
| NAT ORFs | 930 (14.8%) | 119 (2.5%) | 48 (1.0%) | 646 (6.4%) | 256 (12.8%) |
| cis Pairs | 369 (26) | 46 (0) | 20 (0) | 45 (0) | 94 (16) |
| trans Pairs | 411 (196) | 19 (2) | 8 (0) | 722 (56) | 235 (234) |
| Combined | 780 (222) | 65 (2) | 28 (0) | 767 (56) | 329 (250) |
The upper five rows of the table show the absolute number of ORFs involved in antisense relationships as well as relative number (compared with the total number of investigated coding-sequences). We distinguish cis antisense relationships, where both transcripts reside at the same genomic locus and trans antisense relationships where the ORFs originate from different loci. Cis/trans ORFs are involved in cis as well as trans relationships. NAT ORFs refer to the overall number of antisense ORFs. Therefore this number is equal to the sum of cis and trans NATs. The lower three rows of the table list the total number of antisense sequence pairs in each species. In parentheses the number of NAT pairs are listed where at least two ORFs from a multigene family are involved in antisense relationships (pairs of NATs).
We classify NAT pairs as cis, if they populate the same genomic locus on the chromosomes, and as trans, if they originate from different loci, with some ORFs having NAT partners belonging to both classes. cis NATs are directly calculated from their genomic coordinates. This allows the detection of cis NATs even for cases, where they exhibit very small overlap lengths of 1–20 bp, that are typically missed by heuristic alignment programs such as BLAST. We remind that with typical UTR sizes (often >50 bp), the true overlap lengths have to be larger than the size measured by the ORF overlap lengths. The ratio of detected cis NATs compared with the absolute number of ORFs in S.cerevisiae and E.cuniculi (with 11.3 and 9.1%) is up to 12 times larger than in the other three species with 1–2% cis NATs (Table 1).
We see no clear relationship between the gene density of the genomes and the number of cis NATs. While the maximal difference of gene density between any two species has a factor of 3.7 (E.cuniculi and N.crassa), the maximal density of cis NATs per kb genomic sequence varies up to a factor of 33 (E.cuniculi and N.crassa).
Trans NATs are also found in all five fungal organisms but again one observes large differences in the number of involved ORFs. While ∼5% of all ORFs in S.cerevisiae (295 overall), N.crassa (561 overall) and E.cuniculi (100 overall) are trans NATs, only 0.2 (9 overall) and 0.6% (29 overall) of all ORFs are trans NATs in S.pombe and A.gossypii, respectively (Table 1). Thus, there are between 8 and 25 times more trans NATs in S.cerevisiae, N.crassa and E.cuniculi than in A.gossypii and S.pombe.
ORFs can participate in cis NAT relations as well as trans NAT relationships. Again, we note large differences among the species. While ∼8 and 10% of all antisense ORFs in S.cerevisiae and E.cuniculi, respectively, participate in cis relationships as well as trans relationships, this is observed for <1% of the NATs in the other three species (Table 1 and Supplementary Table 1).
The influence of multigene families on the number of NATs is very different among the species. Based on the set of paralogous ORFs a NAT was considered to be a multigene-family-NAT if at least one other member of the family is also a NAT. While in S.cerevisiae and E.cuniculi many NAT pairs are members of a multigene family (28.5 and 76%, respectively, of all NAT pairs), only a minor number of NAT pairs are influenced by multigene families in N.crassa (7.3%), A.gossypii (3.1%) and S.pombe (0%). Interestingly, many of the S.cerevisiae and E.cuniculi NATs, which are both in cis and trans relationships, are also members of multigene families (35 of 73, and 26 of 26 NATs, respectively).
Characteristics of overlap regions
The mean overlap length of cis NATs ranges from 266.9 bp for S.cerevisiae, 218.9 bp for N.crassa, 135.0 bp for E.cuniculi and 88.9 bp for A.gossypii down to 34.0 bp for S.pombe (Table 2). When the overlap lengths of trans NATs are compared, two groups can be detected. The first group consists of S.cerevisiae, A.gossypii and E.cuniculi with mean overlap lengths of 190.6, 184.3 and 196.2 bp, respectively. The second group consists of N.crassa and S.pombe with smaller mean overlap lengths of 118.1 and 132.8 bp, respectively (Table 2). Since cis NATs share the same genomic locus, the antisense regions have a 100% identity. Trans NATs on the other hand do not share the same genomic region; thus, a much lower identity of the antisense region is feasible in principle. Though a low cut-off value (10−9) was used in the WU-BLAST searches, a mean identity of 78–89% for all trans NATs in each of the five organisms is observed (Table 2).
Table 2.
Characterization of antisense overlap region
| Species | Identity | LOL (bp) | LCS (bp) | Corr(LOL/LCS) | |
|---|---|---|---|---|---|
| trans | cis | trans | trans | ||
| S.cerevisiae | 89% (8%) | 266.9 (151.9) | 190.6a (154.1) | 58.7a (76.3) | 0.65a |
| A.gossypii | 78% (3%) | 88.9 (205.6) | 184.3 (64.1) | 14.7 (3.4) | −0.01 |
| S.pombe | 83% (4%) | 34.0 (36.5) | 132.8 (38.3) | 25.0 (18.8) | −0.44 |
| N.crassa | 84% (6%) | 218.9 (149.8) | 118.1 (52.3) | 20.4 (9.1) | −0.49 |
| E.cuniculi | 85% (8%) | 135.0 (150.3) | 196.2b (84.7) | 45.3b (74.8) | 0.50 |
The table lists the mean identity of trans NATs, mean overlap-length (LOL) of cis and trans NATs, mean longest common stretch of identical residues (LCS) and Pearson correlation coefficient of overlap-length and LCS. (Numbers in parentheses indicate their respective standard deviation.)
aSubtracting all ORFs from multigene families (Table 1) results in a mean LOL = 124 bp, a mean LCS = 25 bp and a corr(LOL/LCS) = 0.16.
bSubtracting all ORFs from multigene families (Table 1) results in one remaining NAT pair with an LOL = 52 bp and an LCS = 52 bp.
