Abstract
The ssu71 alleles of the TFG1 gene, which encodes the largest subunit of TFIIF, were isolated as suppressors of a TFIIB defect that affects the accuracy of transcription start site selection in the yeast Saccharomyces cerevisiae. Here we report that ssu71-1 also suppresses the cell growth and start site defects associated with an altered form of the Rpb1 subunit of RNA polymerase II (RNAP II). The ssu71-1 and ssu71-2 alleles were cloned and found to encode single amino acid replacements of glycine-363, either glycine to aspartic acid (G363D) or glycine to arginine (G363R). Two other charged replacements, G363E and G363K, were constructed by site-directed mutagenesis and suppress both TFIIB E62K and Rpb1 N445S, whereas neither G363A nor G363P exhibited any effect. G363 is phylogenetically conserved and its counterpart in human TFIIF (RAP74 G112) is located within the RAP74/RAP30 dimerization domain. We propose that the TFIIF dimerization domain is located in proximity to the B-finger of TFIIB near the active center of RNAP II where the TFIIB–TFIIF–RNAP II interface plays a key role in start site selection.
INTRODUCTION
Promoter-dependent transcription by RNA polymerase II (RNAP II) requires a set of general transcription factors that include the TATA-binding protein (TBP), TFIIB, TFIIE, TFIIF and TFIIH (1–3). TBP binds to the TATA element of promoter DNA to nucleate assembly of the preinitiation complex (PIC), followed by binding of TFIIB both upstream and downstream of TATA. The DNA–TBP–TFIIB ternary complex forms the binding site for RNAP II, which enters the complex in association with TFIIF. TFIIE and TFIIH complete PIC assembly and are required for ATP-dependent promoter melting by RNAP II (4). These steps define the pathway of PIC assembly in vitro. A similar pathway might be followed in vivo as PIC formation has been shown to proceed via formation of structural intermediates in yeast (5).
RNAP II comprises 12 subunits, encoded by the RPB1–RPB12 genes in Saccharomyces cerevisiae (6,7). The Rpb1, Rpb2, Rpb3 and Rpb11 subunits form the catalytic core of RNAP II and are the counterpart of the bacterial α2ββ′ RNAP core enzyme. High-resolution X-ray structures of 10- (lacking Rpb4/Rpb7) and 12-subunit yeast RNAP II complexes have been solved (8–10), as have minimal transcript elongation complexes (11–13). These structures, in combination with high-resolution structures of bacterial RNAPs (14), offer insight into the mechanism of transcription and a framework for the interpretation of a wealth of genetic and biochemical data.
TFIIB plays a key role in transcription initiation. The SUA7 gene, which encodes yeast TFIIB, was initially identified by mutations that alter start site selection (15). The sua7-1 and sua7-3 alleles shift initiation at the CYC1 and ADH1 promoters downstream of normal and encode, respectively, glutamate-62 to lysine (E62K) and arginine-78 to cysteine (R78C) replacements (16). TFIIB E62 replacements do not affect PIC assembly, but abolish transcription in vitro by adversely affecting RNAP II promoter clearance (5,17,18). The N-terminal domain of TFIIB forms a zinc-ribbon that interacts with TFIIF and the ‘dock’ domain of RNAP II (19,20). A phylogenetically conserved domain lies immediately downstream of the zinc-ribbon and is critical for accurate start site selection in yeast (2), human (21) and Archaea (22). The recent high-resolution X-ray structure of a yeast RNAP II–TFIIB complex defined the conserved domain as a ‘B-finger’ that encompasses E62 and R78. The B-finger projects into the RNAP II active center via the ‘saddle’ between the ‘clamp’ and ‘wall’ domains (20). This structure suggests that steric clash between the B-finger and nascent RNA on the saddle accounts for abortive initiation (20), a proposal consistent with the effect of the R78C replacement on promoter clearance (18).
The same genetic selection that yielded the sua7 mutants yielded the sua8 alleles of RPB1 (23). The sua7 and sua8 mutants confer similar growth defects, show identical effects on start site selection and a double sua7-1 sua8-1 mutant is synthetically lethal. These results define a functional interaction between Rpb1 and TFIIB, and imply that this interaction is critical for accurate initiation. The sua8-1 allele encodes an asparagine-445 to serine (N445S) replacement in the active center of Rpb1, which is near the B-finger in the RNAP II–TFIIB complex (20,23).
TFIIF also affects start site selection. Yeast TFIIF is composed of three subunits: Tfg1, Tfg2 and Tfg3. Tfg1 and Tfg2 are counterparts of human TFIIF subunits RAP74 and RAP30; Tfg3 has no counterpart in mammalian TFIIF, but is identical to the conserved TAF14 subunit of TFIID (24). A role for TFIIF in start site selection was uncovered in a genetic selection for suppressors of the cold-sensitive growth defect associated with the sua7-1-encoded TFIIB E62K replacement: two alleles of tfg1 (ssu71-1 and ssu71-2) not only suppressed the sua7-1 cell growth defect, but also partially restored the normal pattern of start site selection (25). More recently, two amino acid replacements in Tfg1 (E346A and W350A) and a single replacement in Tfg2 (L59K) were reported to shift initiation at the ADH1 gene upstream of normal in a SUA7 wild-type strain (26). Thus, TFIIB and TFIIF are critical determinants of start site selection in S.cerevisiae. However, the mechanism(s) by which altered forms of these factors affect start site selection remains unresolved.
