Abstract
Human vaults are the largest cytoplasmic ribonucleoprotein and are overexpressed in cancer cells. Vaults reportedly function in the extrusion of xenobiotics from the nuclei of resistant cells, but the interactions of xenobiotics with the vault-associated proteins or non-coding RNAs have never been directly observed. In the present study, we show that vault RNAs (vRNAs), specifically the hvg-1 and hvg-2 RNAs, bind to a chemotherapeutic compound, mitoxantrone. Using an in-line probing assay (spontaneous transesterification of RNA linkages), we have identified the mitoxantrone binding region within the vRNAs. In addition, we analyzed the interactions between vRNAs and mitoxantrone in the cellular milieu, using an in vitro translation inhibition assay. Taken together, our results clearly suggest that vRNAs have the ability to bind certain chemotherapeutic compounds and these interactions may play an important role in vault function, by participating in the export of toxic compounds.
INTRODUCTION
Vaults are barrel-shaped particles with a mass of 13 MDa and represent the largest ribonucleoprotein complex of eukaryotic cells identified thus far. They exist in diverse eukaryotic species, including mammals, avians and amphibians, and their considerable abundance and striking evolutionary conservation argue for an important cellular function (1). Vault particles appear to reside in both the cytoplasm and the nucleus (2,3) and have been implicated in intracellular (4) and nucleocytoplasmic transport (2,3). Several lines of evidence suggest that vaults may play an important role in intracellular detoxification processes, and thus function in the multidrug resistance (MDR) of cancer cells (5,6).
The human vault constitutes of three proteins [major vault protein (MVP), telomerase-associated protein 1 (TEP1) and vault poly(ADP–ribose)polymerase (VPARP)] and three non-coding RNAs (hvg-1, hvg-2 and hvg-3). These RNAs represent <5% of the mass of the vault complex and share ∼84% sequence identity, but their lengths differ. Human hvg-1 RNA is 98 bases in length, and the other two RNAs are 88 bases long. In humans, the genes for these RNAs are located on chromosome 5.
A previous study on several non-P-glycoprotein MDR cell lines with increased MVP levels demonstrated that the vault RNAs (vRNAs) and MVP are coordinately regulated, and suggested that the entire vault particle is up-regulated in MDR and that a threshold level of vaults is required to impart MDR (7). In a clinical scenario, the elevated expression of MVP was observed in cell lines resistant to various classes of chemotherapeutic compounds, including doxorubicin and mitoxantrone (8). Support for the role of vaults in the extrusion of anthracyclines from the nuclei of resistance cells was reported by Ohno et al. (9). However, to date, no vault components that participate or interact directly with the chemotherapeutic compound(s) have been identified, and thus the function of vaults has remained enigmatic.
RNA molecules are involved in many important metabolic processes, including peptide bond formation and replication (10,11). In recent years, non-coding RNAs have been shown to participate in complex communication pathways in gene regulation and cell development (12,13). The vRNAs isolated previously from different species were predicted to fold in a similar fashion, despite their differences in length, indicating that the association of vRNAs with vaults is not fortuitous (14). The vRNA ends (5′ and 3′) form stem-like structures, but the internal sequences of these RNAs appear to fold differently. These predictions suggest that vRNAs have a fundamental role in the function of the vault particle, possibly through RNA–ligand, RNA–RNA or RNA–protein interactions. Here, we provide the first evidence that vault-associated RNAs interact directly with chemotherapeutic compounds, suggesting that vRNAs play important roles in vault function in the cells.