Because trans NATs exhibit an overall mean identity of ∼84%, a mismatch every eight or nine bases is expected by chance. We define the LCS as the longest chain of continuous matching base pairs within the antisense region. The mean LCS of trans NATs in S.cerevisiae and E.cuniculi is three to four times longer than in N.crassa, A.gossypii and S.pombe (Table 2). We also computed the Pearson correlation coefficient of the absolute overlap lengths and the LCS of all trans NATs within each species (Table 2). We find a high correlation of 0.65 in S.cerevisiae and 0.5 in E.cuniculi, while the correlation coefficients in A.gossypii, N.crassa and S.pombe (−0.01, −0.49 and −0.44, respectively) indicate a non-correlated or even anti-correlated distribution of LCS and overlap-length.
The length of overlap (LOL) and the LCS is strongly affected by multigene families in S.cerevisiae and E.cuniculi: when removing multigene-family-NATs a considerable difference between the species is no longer observed (see footnotes of Table 2).
Next we analyzed the genomic arrangement of the overlap regions. We distinguished three regions of overlaps (5′, inside and 3′; Methods) and we calculated the distribution of the antisense regions along each transcript (Table 3). Almost all overlap regions are biased to the ends of the ORFs. A preference of 5′-overlaps is detected for both cis and trans NATs from S.cerevisiae and E.cuniculi. A mixed or opposite picture is seen for the remaining three species: e.g. the trans NATs in N.crassa are much more biased to 3′-overlaps with 53% of all antisense ORFs having 3′-overlaps compared with 15% of all antisense ORFs having 5′-overlaps. The odd distribution of overlap regions along NATs compared between S.cerevisiae, E.cuniculi and N.crassa is not affected by the effect of multigene families (data not shown).
Table 3.
Genomic arrangement of NATs
| Overlap region | S.cerevisiae | A.gossypii | S.pombe | N.crassa | E.cuniculi | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| cis | trans | cis | trans | cis | trans | cis | trans | cis | trans | |
| 5′ | 319 (43%) | 337 (41%) | 37 (40%) | 19 (50%) | 4 (10%) | 2 (12%) | 36 (40%) | 213 (15%) | 130 (69%) | 215 (46%) |
| Inside | 209 (28%) | 266 (32%) | 2 (2%) | 10 (26%) | 0 (0%) | 8 (50%) | 36 (40%) | 465 (32%) | 19 (10%) | 170 (36%) |
| 3′ | 210 (28%) | 219 (27%) | 53 (58%) | 9 (24%) | 36 (90%) | 6 (38%) | 18 (20%) | 766 (53%) | 39 (21%) | 85 (18%) |
Listed are the number of NATs (in parentheses relative to overall number of NATs) whose antisense region is located either 5′, inside or 3′ in transcript (see Methods for definition of 5′, inside and 3′ overlaps).
Orthologous antisense-transcripts
There are many examples of conserved functional entities in genomes, such as miRNAs or transcriptionally active regions, where the conservation is directly shown or at least believed to be due to functional constraints. Therefore we enquired to which extent ORFs involved in antisense relationships are conserved among the five fungal species.
To analyze conserved ORFs involved in antisense relationships we first determined the number of ORFs that have an orthologous sequence in any of the four other species. From this we computed the mean number of ORFs having orthologous sequences for every species. We notice that this varies by a factor of two between the species, ranging from 25.8% for ON* and 35.2% for OE*, to 50.3% for OC*, 50.8% for OP* and 56.6% for OA* (for notation see Methods). The highest pairwise number was detected between the close relatives A.gossypii and S.cerevisiae: 88.9% of the ORFs from A.gossypii have an ortholog in S.cerevisiae.
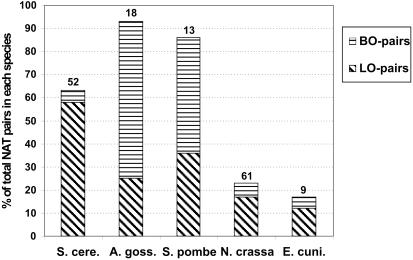
Next we analyzed the degree of conservation of NAT pairs across the species. Here, we distinguished between pairs of NATs where both ORFs have orthologs in another species (BO-pair) and pairs of NATs where only one ORF has an ortholog in another species (LO-pair). Figure 1 illustrates that for many pairs of NATs only one of the two transcripts has an ortholog (i.e. are LO-pairs). For example, of 780 NAT pairs in S.cerevisiae, 493 have an ortholog: 453 are LO-pairs, while only 40 are BO-pairs. Exceptions are found for A.gossypii and for S.pombe where most orthologous NATs are BO-pairs (68 and 58%, respectively). For a complete list of all ortholog assignments for cis and trans NATs see Supplementary Table 2.
Figure 1.
Characterization of identified orthologous NATs. We distinguish LO-pairs, where only one ORF of the NAT pair has an ortholog, and BO-pairs, where both ORFs have an ortholog. 100% refer to the total number of NAT pairs found in each species. The number on the tip of the columns depicts the total number of orthologs which are themselves found to be involved in antisense relationships.
Evolutionary conserved NATs
We further enquired whether antisense relationships of ORFs are evolutionarily conserved. There are no NATs that are conserved as NATs in all species. We found two pairs of fully conserved NATs in S.cerevisiae and A.gossypii, and they are both at syntenic positions. In addition, we observed that the orthologs of NAT ORFs are rarely antisense transcripts themselves (Figure 1).