In an effort to elucidate the mechanism of start site selection by yeast RNAP II and to determine how mutations in components of the transcription initiation machinery alter start sites, we have defined the ssu71-encoded TFIIF mutations and analyzed these replacements in the context of crystal structures of yeast RNAP II–TFIIB and human TFIIF complexes. Our results suggest that the Tfg1–Tfg2 dimer interface of TFIIF is located within the active center of RNAP II initiation complex.
MATERIALS AND METHODS
Yeast strains and nomenclature
The yeast strains used in this study are listed in Table 1. Plasmid-shuffle strains YMH904 (SUA7 tfg1::kanMX4 [TFG1-URA3]), YMH910 (sua7-1 tfg1::kanMX4 [TFG1-URA3]) and YMH916 (sua8-1 tfg1::LEU2 [TFG1-URA3]) were derived from isogenic strains T16 (SUA7), YDW546 (sua7-1) and YDW383 (sua8-1), respectively. The TRP1 gene was first disrupted by one-step disruption with either trp1::hphMX4 (T16, YDW546) or trp1::kanMX6 (YDW383), which were generated by PCR amplification of the drug-resistant cassettes pAG34-hphMX4 (27) or pFA6a-kanMX6 (28) with primer pairs carrying TRP1 5′- and 3′-tails. Strains were then transformed with plasmid pM435 (TFG1-URA3), followed by one-step disruption of the chromosomal TFG1 gene either with tfg1::LEU2 (sua8-1) using a tfg1::LEU2 DNA fragment that was generated by KpnI–SphI digestion of plasmid pM521 or with tfg1::kanMX6 (WT and sua7-1), which was generated from pFA6a-kanMX6 using TFG1-tailed primer pairs. Derivatives of YMH904, YMH910 and YMH916 that carry plasmid-borne TFG1+ or tfg1 alleles encoding G363 replacements were generated by transformation of the respective strains with either pM441 (TFG1+-TRP1) or site-directed derivatives of pM441 encoding TFG1 G363 replacements, followed by counter-selection of pM435 (TFG1-URA3) on 5-FOA medium. Strain YZS96 (sua8-1 tfg1::LEU2 [tfg1-G363D]) was constructed by transformation of YDW383 with plasmid pM482 [tfg1-G363D-URA3], followed by one-step disruption of TFG1, using plasmid pM521, as described above.
Table 1.
List of yeast strains
| Strain | Genotype* | Reference |
|---|---|---|
| T16 | MATα cyc1-5000 cyc7-67 leu2-3,112 ura3-52 cyh2 | (15) |
| YDW546 | MATα cyc1-5000 cyc7-67 leu2-3,112 ura3 -52 cyh2 sua7-1 | (15) |
| YDW383 | MATα cyc1-5000 cyc7-67 leu2-3,112 ura3 -52 cyh2 sua8-1 | (23) |
| YMH71-9C | MATacyc1-5000 cyc7-67 trp5-48 his5-2 ura3-52 sua7-1 | (16) |
| YZS14 | MATacyc1-5000 cyc7-67 trp5-48 his5-2 ura3 -52 sua7 -1 ssu71-1 | (25) |
| YZS45 | MATacyc1-5000 cyc7-67 trp5-48 his5-2 ura3 sua7-1 ssu71 -2 | (25) |
| YZS96 | MATα cyc1-5000 cyc7-67 leu2-3,112 ura3-52 cyh2 sua8-1 tfg1::LEU2 [pM482: ssu71-1-URA3] | this study |
| YMH904 | MATα cyc1-5000 cyc7-67 ura3-52 leu2 -3,112 cyh2trp1::hphMX4tfg1::kanMX4 [pM435: TFG1+-URA3 -CEN] | this study |
| YMH905 | MATα cyc1-5000 cyc7-67 ura3-52 leu2-3,112 cyh2 trp1::hphMX4 tfg1::kanMX4 [pM441: TFG1+-TRP1-CEN] | this study |
| YMH906 | MATα cyc1-5000 cyc7-67 ura3-52 leu2 -3,112 cyh2 trp1::hphMX4 tfg1::kanMX4 [pM1874: tfg1-G363D-TRP1-CEN] | this study |
| YMH907 | MATα cyc1-5000 cyc7-67 ura3-52 leu2-3,112 cyh2 trp1::hphMX4 tfg1::kanMX4 [pM1876: tfg1-G363E-TRP1-CEN] | this study |
| YMH908 | MATα cyc1-5000 cyc7-67 ura3-52 leu2-3,112 cyh2 trp1::hphMX4 tfg1::kanMX4 [pM1877: tfg1-G363K-TRP1-CEN] | this study |
| YMH909 | MATα cyc1-5000 cyc7-67 ura3-52 leu2-3,112 cyh2 trp1::hphMX4 tfg1::kanMX4 [pM1875: tfg1-G363R-TRP1-CEN] | this study |
| YMH910 | MATα_cyc1-5000 cyc7-67 ura3-52 leu2-3,112 cyh2 sua7-1 trp1::hphMX4 tfg1::kanMX4 [pM435: TFG1+-URA3-CEN] | this study |
| YMH911 | MATα_cyc1-5000 cyc7-67 ura3-52 leu2-3,112 cyh2 sua7-1 trp1::hphMX4 tfg1::kanMX4 [pM441: TFG1+-TRP1-CEN] | this study |
| YMH912 | MATα_cyc1-5000 cyc7-67 ura3-52 leu2-3,112 cyh2 sua7-1 trp1::hphMX4 tfg1::kanMX4 [pM1874: tfg1-G363D-TRP1-CEN] | this study |
| YMH913 | MATα_cyc1-5000 cyc7-67 ura3-52 leu2-3,112 cyh2 sua7-1 trp1::hphMX4 tfg1::kanMX4 [pM1876: tfg1-G363E-TRP1-CEN] | this study |