MATERIALS AND METHODS
RNA synthesis
The 98mer hvg-1 RNA and the 88mer of hvg-2 and hvg-3 were enzymatically synthesized by in vitro transcription, using T7 RNA polymerase on a synthetic DNA template. The templates 5′-CTTTAGCTCAGCGGTTACTTCGACAGTTCTTTAATTGAAACAAGCAACCTGTCTGGGTTGTTCGAGACCCGCGGGC-3′ (hvg-1), 5′-CTTTAGCTCAGCGGTTACTTCGAGTACATTGTAACCACCTCTCTGGGTGGTTCGAGACCCGCGGGTGCTTTCCAGCTCTTTT-3′ (hvg-2) and 5′-CTTTAGCTCAGCGGTTACTTCGCGTGTCATCAAACCACCTCTCTGGGTTGTTCGAGACCCGCGGGCGCTCTCCAGCCCTCTT-3′ (hvg-3) were synthesized. To generate the double-stranded DNA and to add the T7 promoter to the template, we synthesized one common forward primer and three individual reverse primers [forward primer, 5′-AGTAATACGACTCACTATAGGGCTGGCTTTAGCTCAGCGGTTACTTC G-3′; and reverse primers, 5′-AAAAGGACTGGAGAGCGCCCGCGGGTCTCGAACAACCCA-3′ (hvg-1), 5′-AAAAGAGCTGGAAAGCACCCGCGGGT-3′ (hvg-2) and 5′-AAGAGGGCTGGAGAGCGCCCGC-3′ (hvg-3)]. Using the template DNA and the primers, we prepared the double-stranded DNA template, which now contained a T7 promoter (in italics), by using a commercial PCR kit (Ex Taq kit, Takara, Japan). The reaction mixture was cycled at 94°C for 1 min 10 sec, 55°C for 50 s, and 68°C for 1 min 10 sec for 10 cycles. The PCR product was precipitated and used for RNA preparation by in vitro T7 transcription. Transcription was performed at 37°C for 3 h, by using AmpliScribe transcription kit. Later, the products were treated with 2 U of DNase I (RNase free) for 10 min at 37°C, to remove the template DNA, and were mixed with an equal volume of 2× urea buffer (7 M urea, 50 mM EDTA, 90 mM Tris-borate containing 0.05% bromophenol blue). The reaction mixtures were denatured at 90°C for 2 min and fractionated on a 10% polyacrylamide gel containing 7 M urea. The RNA band was excised, and the RNA was eluted from the gel. The RNAs were concentrated by vacuum filtration, redissolved in water and then quantitated by the absorbance at 260 nm.
Circular Dichroism (CD) analysis
Circular dichroism titrations were carried out at 25°C on a J-720 spectropolarimeter (JASCO). The starting volume in the 1 cm path length cell was 3 ml, with 1 µM of RNA in a buffer containing 20 mM MgCl2, 50 mM Tris–HCl (pH 8.3 at 25°C) and 100 mM KCl. Control experiments included spectra obtained in the presence of only drug or buffer. Spectra were collected immediately after the addition of a stock solution of mitoxantrone or doxorubicin to a final concentration of 4 or 16 µM. Each concentration was titrated five times.
In-line probing of RNA
RNAs were synthesized enzymatically from the corresponding PCR DNA templates by in vitro transcription using T7 polymerase and were labeled at the 3′ end (15). The labeled RNA was incubated for 40 h at 25°C, in a buffer containing 20 mM MgCl2, 50 mM Tris–HCl (pH 8.3 at 25°C) and 100 mM KCl, in the absence or in the presence of drug concentrations ranging from 2 to 8 µM. After each incubation, the spontaneously cleaved products were resolved by 12% denaturing (8 M urea) PAGE followed by autoradiography.
Post-transcriptional inhibition assay
We used the rapid translation system (RTS) of the pIVEX GST fusion vector (Roche), which constitutively expresses the reporter gene from a T7 promoter, for complete in vitro translation analyses. In order to reduce the length of the intervening sequences between the T7 promoter and the start codon of glutathione S-transferase (GST) protein to <100 nt, upon inserting the hvg-RNAs, we deleted some regions between the T7 promoter, the ribosome binding site and the start codon from the vector, using the NaeI and BsmI restriction sites. These regions were regenerated by the insertion of the truncated regions containing 20–77 nt of hvg-1 and 20–67 nt of hvg-2 and hvg-3. Using the corresponding forward and reverse primers [forward primers, 5′-GGTGATGCCGGCAAATTAATACGACTCACTATAGGGTTACTTCGACAGTTCTTT-3′ (hvg-1), 5′-GGTGATGCCGGCAAATTAATACGACTCACTATAGGGTTACTTCGAGTACATTGT-3′ (hvg-2), 5′-GGTGATGCCGGCAAATTAATACGACTCACTATAGGGTTACTTCGCGTGTCATCA-3′ (hvg-3) and reverse primers, 5′-GGGGAATGCTACATGGTATATctccttCTTAAAGTTAAACAAGGGTCTCGAACAACCCAG-3′ (hvg-1), 5′-GGGGAATGCTACATGGTATATctccttCTTAAAGTTAAACAAGGGTGTCGAACCAGCCAG-3′ (hvg-2) and 5′-GGGGAATGCTACATGGTATATctccttCTTAAAGTTAAACAAGGGTCTCGTTCTTCCCAG-3′ (hvg-3)], we amplified the appropriate vRNA sequences from the vRNA templates used in the above studies. The resulting fragments were inserted into the pIVEX-GST expression vector using the NaeI and BsmI restriction sites (underlined). The fragments included the T7 promoter (in italics), the ribosome binding site (in lowercase letters) and the initiation codon (in bold). Using the similar sequences, hvg-1, with mutations at nt 53, 54, 58 and 59, was also engineered into the same vector system. The modified pIVEX-GST fusion vectors inserted with the vRNAs were translated in vitro in the presence of mitroxantrone, using the rapid translation system RTS 100 Escherichia coli HY (Roche), according to the manufacturer's instructions. The [35S]methionine-labeled proteins were generated by a coupled transcription/translation system and were used as probes to detect vRNA interactions with mitoxantrone. The gene products were identified by autoradiography following SDS–PAGE (Daiichi Pure Chemicals Co., Ltd) and were quantitated (radioactivity) using a Bio image analyzer, BAS 2500 (Fuji Film), after drying the gels.