The number of conserved cis NATs is generally low and we hypothesized that some cis NATs are still conserved as adjacent but non-overlapping ORFs on the chromosomes. To test this hypothesis, we determined the distance of adjacent orthologous ORFs. The majority of orthologs of cis NATs are >1000 bp apart from each other. For the cis NATs of A.gossypii, we found 14 syntenic regions, all in S.cerevisiae which are on average 266 bp apart. There are only two further cis NAT pairs, whose orthologous ORFs are <1000 bp apart from each other (one cis NAT in S.cerevisiae with a syntenic region in A.gossypii and one cis NAT in S.pombe with a syntenic region in A.gossypii).
Functional annotation of antisense transcripts
We used the annotations provided by the GO consortium to annotate all NATs from each species by means of the three classes of the ontology (function, process and component). Since only mappings of GO-terms to gene products for S.cerevisiae and S.pombe are available, we used orthology from ORFs of A.gossypii, N.crassa and E.cuniculi to the ORFs from S.cerevisiae to assign GO-terms to gene products in these species. Of 1059 S.cerevisiae ORFs which have an ortholog in E.cuniculi 594 ORFs are GO-annotated. Similarly, of 3304 N.crassa orthologs 2560 ORFs are GO-annotated and of 4937 A.gossypii orthologs 4361 ORFs have a GO-annotation. To compare the GO categories between each species, we mapped the full ontology to the GO slim terms provided by the Saccharomyces Genome Database (SGD) (http://www.yeastgenome.org). The GO slim terms are cut-down versions of the gene ontologies containing a subset of the terms of the whole ontology. They give a broader overview of the content without the detail of the specific fine grained terms.
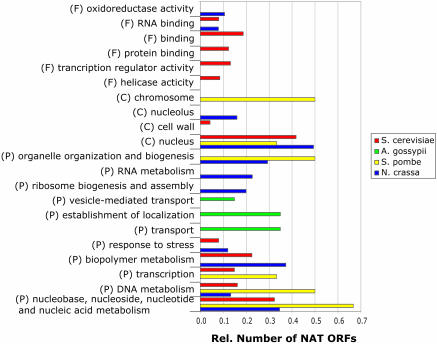
The GO analyses of cis and trans NATs are summarized in Figures 2 and 3, respectively. To ensure significance we only list every GO term shared by ORFs having P-values <0.1 (Methods). Though there are specific annotations of GO terms for each species, there are also GO terms that are shared between species and even between cis and trans NATs. Almost all terms shared by more than one species are also shared between cis and trans NATs. Examples of the process ontology are the terms ‘nucleobase, nucleoside, nucleotide and nucleic acid metabolism’, which are in common by S.cerevisiae, S.pombe and N.crassa and by cis and trans NATs. Another example is the term ‘transcription’, annotated for S.cerevisiae, S.pombe and E.cuniculi and shared between cis and trans NATs at least in S.cerevisiae. From the annotation with the component ontology it can be seen that high numbers of NAT gene products localize to the nucleus, with 30–50% of the annotated trans NATs from S.cerevisiae, S.pombe and N.crassa, respectively, and 33 and 70% of the annotated cis NATs from S.cerevisiae and E.cuniculi, respectively. Removing any multigene-family-NAT does not significantly alter the results of the GO analysis. Changes are very specific, e.g. terms like ‘helicase activity’ no longer appear due to the removed helicases encoded by subtelomeric repeats in S.cerevisiae.
Figure 2.
GO annotation of cis NATs in five fungal species. For each GO category, biological process (P), cellular component (C) and molecular function (F), the number of NATs annotated with the shown GO slim term relative to the total number of GO-annotated NATs in each species is plotted.
Figure 3.
GO annotation of trans NATs in four fungal species. For each GO category, biological process (P), cellular component (C) and molecular function (F), the number of NATs annotated with the shown GO slim term relative to the total number of GO-annotated NATs in each species is plotted.
For cis NATs of S.cerevisiae significant functional groups are found. Here the biggest group consists of proteins that are involved in ribonucleoprotein complexes, with 32 participating ORFs (P-value < 0.1). An important subset of this group consists of 17 ORFs, all of them are constituents of the ribosome (P-value < 0.01). Highly abundant are also functional groups related to transcription factor activity (36 ORFs, P-value < 0.1) or chromosome organization (26 ORFs, P-value < 0.001).
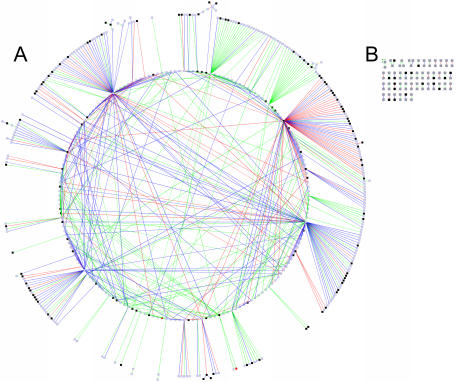
Analysis of the antisense interaction network
We analyzed the network of S.cerevisiae NATs in detail. Most of the cis NATs have unique dual relationships (313 pairs), 28 cis ORFs have two different NAT partners and one has four different NAT partners. In contrast, trans NATs are involved in a much more complex network (Figure 4). There are many trans NATs involved in large networks of relationships, resulting in some bigger subgraphs (Figure 4A–C) and smaller subgraphs (Figure 4D–G) and some mostly dual relationships (Figure 4H). The subgraphs in Figure 4B–G are composed of NATs that are mostly members of multigene families, while the dual relationships (Figure 4H) and the largest subgraph (Figure 4A) contain almost only NATs that are devoid of multigene families.
Figure 4.
Network of trans antisense relationships in S.cerevisiae. An ORF is represented by a vertex and an edge is drawn if the two corresponding ORFs form an antisense relationship. Vertices representing ORFs that are also involved in cis relationships are shown in red color. NATs that have an ortholog in one of the other four fungal species are shown in black color. Vertices representing NATs with no ortholog are colored blue. Edges are colored according to the LOL of the antisense regions: green, 40 bp < LOL < 100 bp;blue, 100 bp < LOL < 160 bp; red, >160 bp. Functional characterization of subgraphs: (A) functionally diverse proteins (transcription/regulation, nucleotide metabolism, chromosome organization—with most of these proteins having nuclear localization); (B) Y′-elements; (C) Ty-elements; (D) Seripauperin-family/DAN-family; (E) Stress-seventy proteins (HSP70); (F) MAL-family, alpha-glucosidases (maltose metabolism); (G) FLO-family (floculation); (H) proteins with diverse or unknown functions.