| YMH914 | MATα_cyc1-5000 cyc7-67 ura3-52 leu2-3,112 cyh2 sua7-1 trp1::hphMX4 tfg1::kanMX4 [pM1877: tfg1-G363K-TRP1-CEN] | this study |
| YMH915 | MATα_cyc1-5000 cyc7-67 ura3-52 leu2 -3,112 cyh2 sua7-1 trp1::hphMX4 tfg1::kanMX4 [pM1875:tfg1-G363R-TRP1-CEN] | this study |
| YMH916 | MATα_cyc1-5000 cyc7-67 ura3-52 leu2-3,112 cyh2 sua8-1 trp1::kanMX tfg1::LEU2 [pM435: TFG1+-URA3 -CEN] | this study |
| YMH917 | MATα_cyc1-5000 cyc7-67 ura3-52 leu2-3,112 cyh2 sua8-1 trp1::kanMX tfg1::LEU2 [pM441: TFG1+- TRP1-CEN] | this study |
| YMH918 | MATα_cyc1-5000 cyc7-67 ura3-52 leu2-3,112 cyh2 sua8-1 trp1::kanMX tfg1::LEU2 [pM1874: tfg1-G363D-TRP1-CEN] | this study |
| YMH919 | MATα_cyc1-5000 cyc7-67 ura3-52 leu2-3,112 cyh2 sua8-1 trp1::kanMX tfg1::LEU2 [pM1876: tfg1-G363E-TRP1-CEN | this study |
| YMH920 | MATα_cyc1-5000 cyc7-67 ura3-52 leu2-3,112 cyh2 sua8-1 trp1::kanMX tfg1::LEU2 [pM1877: tfg1-G363K-TRP1-CEN] | this study |
| YMH921 | MATα_cyc1-5000 cyc7-67 ura3-52 leu2-3,112 cyh2 sua8-1 trp1::kanMX tfg1::LEU2 [pM1876: tfg1-G363R-TRP1-CEN] | this study |
*The symbol ssu71 or tfg1 denotes alleles of TFG1.
Standard yeast nomenclature is used throughout to designate genotypes and phenotypes. The symbol ssu71 denotes the original alleles of the TFG1 gene (25). The symbols Csm− (cold sensitive) and Tsm− (heat sensitive) refer to impaired growth on yeast extract/peptone/dextrose (YPD) medium at 16 and 37°C, respectively, relative to growth on YPD at 30°C. YPD, synthetic complete (SC) and SC lacking inositol (SC–Ino) media were prepared using standard recipes (29).
Site-directed mutagenesis of TFG1
Site-directed mutations in the TFG1 gene were made by the QuickChange mutagenesis kit (Stratagene) using plasmid pM441 (TFG1-TRP1) as template DNA and complementary primer pairs that encode amino acid replacements at Tfg1 position G363. All sequence changes were confirmed by DNA sequence analysis using TFG1-specific primers.
Determination of transcript start site sites
Primer extension was performed as described previously using total RNA and the ADH1-specific primer oIP87 (15). Primer extension products were resolved in a 6% polyacrylamide DNA sequencing gel and visualized by autoradiography. Radioactive signals were quantified using ImageQuant software (Molecular Dynamics).
Retrieval of the ssu71 alleles by gap repair
The ssu71-1 and ssu71-2 alleles were cloned by gap repair from strains YZS14 and YZS45, respectively (30). Plasmid pM436 (TFG1-LEU2-2µ) was digested with BamHI and BglII, thereby creating a sequence gap in the TFG1 open reading frame (ORF), and introduced into YZS14 and YZS45. Leu+ transformants were isolated and shown to retain the Csm+ and Tsm− suppressor phenotypes. Plasmid DNA was isolated from each strain and confirmed to include the tfg1 BamHI–BglII DNA fragment; these plasmids were designated pM484 (ssu71-1) and pM538 (ssu71-2). pM484 and pM538 were reintroduced into YZS14 and YZS45 and the resulting transformants retained the suppressor phenotypes, consistent with having isolated the ssu71 suppressor mutations. The entire BamHI–BglII DNA fragments of pM484 and pM538 were sequenced and a single base pair substitution encoding different amino acid replacements of the same residue (G363D and G363R) were identified for the two clones. The TFG1 ORFs flanking the BamHI–BglII DNA fragment of each allele were subsequently amplified by PCR from YZS14 and YZS45 genomic DNA and sequenced in their entirety. No other sequence changes were found.
Protein sequence and structural analyses
Protein sequence comparisons were performed using the BLAST algorithm (http://www.ncbi.nlm.nih.gov/BLAST). TFIIF crystallographic structural information was acquired through the Protein Data Bank (PDB) (http://www.rcsb.org/pdb/). Amino acid replacements at position G112 of RAP74 were visualized using PyMol.