RESULTS AND DISCUSSION
MDR, a major cause of cancer treatment failure, is a phenomenon whereby cancer cells develop broad resistance to a wide variety of chemotherapeutic drugs (16). Cells utilize several mechanisms to prevent cytotoxic drugs from reaching their target sites, including protein modification of the drugs, up-regulation of plasma membrane pumps and reduction of the nuclear-cytoplasmic ratio by increasing drug export from the nucleus. Multidrug resistant cell lines have been shown to utilize these routes to gain resistance to various drugs. The unexpected role of vaults in MDR is supported by the following evidence: (i) vaults and their components are overexpressed in cells treated with cytotoxic compounds; (ii) vaults are involved in nucleo-cytoplasmic transport; (iii) compounds that interact with the vaults reside within the vaults. Despite these findings, neither vaults nor their components were directly shown to bind the drugs. In order to test this possibility, we have analyzed the ability of various chemotherapeutic compounds to interact with vRNAs, as described below.
Analysis of vRNA binding to the mitoxantrone by CD
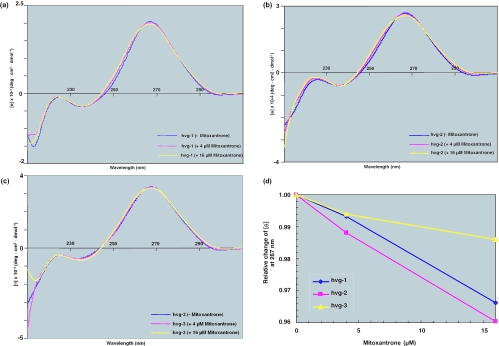
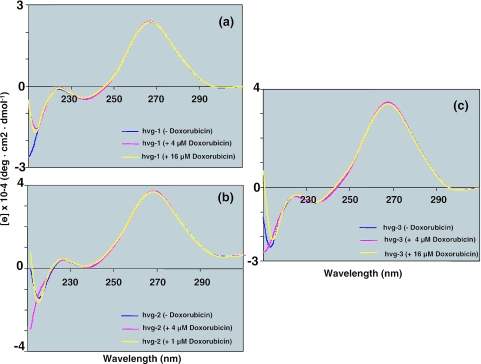
In the present study, we tested two chemotherapeutic compounds, mitoxantrone and doxorubicin, for their ability to interact with the non-coding RNAs associated with the vault complex. As a preliminary assessment, we conducted CD analyses and studied the binding region by an in-line probing assay. The CD spectra were collected immediately after the addition of mitoxantrone, to final concentrations of 4 and 16 µM, to a buffered solution of the RNA. The final spectrum was obtained after subtracting the spectra of the buffer blank and the drug. When mitoxantrone was added, a decrease in the intensity at ∼265 nm and an increase in the intensity at ∼255 nm were observed in the hvg-1 (Figure 1a) and hvg-2 spectra (Figure 1b), but not in the hvg-3 spectra (Figure 1c). In order to see the distinct spectral changes, we have also plotted the mitoxantrone dose-dependent response at 267 nm (Figure 1d). This indicates that hvg-1 and hvg-2, but not hvg-3, interact with mitoxantrone, which presumably results in structural changes at the drug binding site. On the other hand, when doxorubicin was added, no spectral change was observed for either hvg-1, hvg-2 or hvg-3 (Figure 2a–c). These studies suggest that hvg-1 and hvg-2 have the ability to bind specifically to mitoxantrone, whereas hvg-3 has either weak or no binding ability (Figure 1d), in spite of the high similarity between these RNAs. Since the secondary structures of the vRNA differ in the regions other than the 5′ and 3′ ends, these unique regions probably account for the differences in the mitoxantrone binding to the vRNAs. On the other hand, doxorubicin does not interact with any of the hvg-RNAs.