As can be seen, some of these NATs have many antisense partners and they are connectors of the complex subgraphs. For example, the ORFs YOR053W and YBR113W have 52 and 31 trans NAT partners, respectively. We call these founders of antisense relations (FARs): if these elements are removed, the subgraphs disintegrate and many antisense relationships are abolished. The creation of FARs is often explained by the fact that many FARs overlap in antisense with one member from a larger protein family onto the same genomic locus (as a cis-relationship). Consequently, these FARs do also serve as founders for the complex trans-relationships with further members of the protein family. An example of sense/antisense ORF transcription can be found in the subgraph (Figure 4E). As first reported by LeJohn (27,28) for the water mold Achlya klebsiana and later for Drosophila auraria (29), the genomic locus for the NAD-specific glutamate dehydrogenase harbors an antisense transcript which codes for a heat shock protein (HSP70). Both transcripts give rise to proteins and examination via hydropathy plots reveals almost perfect mirror images between the deduced aminoacid sequences of the sense/antisense transcripts (27). The ORF YAL004W, described as having strong similarity to A.klebsiana glutamate dehydrogenase, has also a predicted cis-antisense transcript belonging to the HSP70 family (YAL005C), recapitulating the concept of FARs: it has also conserved trans-antisense relationships with further members of the HSP70 family.
The same principle holds for nearly all subnetworks found in S.cerevisiae. As noted above, two of the big networks and four of the small networks represent larger protein families (most of them are localized in subtelomeric regions). Most ORFs shown in Figure 4B are annotated Y′-elements, containing a helicase-encoding ORF which is expressed only during meiosis and in telomerase-deficient cells (30). The subgraph (Figure 4D) consists of proteins sharing the PAU domain, forming two families which are activated during anaerobic processes and involved in the formation of the cell wall (DAN family) (31) or active during alcoholic fermentation (Seripauperin family) (32).
The largest and most diverse network found for S.cerevisiae is shown in Figure 4A. It consists in total of 137 NATs and is mostly devoid of NATs from multigene families. The 137 NATs were analyzed by means of the GO-annotation as described above. Out of 137 NATs, 69 are annotated components of the ‘nucleus’. The translation products of 51 NATs are involved in ‘nucleobase, nucleoside, nucleotide and nucleic acid metabolism’, most of them are involved in ‘transcription’ (32 NATs) exhibiting ‘transcription regulator activity’ (28 NATs).
Analysis of the trans networks of the other four species remains challenging due to missing annotation. Most ORFs are annotated as ‘predicted proteins’ or ‘hypothetical proteins’. Nevertheless upon visual inspections many topological motifs are recognized, which are similar to those of the trans network of S.cerevisiae. The trans network of N.crassa is shown in Figure 5.
Figure 5.
Network of trans antisense relationships in N.crassa. See Figure 4 for explanation. (A) Single connected network of trans NATs; (B) simple relationships.
Transcribed NATs
Hurowitz et al. (33) analyzed almost all budding yeast ORFs by means of correlating their assumed transcript length with the individual measurements from microarray experiments, thus ensuring that every measurement on the array is due to a transcript of expected size (virtual northern technique). Of special interest are questionable ORFs. A list of 820 questionable ORFs, representing a curation of experimental and computational data from the following articles (34–39), was distributed by the SGD (http://www.yeastgenome.org). Hurowitz et al. confirmed the transcription of 192 ORFs of these ORFs, indicating that these 192 ORFs are likely to represent bona fide genes.
We compared the set of confirmed questionable ORFs of Hurowitz et al. with the set of S.cerevisiae ORFs involved in antisense relationships as predicted by our study. In addition, we used all unambiguous assignments of yeast ESTs [downloaded from SGD (ftp://genome-ftp.stanford.edu/pub/yeast/sacchDB/)] to ORFs. Of 369 cis NAT pairs 152 NAT pairs are confirmed either by the virtual northern dataset of Hurowitz et al. or by the EST mapping or by direct experimental evidence given by SGD. Correspondingly, of 411 trans NAT pairs 206 NAT pairs are confirmed by either of the three data sources as aforementioned (for a detailed listing see Supplementary Table 3).
DISCUSSION
We have performed a genomewide screen of antisense transcripts based on annotated ORFs in four ascomycete and one microsporidian fungi. The number of ORFs exhibiting antisense relationships varies widely between all investigated species, even for the two closely related species S.cerevisiae and A.gossypii. This finding is remarkable considering the large number of orthologs between A.gossypii and S.cerevisiae and the strong synteny that is shown for both genomes (10). One explanation is that A.gossypii lacks any transposons and subtelomeric repeats, which are a major source of antisense transcripts in S.cerevisiae. But even when all effects of multigene families or repetitive families are removed, there are profound differences between the genomes. The high variation in the total number of cis NATs in the five species contrasts with the general idea that the compactness of genomes influences the number of overlapping ORFs and shows that there is more than one factor affecting the total number of overlapping ORFs in a genome.
Several investigations of antisense transcripts in human and mouse have been published (6–8). The results of these analyses clearly depend on the number and kind of antisense transcripts found and the databases used in the mapping procedures. For example Shendure et al. (6) found 217 cis NAT pairs in human, while Yelin et al. (7) reported 2667 cis NAT pairs. Although the set of human antisense transcripts found by Yelin et al. contains also non-coding transcripts, they detected ∼400 NATs with overlaps in coding regions (1.1% of 30 000 human genes). Strikingly we found similar numbers of cis NATs in A.gossypii, S.pombe and N.crassa (1.9, 0.8 and 0.9%, respectively), but not in S.cerevisiae and E.cuniculi, where we found much higher numbers of cis NATs (11.3 and 9.1%, respectively). It is noteworthy that many previously overlooked ORFs in S.cerevisiae (40) show an unexpectedly high number of partial or complete overlaps with known ORFs, supporting the idea of the high occurrence of cis NATs in S.cerevisiae.
