Abstract
The yeast genome encodes seven oxysterol binding protein homologs, Osh1p–Osh7p, which have been implicated in regulating intracellular lipid and vesicular transport. Here, we show that both Osh6p and Osh7p interact with Vps4p, a member of the AAA (ATPases associated with a variety of cellular activities) family. The coiled-coil domain of Osh7p was found to interact with Vps4p in a yeast two-hybrid screen and the interaction between Osh7p and Vps4p appears to be regulated by ergosterol. Deletion of VPS4 induced a dramatic increase in the membrane-associated pools of Osh6p and Osh7p and also caused a decrease in sterol esterification, which was suppressed by overexpression of OSH7. Lastly, overexpression of the coiled-coil domain of Osh7p (Osh7pCC) resulted in a multivesicular body sorting defect, suggesting a dominant negative role of Osh7pCC possibly through inhibiting Vps4p function. Our data suggest that a common mechanism may exist for AAA proteins to regulate the membrane association of yeast OSBP proteins and that these two protein families may function together to control subcellular lipid transport.
Keywords: AAA ATPase, lipid transport, oxysterol binding protein (OSBP)
Introduction
The oxysterol binding protein (OSBP) and its related proteins (ORP, for OSBP-related protein) constitute a large conserved family of proteins in eukaryotes (Lehto et al, 2001; Olkkonen and Levine, 2004). OSBP was first identified as a high-affinity cytosolic receptor for oxysterols, such as 25-hydroxycholesterol (Kandutsch et al, 1984; Dawson et al, 1989a, 1989b). Homologs to OSBP have subsequently been isolated in most eukaryotes including 12 members in humans (Lehto et al, 2001) and seven members in the budding yeast (Beh et al, 2001). These proteins all share a conserved ∼400-amino-acid OSBP-related domain (ORD) found at the C terminus of OSBP, which has been shown to bind oxysterols and contains an ‘OSBP' fingerprint ‘EQVSHHPP'. Members of the OSBP and ORP family vary in length: the short ORPs comprise primarily the ORD whereas the long ones often contain more than 1000 amino acids and possess other functional domains including a Pleckstrin homology (PH) domain and ankyrin repeats.
Because of its high affinity for oxysterols and the potency of oxysterols as feedback regulators of cholesterol homeostasis, OSBP was proposed to mediate the transcriptional control of the mevalonate pathway (Taylor et al, 1984). However, in response to oxysterol loading, OSBP relocates to the Golgi, instead of the nucleus (Ridgway et al, 1992). It was further demonstrated that the translocation of OSBP to Golgi depends on cellular cholesterol content (Storey et al, 1998). Recent studies in both mammalian cells and yeast have linked OSBP and ORPs with cellular sterol, sphingolipid and glycerolipid metabolism: (1) Overexpression of OSBP in Chinese hamster ovary cells resulted in a 50% reduction in sterol ester synthesis and an 80% increase in cholesterol biosynthesis, and also enhanced the stimulatory effects of 25-hydroxycholesterol on sphingomyelin synthesis (Lagace et al, 1997, 1999). (2) The yeast genome encodes seven ORPs, Osh1p–Osh7p (OSH standing for OSBP homolog). None of the single OSH genes is essential but deletion of all seven genes resulted in lethality, accompanied by a 3.5-fold increase in cellular ergosterol levels (Beh et al, 2001). Most recent work showed that intracellular sterol distribution was also altered upon elimination of all OSH function (Beh and Rine, 2004). (3) The yeast Sec14p is a phospholipid transfer protein, which is required for Golgi-derived vesicular transport. The deletion of OSH4/KES1, but none of the other OSH genes, bypasses the essential requirement for Sec14p (Fang et al, 1996). This implies that Osh4p may negatively regulate Sec14-dependent protein secretion, possibly through modulating local lipid composition and thereby affecting the activity of an adenosine diphosphate-ribosylation factor (ARF) (Li et al, 2002). Collectively, these data suggest that OSBP and ORPs influence cellular lipid metabolism and membrane dynamics, but insights into the molecular function of these proteins are still lacking.