Figure 1.
(a) CD spectra of hvg-1 with different concentrations of mitoxantrone (drug concentrations in parentheses). Circular dichroism titrations were carried out at 25°C on a J-720 spectropolarimeter (JASCO). The starting volume in the 1 cm path length cell was 3 ml, with 1 µM of RNA in a buffer containing 20 mM MgCl2, 50 mM Tris–HCl (pH 8.3 at 25°C) and 100 mM KCl. RNAs were synthesized from the corresponding PCR DNA templates by in vitro transcription using T7 RNA polymerase. (b) CD spectra of hvg-2 with different concentrations of mitoxantrone. (c) CD spectra of hvg-3 with different concentrations of mitoxantrone. (d) Relative CD spectral changes of vRNAs at 267 nm with different concentrations of mitoxantrone.
Figure 2.
CD spectra of (a) hvg-1, (b) hvg-2 and (c) hvg-3 with different concentrations of doxorubicin.
Identification of a mitoxantrone binding region in vRNAs by an in-line probing assay
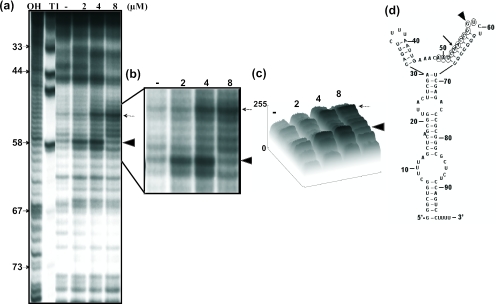
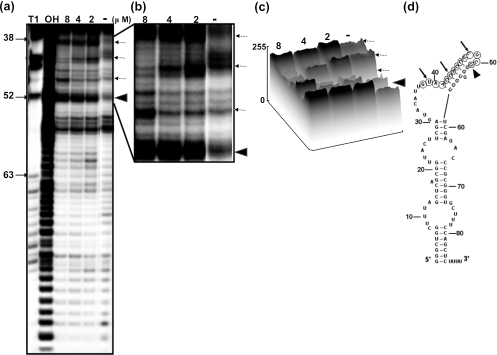
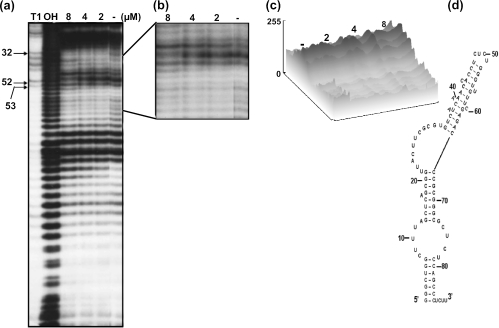
Next, we wished to determine the mitoxantrone binding region in these vRNAs, using an in-line probing assay, which relies on the structure-dependent spontaneous cleavage of RNA (13,17). This sensitive assay has been successfully used to map the binding sites of small molecules in RNA (13). In this assay, the inter-nucleotide linkages between the bases involved in secondary structure interactions are less sensitive to spontaneous hydrolysis than those in the unstructured regions. A significant alteration in the RNA structure upon ligand binding is reflected by a change in the cleavage pattern (18). Using this assay, we monitored the spontaneous cleavage of vRNA in the presence or in the absence of the corresponding ligands (mitoxantrone and doxorubicin). Among the three vRNAs tested with mitoxantrone and doxorubicin, we detected an enhancement in the spontaneous cleavage of hvg-1 upon the addition of mitoxantrone (2–4 µM) (Figure 3a). We observed that the hvg-1 RNA region spanning nt 49–59 was particularly sensitive to increasing concentrations of mitoxantrone (close view, Figure 3b). Using the ImageJ program (free software from http://rsb.info.nih.gov), we carried out a quantitative analysis of the hydrolyzed products obtained in the absence and in the presence of mitoxantrone, in the above region. The band (G58 nt) marked with an arrowhead displayed enhanced cleavage as it attained a saturation point at 2–4 µM of mitoxantrone and declined thereafter. Similarly, the band (A53 nt) marked with an arrow intensified with increasing concentrations of mitoxantrone, representing about a 3-fold enhancement of cleavage in the presence of 4 µM of mitoxantrone, as compared with the drug-free profile (Figure 3c). The cleavage positions at G58 and A53 were not increased significantly at concentrations higher than 4 µM mitoxantrone, suggesting that the drug binds to hvg-1 with an equilibrium dissociation constant (Kd) in the sub-micromolar range. These positions are indicated on the predicted secondary structure of hvg-1 