Though there are prokaryotic examples of trans-encoded antisense transcripts (41), not many examples of functional trans NATs in eukaryotes have been described. Generally, any requirements for functional trans NATs are still unknown. Rosok et al. (20) introduced a technical system to identify endogenous human mRNAs with long complementary regions to known transcripts, based on their capability to form stable RNA/RNA duplexes in vitro. Similarities of 90% in the complementary regions were observed. This is the level of similarity found by us in the overlap regions of trans NATs. Korneev et al. (42) described the trans-regulation of a neural nitric oxidase synthase (nNOS) by an antisense RNA transcribed from an nNOS pseudogene. The overlap region of nNOS and its antisense RNA is 139 bp in length with a similarity of 87%, and it has been shown that these two transcripts form stable duplexes in vivo. These reports show that NATs with overlap regions of 130 bp are capable of forming stable duplexes. Such overlap lengths and corresponding identities are detected for many cis NATs in S.cerevisiae, E.cuniculi and N.crassa as well as for trans NATs in S.cerevisiae and E.cuniculi, denoting them as possible candidates to form RNA/RNA duplexes in vivo.
Another important parameter for duplex formation is the accessibility of complementary sequences with respect to their own secondary and tertiary structures. The initial presentation of three to four bases suffices in many cases to provide the scaffold for rapid interaction with complementary RNA. For instance, the analysis of the ubiquitous YUNR RNA recognition motif showed that the first step in forming RNA/RNA-duplexes involves very few accessible nucleotides (43). Based on this observation we introduced the LCS as the longest chain of continuous matching base pairs in the antisense region, which is a potential target for duplex formation. The antisense overlap region of nNOS and its antisense pseudogene exhibit an LCS of 18 bp (42). Antisense transcripts in S.cerevisiae and E.cuniculi exhibit even much longer LCS; their average LCS' are ∼60 and 45 bp, respectively. Antisense transcripts in N.crassa, A.gossypii and S.pombe, on the other hand, have LCS of roughly one-third of those found in S.cerevisiae and E.cuniculi. This difference, however, is no longer existing when removing multigene-family-NATs in S.cerevisiae and E.cuniculi. This, along with the strong anticorrelation of LCS in N.crassa and S.pombe, indicates, that there are potent molecular mechanisms to prevent longer, perfect duplexes in these two organisms.
In that respect it is interesting that there are many reports from N.crassa and S.pombe regarding RNAi, whereas it is known that S.cerevisiae has completely lost all known components of RNAi (19). In E.cuniculi and A.gossypii, there is also no Dicer or Argonaute homolog encoded in the genome (S. Steigele, personal observation). It could be speculated that an organism with no endogenous response to double-stranded RNA (dsRNA), such as S.cerevisiae and E.cuniculi, tolerates more antisense transcripts by chance than an organism such as N.crassa and S.pombe that would immediately respond with transcriptional silencing or chromatin remodeling. However, the low number of NATs in A.gossypii, which is also devoid of RNAi, points to more yet unknown factors determining the tolerance against dsRNA in an organism. To ultimately evaluate this hypothesis, more comparative studies of a larger spectrum of species with or without the capability to perform RNAi are necessary.
The analysis of orthologous sequences illustrates that many of the described NATs belong to a class of proteins with a marginal conservation, at least on the primary sequence level. When dealing with such orphan sequences (ORFans) it is often impossible to decide whether these sequences are transcribed and translated or if they are spuriously marked as ORFs. Besides the possibility of the de novo generation of coding sequences (44), it is shown that ORFans in Drosophila melanogaster are rapidly evolving genes, loosing their similarity to known proteins in a very short time span (45), even if they are related to them. In light of the short evolutionary distance between human and mouse and the very distant evolutionary time of the whole ascomycete phylum (46), it could be expected that more unique proteins have arisen in ascomycetes compared with proteins found in human or mouse. A detailed analysis of yeast evolution by Dujon et al. (47) supports this idea. Furthermore, these authors report that the hemiascomycete phylum appears much more diverse than the entire chordate phylum. This suggests that the high number of ORFans is not surprising for the hemiascomycete phylum, but it is surprising and it has not been shown so far that an unusually high number of these ORFans are recruited to antisense relationships.
Generally, it is unclear why NATs even from closely related species are less conserved than other genomic attributes. This seems to be a general observation independent of the chosen species. For example, a large-scale analysis for mammalian gene pairs showed that many overlapping human protein-coding genes lack any mouse ortholog (and vice versa) (8). This shows that even for human and mouse, which are at least 80 million years apart, hundreds of unique species-specific antisense pairs are found. All these analyses are based on the comparison of the primary sequence structures. However, there are some well-known examples of NATs with elaborate secondary structures, such as the pattern of partial duplexes known from the stable four-way junction of CopA/CopT, determining the effective structure of this antisense system (48). It is thus, of course, possible that a higher number of conserved NATs could be detected when taking features like the secondary structure of the mRNAs into account.
Another potential mechanism to produce unique NATs is the mechanism of overprinting (49), in which an existing nucleotide sequence is translated de novo into different reading frames. This is particularly interesting in the case of cis NATs. For the majority of NATs only one NAT has an ortholog and many of the ORFan antisense partners could have been generated by virtue of the overprinting effects. Our analysis detected the highest number of conserved NATs betweeen S.cerevisiae and N.crassa along with the interesting finding that many orthologs of cis NATs in S.cerevisiae are trans NATs in N.crassa and vice versa. It could be speculated that these NATs are remnants of earlier overprinting effects, with a pronounced selective pressure in N.crassa separating these ORFs.