RESULTS
The ssu71 suppressors encode Tfg1 G363 amino acid replacements
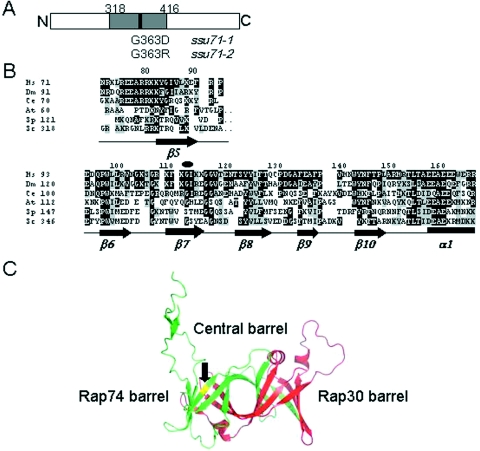
Two independent ssu71 mutants were isolated as suppressors of the Csm− phenotype associated with sua7-1 (25). The ssu71-1 and ssu71-2 alleles were retrieved from strains YZS14 and YZS45 by gap repair. DNA sequence analysis of the entire ssu71 ORF revealed that both alleles are the result of single base pair substitutions encoding different amino acid replacements at the same position in the Tfg1 subunit of TFIIF: ssu71-1 encodes glycine-363 to aspartic acid (G363D), whereas ssu71-2 encodes glycine-363 to arginine (G363R) (Figure 1A). These two different replacements of the same residue underscore the importance of position G363 and point to this region of Tfg1 as playing an important role in the functional interaction between TFIIB, TFIIF and RNAP II during transcription start site selection.
Figure 1.
Depiction of the ssu71 suppressor mutations. (A) The ssu71-1 and ssu71-2 alleles encode G363D and G363R replacements, respectively. The gray box denotes the conserved sequence (residues 318–416) shown in B. (B) Sequence alignment of the conserved region of the Tfg1/RAP74 subunit of TFIIF. Hs, human; Dm, Drososphila melanogaster; Ce, Caenorhabditis elegans; At, Arabidopsis thaliana; Sp, S.pombe; and Sc, S.cerevisiae. Secondary structures from the human RAP74 protein are indicated below the sequences. The conserved G363 residue of Tfg1 lies within the β7-sheet and is denoted by the black oval. Black shading indicates identity with the human sequence; gray shading denotes similarity. This sequence was reproduced from Figure 1 of Funk et al. (33), with permission. (C) RAP74 G112 (Tfg1 G363) lies within the triple barrel structure of the RAP74–RAP30 interaction domain. RAP30 (residues 2–219) is depicted in red; RAP74 (residues 2–172) in green; residue G112 is highlighted in yellow and marked by the black arrow. This structure was created using PyMol based on the X-ray structure of human RAP74/RAP30 (34).
Tfg1 G363 lies at the Tfg1/Tfg2 interface
The Tfg1 subunit of yeast TFIIF shows limited amino acid sequence similarity to the RAP74 subunit of human TFIIF (24,25). However, alignments based on hydrophobic cluster analysis suggested that Tfg1 residues 312–441 are structurally related to region I near the N-terminus of RAP74 (residues 66–187) (31). This alignment is also supported by domain searches using the PFam database (32). PFam analysis identified a single domain common to S.cerevisiae Tfg1 and its counterpart in Schizosaccharomyces pombe protein T41039; no other common domains were identified (33). These two regions correspond to residues 318–416 of Tfg1 and residues 121–206 of T41039 (Figure 1B). Accordingly, Tfg1 G363 lies within the most phylogenetically conserved region Tfg1 and corresponds to G112 of human RAP74.
An X-ray structure for the RAP74(2-172)/RAP30(2-119) dimerization domain has been solved to 1.7 Å resolution for human TFIIF (34). This complex forms a novel ‘triple barrel’ that has been proposed to form loops and extensions important for interaction with other components of the PIC. The RAP74/RAP30 dimer interface comprises β-sheets β1, β2, β3, β6, β7 and β8 of RAP74 and the corresponding β-sheets of RAP30 (Figure 1C) (34). G112 of human RAP74 lies within the β7-sheet at the RAP74/RAP30 interface (Figure 1B and C). Thus, sequence alignments and comparison with the human RAP74/RAP30 crystal structure place G363 at the interface between Tfg1 and Tfg2. These results suggest that the Tfg1/Tfg2 dimerization region functionally interacts with TFIIB to affect transcription start site selection.
Tfg1 G363D suppresses the Rpb1 N445S replacement in the RNAP II active center
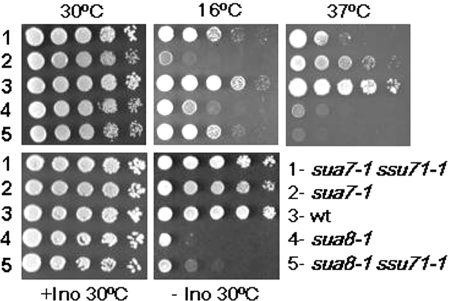
Based on the similar growth phenotypes and start site defects associated with sua7 and sua8 mutants, we asked whether ssu71-1 would also suppress sua8-1. The sua8-1 allele encodes an Rpb1 N445S replacement within the active center of RNAP II (23). An isogenic sua8-1 TFG1/SSU71+ and sua8-1 ssu71-1 strain pair was constructed as described in the Materials and Methods. The ssu71-1 allele clearly suppressed sua8-1, resulting in comparable growth of the sua7-1 ssu71-1 and sua8-1 ssu71-1 strains at 16°C (Figure 2, 16°C panel). In contrast to sua7 mutants, sua8 mutants are Ino− and this phenotype was also partially suppressed by ssu71-1 (rows 3–5, +Ino and −Ino panels). Although ssu71-1 acts as a suppressor of the Csm− phenotype of sua7-1, it confers a synthetic Tsm− phenotype in combination with sua7-1 at 37°C) (rows 1–3, 37°C panel). The sua8-1 ssu71-1 double mutant also exhibited a synthetic Tsm− phenotype, although this effect is more difficult to discern owing to the inherent Tsm− phenotype of the sua8-1 primary mutant (rows 4 and 5, 37°C panel). Thus, Tfg1 G363D suppresses the growth phenotypes associated with defects in both TFIIB and Rpb1. Moreover, the location of TFIIB E62 and Rpb1 N445 within the crystal structures of RNAP II and the RNAP II–TFIIB complex imply that the Tfg1/Tfg2 interface of TFIIF is positioned within the active center of the RNAP II PIC.