Recent advances, however, provide exciting new evidence suggesting that OSBP and its homologs may function to transport lipids between cellular membranes. OSBP interacts with VAMP-associated protein-A (VAP-A), a syntaxin-like protein implicated in endoplasmic reticulum (ER)/Golgi vesicular transport and this interaction appears to regulate protein and ceramide transport from the ER to the Golgi apparatus (Wyles et al, 2002). OSBP and some of its homologs can target different cell membranes: Osh1p associates with Golgi via its PH domain and the nuclear-vacuole (NV) junction via its ankyrin repeats (Levine and Munro, 1998, 2001). Interestingly, Osh1p also interacts with Scs2p, a resident ER protein and a homolog of mammalian VAP-A, through an FFAT (two phenylalanines in an acid tract) motif, which is well conserved in several seemingly unrelated lipid binding proteins including GPBP (Good Pasture antigen-binding protein) (Loewen et al, 2003). A recent landmark study proved that a truncated form of GPBP, GPBP26, or CERT is required for the nonvesicular ceramide transport from ER to the Golgi in an ATP-dependent manner (Hanada et al, 2003). In order to fulfill its ceramide carrier function, CERT has a PH domain for Golgi targeting, an FFAT motif for ER targeting and the START domain for ceramide binding and extraction (Hanada et al, 2003; Munro, 2003). While direct evidence is lacking, it is probable that with a domain structure very similar to CERT, the OSBP and certain ORPs may play a role in moving lipids between membranes, most likely at membrane contact sites (MCS) (Levine, 2004).
Although the exact molecular function of OSBP and ORPs remains obscure at present, it is rather clear that this family of proteins work at cellular membranes. To carry out their functions, these proteins must be able to interact with membranes in a highly regulated manner. In this study, we established for the first time a link between ORPs and AAA ATPases (ATPase associated with a variety of cellular activities): a family of ATPases with diverse cellular activities. The key mode of action of the AAA family members is energy-dependent unfolding of proteins and disassembly of protein complexes (Patel and Latterich, 1998). Here, we provide evidence that the membrane association of Osh proteins in yeast is regulated by AAA ATPases, which probably exert their effects by breaking the interaction between Osh proteins and their membrane receptors. We further propose that members of these two protein families work in concert to regulate lipid transport.
Results
Characterization of Osh6p and Osh7p
The yeast genome encodes seven OSBP homologs, Osh1p–Osh7p. Osh1p–3p are long ORPs whereas Osh4p–7p comprise only the ORD. Based on sequence homology, the Osh proteins can be further divided into four subfamilies: Osh1p and Osh2p; Osh3p; Osh4p and Osh5p; Osh6p and Osh7p (Beh et al, 2001). Within each subfamily, members are at least 55% identical whereas the identity is less than 30% between subfamilies. Our primary interest is in Osh6p and Osh7p as a result of a two-hybrid screening described below. To localize Osh6p and Osh7p, we constructed OSH6-GFP and OSH7-GFP chimera on CEN plasmids: pRS316OSH6-GFP or pRS316OSH7-GFP. To ascertain whether these GFP-tagged proteins are functional as native proteins in vivo, each plasmid was introduced into strain JRY6326 and grown on yeast minimal (YM) plates containing 2 mM methionine for 72 h. JRY6326 is a strain deleted for all OSHs, with conditional expression of OSH2 from the MET promoter. Since loss of all OSH proteins is lethal, JRY6326 is inviable when plated on medium containing methionine, which suppresses OSH2 expression. Reintroduction of any single OSH into JRY6326 cells restores cell growth in the presence of methionine (Beh et al, 2001). As expected, Osh6p-GFP and Osh7p-GFP restored JRY6326 cell viability (data not shown), suggesting that these GFP fusions behave the same as native protein in terms of cell growth. In addition, the GFP fusion proteins remain intact in the cell as detected by Western blotting (Supplementary Figure S1).
Next, we examined the cellular distribution of Osh6p-GFP and Osh7p-GFP in live cells using fluorescence microscopy. Both Osh6-GFP and Osh7p-GFP were diffusely distributed in the cytoplasm, whereas Osh4p-GFP was found in a few punctate structures, which probably represent the Golgi (Li et al, 2002) (Figure 1A). Occasionally, Osh6p-GFP can also be weakly seen in patches near cell periphery and in small punctate structures but we cannot determine whether these structures represent the plasma membrane (PM), ER or early endosomes.