Many ORFans are really transcribed, as shown for S.cerevisiae (50), and this suggests that these ORFans could be utilized to control the gene expression of their antisense partners at many levels (transcription, maturation, transport, stability and translation) (2). For example, the mRNAs of yeast are known to involve major control elements (51) which act through specific secondary (tertiary) RNA structures. There is an emerging number of RNA based regulation enlarging the ways of classical gene regulation. Isaacs et al. (52) showed that artifical riboregulators in the 5′-UTR of mRNAs are sensible to short complementary RNAs, which upon binding are competent to switch on/off the translation of the corresponding protein. A related mechanism acting on the stability of mRNAs is the interaction of short modifier RNAs with AU-rich elements (53). The basis of all these mechanisms are complementary RNAs perturbing or preserving a structural conformation of mRNA. It is conceivable that many antisense transcripts act in a transient fashion as modifiers of RNA secondary structures. One nice prediction of these models is that modifier RNAs evolve very fast and do not leave any phylogenetic footprints (54), perhaps one reason for the generally low conservation of antisense relationships we and others have seen.
The networks we describe have some interesting features in common. For instance, the trans NAT networks we described have some nodes with a high connectivity and many others with a low connectivity, similar to social and communication networks (55). One simple explanation for the emergence of the yeast network comes from the observation that the distribution of protein family sizes in yeast follows a power law (56), with a very small number of proteins having a large number of paralogs and vice versa. If many antisense relationships occur randomly in this set, it could explain the emergence of the complex network by simply displaying the underlying power-law distribution from the family sizes of the involved proteins. Nevertheless, this does not explain the high interconnectivity between each unit of the network, demanding for some yet unknown consolidating forces. The FARs in S.cerevisiae or E.cuniculi serve as an example. They have genomic overlaps with one member of a conserved protein family, resulting in trans relationships with further members of the protein family (multigene family). Although our model explains the networks from S.cerevisiae and E.cuniculi, it fails to explain the pronounced network of N.crassa, which is known to have a very low fraction of multigene families and no known repetitive elements.
In light of all these differences the functional annotation based on the GO revealed interesting results. We found significant groupings of specific terms between all species for both cis and trans NATs. Despite the diverse function of most NATs, many NATs are found to be related to aspects of nucleic acid metabolism and many of these proteins are involved in the regulation of transcription. Additionally, an overwhelming proportion of all proteins is specifically localizing to the nucleus, which was also recently found for human cis NATs (57). Lehner et al. (5) similarly categorized a high number of all human antisense transcripts to be involved in ‘nucleic acid binding’ and ‘transcription factor activity’.
In contrast to trans NATs, where most groups of ORFs sharing GO-terms are easily explained by virtue of the FARs-effects, any functional grouping of ORFs related to cis NATs is remarkable. In S.cerevisiae 32 proteins are involved in ‘ribonucleotide complexes’, comprising 17 proteins that are structural constituents of the ribosome and 11 proteins that are part of the ‘RNA-splicing’ machinery. This noticeable accumulation of functionally related NATs points to regulative constraints, which are important for the assembly and/or coordinated function of these proteins: in the case of ribosomes it is well known that the rate of accumulation of each ribosomal protein is carefully regulated by the yeast cell to provide the equimolar ratio necessary for the assembly of the ribosome (58).
Our analysis has shown that antisense transcripts are common in the species investigated in this study. They are less conserved than overlapping ORFs from bacteria. The functional characterization of antisense transcripts demonstrates an interesting accumulation of specific functional groups, as shown before for other species. The networks we have found have the potential to give important hints about the function and evolution of natural antisense regulated systems.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
Supplementary Material
Acknowledgments
We thank Andreas Schwienhorst for reading and commenting an earlier version of this manuscript. We are also grateful to Peter Stadler and Stefanie Bette for discussions. We thank Peter Wills for reading and commenting the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft, AZ BIZ 1/1-3. Funding to pay the Open Access publication charges for this article was provided by Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
REFERENCES
- 1.Knee R., Murphy P.R. Regulation of gene expression by natural antisense RNA transcripts. Neurochem. Int. 1997;31:379–392. doi: 10.1016/s0197-0186(96)00108-8. [DOI] [PubMed] [Google Scholar]
- 2.Vanhee-Brossollet C., Vaquero C. Do natural antisense transcripts make sense in eukaryotes. Gene. 1998;211:1–9. doi: 10.1016/s0378-1119(98)00093-6. [DOI] [PubMed] [Google Scholar]
- 3.Tufarelli C., Stanley J.A.S., Garrick D., Sharpe J.A., Ayyub H., Wood W.G., Higgs D.R. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nature Genet. 2003;34:157–165. doi: 10.1038/ng1157. [DOI] [PubMed] [Google Scholar]