RNA (Figure 3d). Similarly, a mitoxantrone-mediated modulation was also observed in hvg-2, with an increased or decreased level of cleaved product near the binding region of nt 38–51 (Figure 4a, close view Figure 4b). In particular, the position highlighted with an arrow-head (U51 nt) displayed enhanced cleavage upon the addition of increasing concentrations of mitoxantrone, representing more than a 3-fold increase as compared with the control (Figure 4c). The bands (U39, C43 and C48) marked with arrows also underwent specific modulation. These residues are highlighted on the predicted secondary structure of hvg-2 RNA (Figure 4d). In contrast, the hvg-3 RNA did not show any significant difference in the cleavage pattern from the ligand-free control (Figure 5a–d). The gel patterns obtained for doxorubicin with hvg-1, hvg-2 and hvg-3 did not exhibit any remarkable cleavage pattern, as compared with the control (data not shown). Taken together, the CD and in-line attack analyses clearly suggest that the hvg-1 and hvg-2 RNAs specifically bind to mitoxantrone, while the hvg-3 RNA does not, and further imply that the ability to recognize various compounds is based on differences in their 3D structures, which might have originated from point mutations among these RNAs. In addition, it appears that two of the vRNAs (hvg-1 and hvg-2) have the ability to interact with mitoxantrone, despite the differences in their primary sequences near the binding region. To evaluate their interactions in a cellular context, we have carried out a post-transcriptional inhibition assay.
Figure 3.
(a) A representative autoradiogram showing an in-line probing assay of interactions between hvg-1 and mitoxantrone. OH-alkali digestion: T1 (specific for G bases). RNAs were prepared by PCR from DNA templates and were labeled at the 3′ end. The labeled RNA was incubated for 40 h at 25°C in a buffer containing 20 mM MgCl2, 50 mM Tris–HCl (pH 8.3 at 25°C) and 100 mM KCl, with drug concentrations ranging from 2 to 8 µM. After each incubation, spontaneously cleaved products were resolved by 12% denaturing (8 M urea) PAGE followed by autoradiography. (b) Enlarged view: arrows and arrow heads indicate sites of enhanced cleavage. (c) Quantitative analysis of cleaved products. (d) Secondary structure of RNA (RNA structure 3.71). The mitoxantrone binding region is indicated by circles, and the specific residues are indicated as in (b).
Figure 4.
(a) A representative autoradiogram showing an in-line probing assay of interactions between hvg-2 and mitoxantrone. (b) Enlarged view: arrows and arrow heads indicate sites of enhanced cleavage. (c) Quantitative analysis of cleaved products. (d) Secondary structure of RNA (RNA structure 3.71). The mitoxantrone binding region is indicated by circles, and the specific residues are indicated as in (b).
Figure 5.
(a) A representative autoradiogram showing the results of an in-line probing assay of hvg-3 and mitoxantrone. (b) Enlarged view. (c) Quantitative analysis of cleaved products. (d) Secondary structure of RNA (RNA structure 3.71).