Figure 2.
Phenotypes associated with the ssu71-1 suppressor of sua7-1 and sua8-1. The top three panels depict cell growth on YPD medium at the indicated temperatures. The bottom panels depict growth on SC medium either in the presence (+Ino) or absence (−Ino) of inositol at 30°C. The 10-fold serial dilutions of each strain were spotted onto culture plates and photographed after 2 (30°C), 3 (37°C) or 5 (16°C) days of incubation. Strains: 1, YZS14 (sua7-1 ssu71-1); 2, YDW546 (sua7-1); 3, T16 (wt); 4, YDW383 (sua8-1); and 5, YZS96 (sua8-1 ssu71-1).
Suppression is specific for Tfg1 G363 charged residue replacements
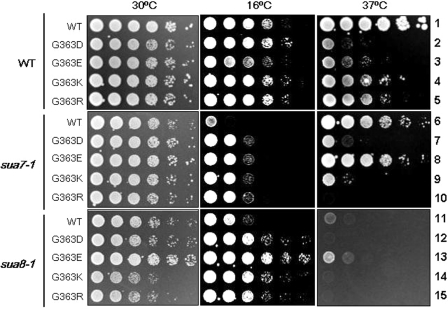
In an effort to define the Tfg1 structural requirements at position 363 for suppression of TFIIB and RNAP II defects, we constructed a TFG1 plasmid-shuffle strain in isogenic SUA7, sua7-1 (E62K) and sua8-1 (N445S) genetic backgrounds and scored site-directed G363 amino acid replacements for suppression of the sua7-1 and sua8-1 growth phenotypes. Results are presented in Figure 3. Consistent with the phenotypes of the original ssu71 suppressor strains, G363D and G363R suppressed the sua7-1 Csm− phenotype (cf. row 6 with rows 7 and 10 at 16°C), confirming that suppression is due solely to these replacements. Furthermore, two other charged replacements, G363E and G363K, also suppressed the sua7-1 Csm− phenotype (rows 8 and 9). The interactions with TFIIB were not identical for all G363 replacements, however, as G363D, G363K and G363R caused synthetic Tsm− growth defects in combination with sua7-1 (rows 7, 9 and 10 at 37°C) whereas G363E exhibited no synthetic growth defect (row 8). These suppressor (Csm−) and synthetic (Tsm−) growth phenotypes are consistent with those of the original YZS14 and YZS45 strains, which were initially selected based on screening Csm+ revertants for a Tsm− phenotype (25). In contrast to the four charged residue replacements of G363, neither G363A nor G363P suppressed the sua7-1 Csm− phenotype (data not shown).
Figure 3.
Growth phenotypes associated with Tfg1 G363 amino acid replacements in the WT, sua7-1 and sua8-1 backgrounds. All strains (Table 1) are isogenic, differing only by the indicated sua7-1 or sua8-1 alleles and Tfg1 G363 amino acid replacements, D, E, K or R.
Although the sua8-1 mutant exhibits a less severe Csm− phenotype than the sua7-1 mutant (Figure 3, cf. rows 1, 6 and 11 at 16°C), this phenotype is nonetheless discernable and is clearly suppressed by all four G363 replacements (cf. row 11 with rows 12–15). Comparable with their effects on sua7-1, the G363D, G363K and G363R replacements also enhanced the sua8-1 Tsm− defect, resulting in no growth at 37°C. In contrast, but identical to its effect on sua7-1, G363E has no effect on sua8-1 at 37°C (cf. rows 11 and 13). These results underscore the functional importance of Tfg1 residue G363 and suggest that suppression by charged residue replacements might be a consequence of forming alternative salt bridges within the RNAP II active center (Discussion).
Effects of Tfg1 G363 amino acid replacements on transcription start site selection
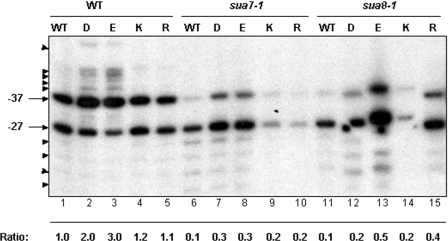
The sua7-1 and sua8-1 alleles cause the selection of transcription start sites to shift downstream of normal in a promoter-specific manner (16,23). We therefore asked whether the Tfg1 G363 amino acid replacements also compensate for this defect. Transcription start sites at the ADH1 gene were mapped by primer extension. Results are shown in Figure 4. The sua7-1 and sua8-1 alleles shifted initiation downstream, as seen by diminished initiation at −37 and enhanced initiation at sites downstream of −27 (cf. lane 1 with lanes 6 and 11). The acidic G363D and G363E replacements suppressed the sua7-1 defect, partially restoring the normal ADH1 start site pattern (lanes 7 and 8). The most notable effects of the basic G363K and G363R replacements were to diminish the overall levels of ADH1 transcripts, although these replacements also weakly compensated for the downstream start site shift associated with sua7-1 (cf. lanes 9 and 10 with lane 6).