- 4.Seitz H., Youngson N., Lin S.-P., Dalbert S., Paulsen M., Bachellerie J.-P., Ferguson-Smith A.C., Cavaille J. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nature Genet. 2003;34:261–262. doi: 10.1038/ng1171. [DOI] [PubMed] [Google Scholar]
- 5.Lehner B., Williams G., Campbell R.D., Sanderson C.M. Antisense transcripts in the human genome. Trends Genet. 2002;18:63–65. doi: 10.1016/s0168-9525(02)02598-2. [DOI] [PubMed] [Google Scholar]
- 6.Shendure J., Church G.M. Computational discovery of sense–antisense transcription in the human and mouse genomes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-9-research0044. RESEARCH0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yelin R., Dahary D., Sorek R., Levanon E.Y., Goldstein O., Shoshan A., Diber A., Biton S., Tamir Y., Khosravi R., et al. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- 8.Veeramachaneni V., Makalowski W., Galdzicki M., Sood R., Makalowska I. Mammalian overlapping genes: the comparative perspective. Genome Res. 2004;14:280–286. doi: 10.1101/gr.1590904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goffeau A., Barrell B.G., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., et al. Life with 6000 genes. Science. 1996;274:546. doi: 10.1126/science.274.5287.546. 563–567. [DOI] [PubMed] [Google Scholar]
- 10.Dietrich F.S., Voegeli S., Brachat S., Lerch A., Gates K., Steiner S., Mohr C., Poehlmann R., Luedi P., Choi S., et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- 11.Wood V., Gwilliam R., Rajandream M.-A., Lyne M., Lyne R., Stewart A., Sgouros J., Peat N., Hayles J., Baker S., et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 12.Galagan J.E., Calvo S.E., Borkovich K.A., Selker E.U., Read N.D., Jaffe D., FitzHugh W., Ma L.-J., Smirnov S., Purcell S., et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 13.Katinka M.D., Duprat S., Cornillot E., Metenier G., Thomarat F., Prensier G., Barbe V., Peyretaillade E., Brottier P., Wincker P., et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 14.Raponi M., Arndt G.M. Dominant genetic screen for cofactors that enhance antisense RNA-mediated gene silencing in fission yeast. Nucleic Acids Res. 2002;310:2546–2554. doi: 10.1093/nar/30.11.2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raponi M., Arndt G.M. Double-stranded RNA-mediated gene silencing in fission yeast. Nucleic Acids Res. 2003;31:4481–4489. doi: 10.1093/nar/gkg484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volpe T., Schramke V., Hamilton G.L., White S.A., Teng G., Martienssen R.A., Allshire R.C. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 2003;11:137–146. doi: 10.1023/a:1022815931524. [DOI] [PubMed] [Google Scholar]
- 17.Cogoni C., Macino G. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional trans-gene silencing in Neurospora crassa. Proc. Natl Acad. Sci. USA. 1997;94:10233–10238. doi: 10.1073/pnas.94.19.10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cogoni C., Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- 19.Aravind L., Watanabe H., Lipman D.J., Koonin E.V. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc. Natl Acad. Sci. USA. 2000;97:11319–11324. doi: 10.1073/pnas.200346997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosok O., Sioud M. Systematic identification of sense–antisense transcripts in mammalian cells. Nat. Biotechnol. 2004;22:104–108. doi: 10.1038/nbt925. [DOI] [PubMed] [Google Scholar]
- 21.Pruitt K.D., Katz K.S., Sicotte H., Maglott D.R. Introducing RefSeq and LocusLink: curated human genome resources at the NCBI. Trends Genet. 2000;16:44–47. doi: 10.1016/s0168-9525(99)01882-x. [DOI] [PubMed] [Google Scholar]
- 22.Sonnhammer E.L.L., Koonin E.V. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 2002;18:619–620. doi: 10.1016/s0168-9525(02)02793-2. [DOI] [PubMed] [Google Scholar]
- 23.Tatusov R.L., Koonin E.V., Lipman D.J. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 24.Pearson W.R., Lipman D.J. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiese R., Eiglsperger M., Kaufmann M. yfiles: visualization and automatic layout of graphs. Proceedings of the Ninth International Symposium on Graph Drawing; Vienna, Austria. 2001. pp. 453–454. [Google Scholar]
- 26.Harris M.A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LeJohn H.B., Cameron L.E., Yang B., Rennie S.L. Molecular characterization of an NAD-specific glutamate dehydrogenase gene inducible by l-glutamine. antisense gene pair arrangement with l-glutamine-inducible heat shock 70-like protein gene. J. Biol. Chem. 1994;269:4523–4531. [PubMed] [Google Scholar]
- 28.LeJohn H.B., Cameron L.E., Yang B., MacBeath G., Barker D.S., Williams S.A. Cloning and analysis of a constitutive heat shock (cognate) protein 70 gene inducible by l-glutamine. J. Biol. Chem. 1994;269:4513–4522. [PubMed] [Google Scholar]
- 29.Konstantopoulou I., Ouzounis C.A., Drosopoulou E., Yiangou M., Sideras P., Sander C., Scouras Z.G. A Drosophila hsp70 gene contains long, antiparallel, coupled open reading frames (lac ORFs) conserved in homologous loci. J. Mol. Evol. 1995;41:414–420. doi: 10.1007/BF00160312. [DOI] [PubMed] [Google Scholar]
- 30.Yamada M., Hayatsu N., Matsuura A., Ishikawa F. Y′-help1, a DNA helicase encoded by the yeast subtelomeric Y′ element, is induced in survivors defective for telomerase. J. Biol. Chem. 1998;273:33360–33366. doi: 10.1074/jbc.273.50.33360. [DOI] [PubMed] [Google Scholar]
- 31.Abramova N., Sertil O., Mehta S., Lowry C.V. Reciprocal regulation of anaerobic and aerobic cell wall mannoprotein gene expression in Saccharomyces cerevisiae. J. Bacteriol. 2001;183:2881–2887. doi: 10.1128/JB.183.9.2881-2887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rachidi N., Martinez M.J., Barre P., Blondin B. Saccharomyces cerevisiae PAU genes are induced by anaerobiosis. Mol. Microbiol. 2000;35:1421–1430. doi: 10.1046/j.1365-2958.2000.01807.x. [DOI] [PubMed] [Google Scholar]