Post-transcriptional inhibition assay to analyze vRNA-mitoxantrone interactions
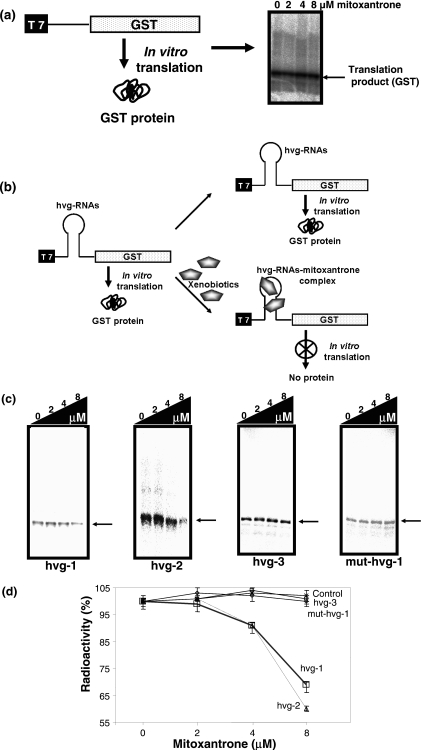
The ability of RNA to interact with small molecules, such as antibiotics and metabolites, has been well documented in the literature (19). Furthermore, Werstuck and Green (20) have shown that appending an RNA motif, which recognizes a small molecule, into the 5′ untranslated region of messenger RNA allowed its translation to be repressed by ligand addition in vitro as well as in vivo. The inhibition of the translation product by these small molecules in this assay was attributed to their ability to interfere with ribosome binding. Using this assay, several small ligands that bind to RNAs have been evaluated and explored for controlling gene expression in living cells (17,20–22). In order to evaluate the observed interactions between vRNAs and mitxantrone in terms of transcription and translation, we employed a post-transcriptional inhibition assay. To evaluate its suitability for our study, we first tested the general ability of the inhibition on the coupled-transcription and translation, using RTS 100 E.coli HY in vitro cell-free system, with increasing amounts of mitoxantrone (2–8 µM), and found that this compound has no influence on the efficiency of yielding the correct translation product (Figure 6a). In addition, the intervening sequences within the template between the T7 promoter and the start codon (AUG) region need to be restricted between 20 and 100 nt, for efficient synthesis of the translation product in a coupled-transcription and translation system (23). Hence, this assay is more suitable for shorter RNA motifs that interact with cognate ligands. To adapt this assay, we truncated the 5′ and 3′ ends from the full-length vRNAs, based on the in-line probing assay, and appended these sequences immediately downstream from the T7 promoter (Figure 6b). The minimal vRNAs thus generated were mini-hvg-1 RNA (59mer, sequences between 20 and 77 nt), mini-hvg-2 RNA (49mer, 20–67 nt) and mini-hvg-3 RNA (49mer, 20–67 nt). Before appending these sequences, the intervening sequence of the vector (pIVEX-GST) between the T7 promoter and start codon, including the ribosome binding site, was removed by digestions with restriction enzymes (see Materials and Methods for details on the vector construction), and then the ribosome binding site was generated together with the minimal vRNAs sequences by cloning. The resulting vectors were pIVEX-GSThvg-1, pIVEX-GSThvg-2 and pIVEX-GSThvg-3. To evaluate the importance of the sequence near the mitoxantrone binding site, based on the in-line probing assay results, we also prepared the hvg-1 RNA mutant with 53A, 54A and 58G, 59U substituted with 53U, 54U and 58C, 59A, respectively (pIVEX-GSTmut-hvg-1). The resulting intervening sequences, upon transcription, between the T7 promoter and the start codon (AUG) are 87, 77, 77 and 87 nt for the vectors pIVEX-GSThvg-1, pIVEX-GSThvg-2, pIVEX-GSThvg-3 and pIVEX-GSTmut-hvg-1, respectively. These vectors yielded 30 000 Da translated products, although pIVEX-GSThvg-1 and pIVEX-GSTmut-hvg-1 produced lower amounts of the translation products, as compared with the other two vectors. This could be due to the extra 10 nt between the T7 promoter and the start codon.
Figure 6.
Post-translation inhibition assay. (a) Synthesis of translated product (30 000 daltons) using pIVEX-GST in the presence of mitoxantrone (2–8 µM) in an E.coli in vitro system. (b) A model system to analyze the interactions between mitoxantrone and vRNAs. (c) Synthesis efficiency of translated products of different vRNAs in the absence and in the presence of mitoxantrone (2–8 µM). The arrow indicates the position of the translated product. (d) Quantitative data of translational products synthesized by the four different vRNAs (values are averages of three experiments).
Using these vectors, we carried out coupled-transcription and translation reactions in the absence and in the presence of various concentrations of mitoxantrone (2–8 µM), and analyzed the amounts of translation products synthesized. The hvg-1 and hvg-2 containing transcripts yielded decreasing amounts of translational products with increasing concentrations of mitoxantrone (Figure 6c). However, the hvg-3 transcript was translated efficiently, even in the presence of 8 µM mitoxantrone. The reduction in the amount of translational product is attributable to an interference with ribosome binding upon mitoxantrone binding, specifically with the hvg-1 and hvg-2 motifs, as observed for antibiotics and metabolites in previous studies (20,24). On the other hand, when the residues of hvg-1 that were involved in the mitoxantrone binding were substituted (53A,54A and 58G,59U to 53U,54U and 58C,59A), the mitoxantrone-dependent inhibition was not observed (Figure 6c). Quantification of these results clearly suggested that the hvg-1 and hvg-2 RNA motifs have the ability to recognize and bind mitoxantrone specifically (Figure 6d).