Figure 4.
Primer extension analysis of ADH1 transcription start sites. All strains (Table 1) are identical to those defined in Figure 3. In the wild-type strain (lane 1), transcription initiates at two sites, −37 and −27 (A of ATG is denoted +1), indicated by the arrows. Start sites that are enhanced in the mutant and suppressor strains upstream of −37 and downstream of −27 are highlighted by arrow heads. The ratio indicates the amount of transcript at −37 relative to −27. Quantification was performed using software from the Alpha Imager (Alpha Innotech). All values were normalized to the −37:−27 ratio for the WT strain, which was defined as 1.0.
Somewhat different effects were observed in the sua8-1 background. In this case, the G363D and G363K replacements weakly compensated for the downstream shift and the principal effect of the G363K replacement was again to diminish overall ADH1 transcript levels (cf. lanes 12 and 14 with lane 11). Conversely, the effects of G363E and G363R on compensation of the downstream start site shift associated with sua8-1 were more pronounced (cf. lanes 13 and 15 with lane 11). Furthermore, G363E consistently enhanced overall ADH1 transcript levels.
In the otherwise wild-type background, the two negatively charged replacements, G363D and G363E, enhanced initiation upstream of normal (cf. lane 1 and lanes 2 and 3), whereas neither of the positively charged replacements had any effect (lanes 4 and 5). Thus, acidic and basic residue replacements of Tfg1 G363 exert differential effects on both the accuracy and level of ADH1 transcription in a manner dependent upon TFIIB and Rpb1. These results support our conclusion that the ‘triple barrel’ structure at the Tfg1/Tfg2 interface lies within the active center of RNAP II where it plays a critical role in the mechanism of transcription initiation.
DISCUSSION
In this study we have defined charged residue replacements of G363 in the largest subunit of TFIIF that are responsible for suppression of the Csm− growth defects and altered initiation pattern associated with TFIIB and Rpb1 defects. These results define a functional interaction among the TFIIB–Tfg1–Rpb1 components of the PIC and implicate the altered regions of these three polypeptides in the mechanism of start site selection.
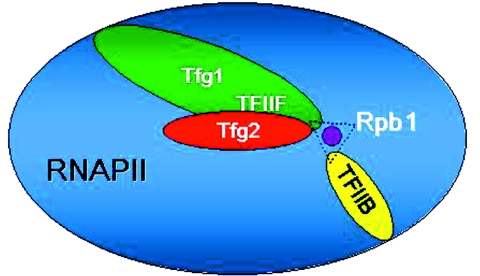
The topological organization of the yeast RNAP II–TFIIF complex has been defined by cryo-electron microscopy (35). The TFIIF electron density was distributed across the surface of RNAP II and other regions of the complex. Tfg1 was deduced to interact with the Rpb4–Rpb7 subcomplex and with the mobile ‘clamp’ that swings over the active center. These images, however, are not of sufficient resolution to define specific contact points of Tfg1 or Tfg2 with the RNAP II–TFIIF structure. Our identification of Tfg1 G363 replacements as suppressors of start site defects associated with amino acid replacements of TFIIB and Rpb1 residues, both of which lie within the active center of the TFIIB–RNAP II complex, combined with comparative sequence information that places G363 at the Tfg1–Tfg2 dimer interface, make a strong case for the TFIIF dimerization region being located within the active center of RNAP II during initiation (Figure 5). Furthermore, these results, and those from the Ponticelli laboratory (26), implicate the triple barrel region of TFIIF as a determinant of start site selection, presumably by interacting with the B-finger–Rpb1 interface.
Figure 5.
Model depicting interactions among TFIIB, TFIIF and Rpb1 at the active center of RNAP II. The sua7-1 (TFIIB E62K) and sua8-1 (Rpb1 N445S) alleles confer similar growth defects and altered patterns of transcription start site selection at ADH1. The ssu71-1 and ssu71-1 alleles of TFG1 encode charged residue replacements of Tfg1 G363, which is predicted to lie at the Tfg1–Tfg2 dimer interface of TFIIF (Figure 1). Based on suppression by Tfg1 G363 replacements of the cell growth and start site defects associated with the TFIIB E62K and Rpb1 N445S replacements (Figures 3 and 4), we place the TFIIF dimer interface at the active center (purple dot) of RNAP II. The dotted black triangle denotes genetic, and presumably physical (Discussion), interactions among Tfg1, TFIIB and Rpb1. This model is consistent with earlier protein–DNA crosslinking data showing Tfg1 and Tfg2 contact points at the transcription start site (36).
High-resolution protein–DNA contacts have been determined for the yeast RNAP II transcription initiation machinery (36). The results from that study revealed that Tfg1 and Tfg2 crosslinked to promoter DNA at the transcription start site. Moreover, the TFIIB R78C replacement, which shifts start site selection in a manner comparable with TFIIB E62K, has no effect on TFIIB–DNA interactions, but instead enhances the crosslinking of Tfg1 and Tfg2 at the start site. These results offer strong biochemical support for our model placing the Tfg1–Tfg2 dimer interface within the active center of RNAP II where it functionally interacts with the B-finger to affect start site selection.