- 33.Hurowitz E.H., Brown P.O. Genome-wide analysis of mRNA lengths in Saccharomyces cerevisiae. Genome Biol. 2003;5:R2. doi: 10.1186/gb-2003-5-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wood V., Rutherford K.M., Ivens A., Rajandream M., Barrell B. A re-annotation of the Saccharomyces cerevisiae genome. Comp. Funct. Genomics. 2001;2:143–154. doi: 10.1002/cfg.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- 36.Brachat S., Dietrich F.S., Voegeli S., Zhang Z., Stuart L., Lerch A., Gates K., Gaffney T., Philippsen P. Reinvestigation of the Saccharomyces cerevisiae genome annotation by comparison to the genome of a related fungus: Ashbya gossypii. Genome Biol. 2003;4:R45. doi: 10.1186/gb-2003-4-7-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kellis M., Patterson N., Endrizzi M., Birren B., Lander E.S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 38.Kessler M.M., Zeng Q., Hogan S., Cook R., Morales A.J., Cottarel G. Systematic discovery of new genes in the Saccharomyces cerevisiae genome. Genome Res. 2003;13:264–271. doi: 10.1101/gr.232903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oshiro G., Wodicka L.M., Washburn M.P., Yates J.R., Lockhart D.J., Winzeler E.A. Parallel identification of new genes in Saccharomyces cerevisiae. Genome Res. 2002;12:1210–1220. doi: 10.1101/gr.226802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar A., Harrison P.M., Cheung K.-H., Lan N., Echols N., Bertone P., Miller P., Gerstein M.B., Snyder M. An integrated approach for finding overlooked genes in yeast. Nat. Biotechnol. 2002;20:58–63. [Google Scholar]
- 41.Gerdes K., Gultyaev A.P., Franch T., Pedersen K., Mikkelsen N.D. Antisense RNA-regulated programmed cell death. Annu. Rev. Genet. 1997;31:1–31. doi: 10.1146/annurev.genet.31.1.1. [DOI] [PubMed] [Google Scholar]
- 42.Korneev S.A., Park J.H., O'Shea M. Neuronal expression of neural nitric oxide synthase (nNOS) protein is suppressed by an antisense RNA transcribed from an NOS pseudogene. J. Neurosci. 1999;19:7711–7720. doi: 10.1523/JNEUROSCI.19-18-07711.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franch T., Petersen M., Wagner E.G., Jacobsen J.P., Gerdes K. Antisense RNA regulation in prokaryotes: rapid RNA/RNA interaction facilitated by a general U-turn loop structure. J. Mol. Biol. 1999;294:1115–1125. doi: 10.1006/jmbi.1999.3306. [DOI] [PubMed] [Google Scholar]
- 44.Boldogkoi Z. Coding in the noncoding DNA strand: a novel mechanism of gene evolution. J. Mol. Evol. 2000;51:600–606. doi: 10.1007/s002390010123. [DOI] [PubMed] [Google Scholar]
- 45.Domazet-Loso T., Tautz D. An evolutionary analysis of orphan genes in Drosophila. Genome Res. 2003;13:2213–2219. doi: 10.1101/gr.1311003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heckman D.S., Geiser D.M., Eidell B.R., Stauffer R.L., Kardos N.L., Hedges S.B. Molecular evidence for the early colonization of land by fungi and plants. Science. 2001;293:1129–1133. doi: 10.1126/science.1061457. [DOI] [PubMed] [Google Scholar]
- 47.Dujon B., Sherman D., Fischer G., Durrens P., Casaregola S., Lafontaine I., Montigny J.D., Marck C., Neuveglise C., Talla E., et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 48.Kolb F.A., Engdahl H.M., Slagter-Jäger J.G., Ehresmann B., Esmann C.E., Westhof E., Wagner E.G., Romby P. Progression of a loop–loop complex to a four-way junction is crucial for the activity of a regulatory antisense RNA. EMBO J. 2000;19:5905–5915. doi: 10.1093/emboj/19.21.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keese P.K., Gibbs A. Origins of genes: “big bang” or continuous creation. Proc. Natl Acad. Sci. USA. 1992;89:9489–9493. doi: 10.1073/pnas.89.20.9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naitou M., Hagiwara H., Hanaoka F., Eki T., Murakami Y. Expression profiles of transcripts from 126 open reading frames in the entire chromosome VI of Saccharomyces cerevisiae by systematic northern analyses. Yeast. 1997;13:1275–1290. doi: 10.1002/(SICI)1097-0061(199710)13:13<1275::AID-YEA172>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 51.McCarthy J.E. Posttranscriptional control of gene expression in yeast. Microbiol. Mol. Biol. Rev. 1998;62:1492–1553. doi: 10.1128/mmbr.62.4.1492-1553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Isaacs F.J., Dwyer D.J., Ding C., Pervouchine D.D., Cantor C.R., Collins J.J. Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol. 2004;22:841–847. doi: 10.1038/nbt986. [DOI] [PubMed] [Google Scholar]
- 53.Meisner N.-C., Hackermüller J., Uhl V., Aszodi A., Jaritz M., Auer M. mRNA openers and closers: modulating AU-rich element-controlled mRNA stability by a molecular switch in mRNA secondary structure. Chembiochem. 2004;5:1432–1447. doi: 10.1002/cbic.200400219. [DOI] [PubMed] [Google Scholar]
- 54.Hackermüller J., Meisner N.-C., Auer M., Jaritz M., Stadler P.F. The effect of RNA secondary structures on RNA-ligand binding and the modifier RNA mechanism: a quantitative model. Gene. 2005;345:3–12. doi: 10.1016/j.gene.2004.11.043. [DOI] [PubMed] [Google Scholar]
- 55.Watts D.J., Strogatz S.H. Collective dynamics of ‘small-world’ networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 56.Qian J., Luscombe N.M., Gerstein M. Protein family and fold occurrence in genomes: power-law behaviour and evolutionary model. J. Mol. Biol. 2001;313:673–681. doi: 10.1006/jmbi.2001.5079. [DOI] [PubMed] [Google Scholar]
- 57.Chen J., Sun M., Kent W.J., Huang X., Xie H., Wang W., Zhou G., Shi R.Z., Rowley J.D. Over 20% of human transcripts might form sense–antisense pairs. Nucleic Acids Res. 2004;32:4812–4820. doi: 10.1093/nar/gkh818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Warner J.R., Mitra G., Schwindinger W.F., Studeny M., Fried H.M. Saccharomyces cerevisiae coordinates accumulation of yeast ribosomal proteins by modulating mRNA splicing, translational initiation, and protein turnover. Mol. Cell. Biol. 1985;5:1512–1521. doi: 10.1128/mcb.5.6.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.