Considering the current study, including the CD analysis, the in-line probing and the translational inhibition assay, it is clear that the hvg-1 and hvg-2 RNAs bind to mitoxantrone. A common asymmetric loop region is found when compared between these two RNAs based on predicted secondary structures and this loop is absent in hvg-3 RNA. However, we could not observe apparent cleavage sites (under in-line probing studies) in this loop region in the presence of the mitoxantrone. Similar tertiary structure to recognize mitoxantrone by these two RNAs (hvg-1 and hvg-2) might be another distinct possibility for binding and extrusion of the drug. The physiological relevance of the interaction of vRNAs with mitoxantrone has not been defined yet. However, it is tempting to speculate about their importance, based on previous observations of the overexpression of vault complexes (containing three hvg-RNAs and three proteins) associated with resistance to chemotherapeutic compounds (25). Despite these observations, neither the vRNAs nor the associated proteins have been implicated or shown to directly interact with chemotherapeutic compounds. On the other hand, vault-associated proteins have been implicated in a cytoskeletal-mediated transport pathway between the nucleus and cytoplasm (6). The studies presented above clearly indicated, for the first time, that the vault-associated non-coding RNAs, such as the hvg-1 and hvg-2 RNAs, have the ability to bind and sequester mitoxantrone. The vault-associated proteins may be involved in export, to prevent the drug from reaching the site of action. Indeed, more studies are required to investigate the binding of vRNAs to other chemotherapeutic compounds. In addition, the generation of vRNA knockout cells may help to reveal the function of vRNAs and their significance in the complex.
Acknowledgments
The authors wish to thank Prof. Uesugi for valuable discussion on CD data. This work was supported by funds from National Institute of Industrial Science and Technology (AIST) to P.K.R.K, and S.C.B.G is supported by Japan Society for the Promotion of Science (JSPS). M.K. was supported by Grants-in-Aid for Scientific Research (Nos 17026011 and 17048009) and the Protein 3000 Project of the Ministry of Education, Culture, Sports, Science and Technology of Japan, and by PRESTO of JST. Funding to pay the Open Access publication charges for this article was provided by AIST, Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.van Zon A., Mossink M.H., Scheper R.J., Sonneveld P., Wiemer E.A.C. The vault complex. Cell. Mol. Life Sci. 2003;60:1828–1837. doi: 10.1007/s00018-003-3030-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chugani D.C., Rome L.H., Kedersha N.L. Evidence that vault ribonucleoprotein particles localize to the nuclear pore complex. J. Cell. Sci. 1993;106:23–29. doi: 10.1242/jcs.106.1.23. [DOI] [PubMed] [Google Scholar]
- 3.Abbondanza C., Rossi V., Roscigno A., Gallo L., Belsito A., Piluso G., Medici N., Nigro V., Molinari A.M., Moncharmont B., Puca G.A. Interaction of vault particles with estrogen receptor in the MCF-7 breast cancer cell. J. Cell Biol. 1998;141:1301–1310. doi: 10.1083/jcb.141.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herrmann C., Golkaramnay E., Inman E., Rome L., Volknandt W. Recombinant major vault protein is targeted to neuritic tips of PC12 cells. J. Cell Biol. 1999;144:1163–1172. doi: 10.1083/jcb.144.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitazono M., Sumizawa T., Takebayashi Y., Chen Z.S., Furukawa T., Nagayama S., Seto K., Aikou T., Akiyama S. Multidrug resistance and the lung resistance-related protein in human colon carcinoma SW-620 cells [see comments] J. Natl Cancer. Inst. 1999;91:1647–1653. doi: 10.1093/jnci/91.19.1647. [DOI] [PubMed] [Google Scholar]
- 6.Scheffer G.L., Schroeijers A.B., Izquierdo M.A., Wiemer E.A., Scheper R.J. Lung resistance-related protein/major vault protein and vaults in multidrug-resistant cancer [In Process Citation] Curr. Opin. Oncol. 2000;12:550–556. doi: 10.1097/00001622-200011000-00007. [DOI] [PubMed] [Google Scholar]