How do Tfg1 G363 replacements by positively or negatively charged residues suppress TFIIB E62K? The E62 residue lies within the B-finger opposite to R78 and appears to form an E62-R78 salt bridge important for maintaining the structure of the B-finger (16,20). E62K and R78C replacements confer similar growth phenotypes and start site defects, implying that the structural integrity of the B-finger is critical for accurate initiation. We suggest that Tfg1 G363 is positioned within the active center of the PIC where charged replacements compensate for disruption of the B-finger E62-R78 salt bridge, perhaps forming a compensatory salt bridge. For example, Tfg1 acidic replacements G363D or G363E might replace TFIIB E62, forming a salt bridge with TFIIB R78. Conversely, basic residue replacements G363K and G363R might form a salt bridge with a different, negatively charged TFIIB residue such as conserved residue D58. In either case, we propose that alternative salt bridges involving Tfg1 G363 replacements partially restore the structural integrity of the B-finger that is critical for accurate start site selection.
How then do Tfg1 G363 replacements compensate for Rpb1 N445S? The X-ray structure of an RNAP II elongation complex revealed that Rpb1 N445 directly contacts the DNA template strand at positions −1/−2, immediately upstream of the active center (11). It is interesting that this is the precise location where Tfg1 and Tfg2 crosslink to promoter DNA in a DNA–TBP–TFIIB–TFIIF–RNAP II complex (36). Moreover, these crosslink sites are altered by the TFIIB R78C replacement. Although this information does not define how Tfg1 G363 charges residue replacements suppress Rpb1 N445S it supports our conclusion that the TFIIF dimerization domain lies within the active center of the RNAP II PIC.
We wish to note that defects distal to the RNAP II active center can also affect start site selection. An altered form of the Rpb9 subunit of RNAP II was identified in the same genetic screen that identified the Tfg1 G363 suppressors (37). Furthermore, deletion of the RPB9 gene shifted transcription start site selection upstream of normal at the ADH1 gene in a manner comparable with the Tfg1 G363 replacements defined here, or with the Tfg1 E364A and W350A replacements defined previously (26,37–39). An Rpb2 replacement (G369S) located near the Rpb9 subunit conferred a similar upstream start site shift and suppressed the TFIIB R78C defect (18). These results define a functional relationship between Rpb9 and its neighboring Rpb2 region with the active center Rpb1–TFIIB–TFIIF interface. Yet in the crystal structure of yeast RNAP II the Rpb2–Rpb9 interface forms part of the upper ‘jaw-lobe’ region of RNAP II, which is distal to the active center (10). This apparent discrepancy can be explained, however, by impaired TFIIF–RNAP II association conferred by deletion of Rpb9 (26,39). Accordingly, Rpb9 is likely to play an indirect role in start site selection by altering interaction of the triple barrel within the active center of RNAP II, effectively mimicking the structural changes associated with the Tfg1 W350, G363 and E364 amino acid replacements.
Acknowledgments
We thank Mariela Reyes-Reyes for preparing PyMol images. This work was supported by fellowships from the University of La Coruña and from the Xunta de Galicia to M.A.F.P and by NIH grant GM39484 to M.H. Funding to pay the Open Access publication charges for this article was provided by NIH GM39484.
Conflict of interest statement. None declared.
REFERENCES
- 1.Orphanides G., LaGrange T., Reinberg D. The general initiation factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 2.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. Nature Struct. Mol. Biol. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim T.K., Ebright R.H., Reinberg D. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science. 2000;288:1418–1422. doi: 10.1126/science.288.5470.1418. [DOI] [PubMed] [Google Scholar]
- 5.Ranish J.A., Yudkovsky N., Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woychik N.A. Fractions to functions: RNA polymerase II thirty years later. Cold Spring Harb. Symp. Quant. Biol. 1998;63:311–317. doi: 10.1101/sqb.1998.63.311. [DOI] [PubMed] [Google Scholar]
- 7.Woychik N.A., Hampsey M. The RNA polymerase II machinery: structure illuminates function. Cell. 2002;108:453–463. doi: 10.1016/s0092-8674(02)00646-3. [DOI] [PubMed] [Google Scholar]
- 8.Armache K.J., Kettenberger H., Cramer P. Architecture of initiation-competent 12-subunit RNA polymerase II. Proc. Natl Acad. Sci. USA. 2003;100:6964–6968. doi: 10.1073/pnas.1030608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bushnell D.A., Kornberg R.D. Complete, 12-subunit RNA polymerase II at 4.1-A resolution: implications for the initiation of transcription. Proc. Natl Acad. Sci. USA. 2003;100:6969–6973. doi: 10.1073/pnas.1130601100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cramer P., Bushnell D.A., Kornberg R.D. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 11.Gnatt A.L., Cramer P., Fu J., Bushnell D.A., Kornberg R.D. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 12.Westover K.D., Bushnell D.A., Kornberg R.D. Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science. 2004;303:1014–1016. doi: 10.1126/science.1090839. [DOI] [PubMed] [Google Scholar]
- 13.Kettenberger H., Armache K.J., Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 14.Murakami K.S., Darst S.A. Bacterial RNA polymerases: the wholo story. Curr. Opin. Struct. Biol. 2003;13:31–39. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]