- 7.Kickhoefer V.A., Searles R.P., Kedersha N.L., Garber M.E., Johnson D.L., Rome L.H. Vaults are up-regulated in multidrug-resistant cancer cell lines. J. Biol. Chem. 1998;273:8971–8974. doi: 10.1074/jbc.273.15.8971. [DOI] [PubMed] [Google Scholar]
- 8.Hu Y., Stephen A.G., Cao J., Tanzer L.R., Slapak C.A., Harrison S.D., Devanarayan V., Dantzig A.H., Starling J.J., Rome L.H., Moore R.E. A very early induction of major vault protein accompanied by increased drug resistance in U-937 cells. Int. J. Cancer. 2002;97:149–156. doi: 10.1002/ijc.1590. [DOI] [PubMed] [Google Scholar]
- 9.Ohno N., Tani A., Uozumi K., Hanada S., Furukawa T., Akiba S., Sumizawa T., Utsunomiya A., Arima T., Akiyama S. Expression of functional lung resistance–related protein predicts poor outcome in adult T-cell leukemia. Blood. 2001;98:1160–1165. doi: 10.1182/blood.v98.4.1160. [DOI] [PubMed] [Google Scholar]
- 10.Wang H.W., Wu H.L., Chen D.S., Chen P.J. Identification of the functional regions required for hepatitis virus replication and transcription by linker-scanning mutagenesis of viral genome. Virology. 1997;239:119–131. doi: 10.1006/viro.1997.8818. [DOI] [PubMed] [Google Scholar]
- 11.Nissen P., Hansen J., Ban N., Moore P.B., Steitz T.A. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 12.Mallory A.C., Dugas D.V., Bartel D.P., Bartel B. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 2004;14:1035–1046. doi: 10.1016/j.cub.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 13.Mandal M., Breaker R.R. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nature Struct. Molecular. Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 14.Kickhoefer V.A., Searles R.P., Kedersha N.L., Garber M.E., Johnson D.L., Rome L.H. Vault ribonucleoprotein particles from rat and bullfrog contain a related small RNA that is transcribed by RNA polymerase III. J. Biol. Chem. 1993;268:7868–7873. [PubMed] [Google Scholar]
- 15.Krol A., Carbon P. A guide for probing native small nuclear RNA and Ribonucleoprotein structures. Methods Enzymol. 1989;180:212–227. doi: 10.1016/0076-6879(89)80103-x. [DOI] [PubMed] [Google Scholar]
- 16.Roninson I.B., editor. Molecular and Cellular Biology of Multidrug Resistance in Tumour Cells. New York: Plenum Press; 1991. pp. 1–406. [Google Scholar]
- 17.Nahvi A., Sudarsan N., Ebert M.S., Zou X., Brown K.L., Breaker R.R. Genetic control by a metabolite binding mRNA. Chem. Biol. 2002;9:1043–1049. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 18.Soukup G.A., Breaker R.R. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hermann T. Chemical and functional diversity of small molecule ligands for RNA. Biopolymers. 2003;70:4–18. doi: 10.1002/bip.10410. [DOI] [PubMed] [Google Scholar]
- 20.Werstuck G., Green M.R. Controlling gene expression in living cells through small molecule-RNA interactions. Science. 1998;282:296–298. doi: 10.1126/science.282.5387.296. [DOI] [PubMed] [Google Scholar]
- 21.Harvey I., Garneau P., Pelletier J. Inhibition of translation by RNA-small molecules interactions. RNA. 2002;8:452–463. doi: 10.1017/s135583820202633x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanson S., Berthelot K., Fink B., McCarthy J.E.G., Suess B. Tetracycline-aptamer-mediated translational regulation in yeast. Mol. Microbiol. 2003;49:1627–1637. doi: 10.1046/j.1365-2958.2003.03656.x. [DOI] [PubMed] [Google Scholar]
- 23.Jain C., Belasco J.G. Rapid genetic analysis of RNA-Protein interactions by translational repression in Escherichia coli. Methods Enzymol. 2000;318:309–332. doi: 10.1016/s0076-6879(00)18060-7. [DOI] [PubMed] [Google Scholar]
- 24.Chiba Y., Sakurai R., Yoshino M., Ominato K., Ishikawa M., Onouchi H., Naito S. S-adenosyl-L-methionine is an effector in the posttranscriptional autoregulation of the cystathionine gamma-synthase gene in Arabidopsis. Proc. Natl Acad. Sci. USA. 2003;100:10225–10230. doi: 10.1073/pnas.1831512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mossink M.H., van Zon A., Scheper R.J., Sonneveld P., Wiemer E.A. Vaults: a ribonucleoprotein particle involved in drug resistance? Oncogene. 2003;22:7458–7467. doi: 10.1038/sj.onc.1206947. [DOI] [PubMed] [Google Scholar]