- 15.Pinto I., Ware D.E., Hampsey M. The yeast SUA7 gene encodes a homolog of human transcription factor TFIIB and is required for normal start site selection in vivo. Cell. 1992;68:977–988. doi: 10.1016/0092-8674(92)90040-j. [DOI] [PubMed] [Google Scholar]
- 16.Pinto I., Wu W.-H., Na J.G., Hampsey M. Characterization of sua7 mutations defines a domain of TFIIB involved in transcription start site selection in yeast. J. Biol. Chem. 1994;269:30569–30573. [PubMed] [Google Scholar]
- 17.Cho E.J., Buratowski S. Evidence that transcription factor IIB is required for a post-assembly step in transcription initiation. J. Biol. Chem. 1999;274:25807–25813. doi: 10.1074/jbc.274.36.25807. [DOI] [PubMed] [Google Scholar]
- 18.Chen B.-S., Hampsey M. Functional interaction between TFIIB and the Rpb2 subunit of RNA polymerase II: implications for the mechanism of start site selection. Mol. Cell. Biol. 2004;24:3983–3991. doi: 10.1128/MCB.24.9.3983-3991.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu W.L., Zeng Q.D., Colangelo C.M., Lewis L.M., Summers M.F., Scott R.A. The N-terminal domain of TFIIB from Pyrococcus furiosus forms a zinc ribbon. Nature Struct. Biol. 1996;3:122–124. doi: 10.1038/nsb0296-122. [DOI] [PubMed] [Google Scholar]
- 20.Bushnell D.A., Westover K.D., Davis R.E., Kornberg R.D. Structural basis of transcription: an RNA polymerase II–TFIIB cocrystal at 4.5 angstroms. Science. 2004;303:983–988. doi: 10.1126/science.1090838. [DOI] [PubMed] [Google Scholar]
- 21.Fairley J.A., Evans R., Hawkes N.A., Roberts S.G. Core promoter-dependent TFIIB conformation and a role for TFIIB conformation in transcription start site selection. Mol. Cell. Biol. 2002;22:6697–6705. doi: 10.1128/MCB.22.19.6697-6705.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bell S.D., Jackson S.P. The role of transcription factor B in transcription initiation and promoter clearance in the archaeon Sulfolobus acidocaldarius. J. Biol. Chem. 2000;275:12934–12940. doi: 10.1074/jbc.275.17.12934. [DOI] [PubMed] [Google Scholar]
- 23.Berroteran R.W., Ware D.E., Hampsey M. The sua8 suppressors of Saccharomyces cerevisiae encode replacements of conserved residues within the largest subunit of RNA polymerase II and affect transcription start site selection similarly to sua7 (TFIIB) mutations. Mol. Cell. Biol. 1994;14:226–237. doi: 10.1128/mcb.14.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henry N.L., Campbell A.M., Feaver W.J., Poon D., Weil P.A., Kornberg R.D. TFIIF–TAF–RNA polymerase II connection. Genes Dev. 1994;8:2868–2878. doi: 10.1101/gad.8.23.2868. [DOI] [PubMed] [Google Scholar]
- 25.Sun Z.W., Hampsey M. Identification of the gene (SSU71/TFG1) encoding the largest subunit of transcription factor TFIIF as a suppressor of a TFIIB mutation in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1995;92:3127–3131. doi: 10.1073/pnas.92.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghazy M.A., Brodie S.A., Ammerman M.L., Ziegler L.M., Ponticelli A.S. Amino acid substitutions in yeast TFIIF confer upstream shifts in transcription initiation and altered interaction with RNA polymerase II. Mol. Cell. Biol. 2004;24:10975–10985. doi: 10.1128/MCB.24.24.10975-10985.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldstein A.L., McCusker J.H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 28.Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 29.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 30.Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 31.Wang B.Q., Burton Z.F. Functional domains of human RAP74 including a masked polymerase binding domain. J. Biol. Chem. 1995;270:27035–27044. doi: 10.1074/jbc.270.45.27035. [DOI] [PubMed] [Google Scholar]
- 32.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Funk J.D., Nedialkov Y.A., Xu D., Burton Z.F. A key role for the alpha 1 helix of human RAP74 in the initiation and elongation of RNA chains. J. Biol. Chem. 2002;277:46998–47003. doi: 10.1074/jbc.M206249200. [DOI] [PubMed] [Google Scholar]
- 34.Gaiser F., Tan S., Richmond T.J. Novel dimerization fold of RAP30/RAP74 in human TFIIF at 1.7 A resolution. J. Mol. Biol. 2000;302:1119–1127. doi: 10.1006/jmbi.2000.4110. [DOI] [PubMed] [Google Scholar]
- 35.Chung W.H., Craighead J.L., Chang W.H., Ezeokonkwo C., Bareket-Samish A., Kornberg R.D., Asturias F.J. RNA Polymerase II/TFIIF structure and conserved organization of the initiation complex. Mol. Cell. 2003;12:1003–1013. doi: 10.1016/s1097-2765(03)00387-3. [DOI] [PubMed] [Google Scholar]
- 36.Chen B.S., Mandal S.S., Hampsey M. High-resolution protein–DNA contacts for the yeast RNA polymerase II general transcription machinery. Biochemistry. 2004;43:12741–12749. doi: 10.1021/bi048993r. [DOI] [PubMed] [Google Scholar]
- 37.Sun Z.W., Tessmer A., Hampsey M. Functional interaction between TFIIB and the Rpb9 (Ssu73) subunit of RNA polymerase II in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:2560–2566. doi: 10.1093/nar/24.13.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hull M.W., Mckune K., Woychik N.A. RNA polymerase II subunit RPB9 is required for accurate start site selection. Genes Dev. 1995;9:481–490. doi: 10.1101/gad.9.4.481. [DOI] [PubMed] [Google Scholar]
- 39.Ziegler L.M., Khaperskyy D.A., Ammerman M.L., Ponticelli A.S. Yeast RNA polymerase II lacking the Rpb9 subunit is impaired for interaction with transcription factor IIF. J. Biol. Chem. 2003;278:48950–48956. doi: 10.1074/jbc.M309656200. [DOI] [PubMed] [Google Scholar]