Figure 1.
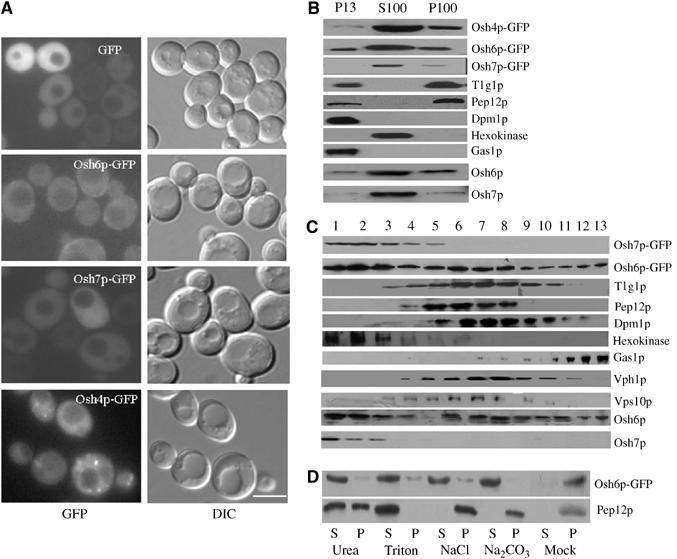
Characterization of the subcellular localization of Osh6p and Osh7p. (A) Fluorescence images of GFP, Osh6p-GFP, Osh7p-GFP and Osh4p-GFP expressed from YCplac111 (CEN)-based plasmids in WT (Y00000) cells. DIC: differential interference contrast. Scale bar: 5 μm. (B) Subcellular fractionation by differential centrifugation. WT (Y00000) cells were lysed and cellular components were fractionated into P13, S100 and P100 by differential centrifugation at 13 000 g and 100 000 g sequentially. GFP fusions were detected using anti-GFP antisera, and native Osh6p/Osh7p were probed with anti-Osh6p/Osh7p antisera. (C) Subcellular fractionation on a continuous sucrose density gradient. WT cells (Y00000) were lysed and the cellular components were separated on a continuous sucrose density gradient ranging from 20 to 50% w/w by centrifugation at 100 000 g. A total of 13 fractions from top to bottom were collected and the distribution of various marker proteins was analyzed by Western blotting with various antibodies as indicated. GFP fusions were detected using anti-GFP antisera, and native Osh6p/Osh7p were probed with anti-Osh6p/Osh7p antisera. (D) Membrane extraction. The pellet fraction of Osh6p-GFP from WT cells was treated with the indicated reagents on ice and centrifuged at 100 000 g again resulting in pellet (designated P) and supernatant (designated S). Mock: treatment with lysis buffer only.
We also performed a series of subcellular fractionation experiments to further analyze the cellular distribution of these proteins. Cell extracts prepared from a wild-type (WT) strain (BY4741) were fractionated by centrifugation at 13 000 g for 10 min, resulting in P13 pellet and S13 supernatant fractions. The S13 was further centrifuged for 1 h at 100 000 g, resulting in P100 pellet and S100 supernatant fractions, which were probed with antibodies against GFP and Osh6/7p, and specific resident proteins of subcellular compartments. As shown in Figure 1B, in addition to a large pool in S100, a small amount of Osh6p was associated with both P13 and P100 fractions. Large membranes such as vacuole, ER, PM and mitochondria associate with P13 predominantly, while Golgi and endosomal membranes are present in both P13 and P100 with varying ratio. Small transport vesicles are associated with the P100 fraction predominantly. The presence of GFP, hexokinase (cytoplasmic), Tlg1p and Pep12p (endosomal), Gas1p (PM), Dpm1p (ER), Osh6p and Osh7p was detected by immunoblotting with specific antisera. As shown in Figure 1B, Osh4p/Kes1p was predominantly associated with P100 with a minute pool in P13. Hexokinase, a cytosolic protein, was observed only in S100. The distribution pattern of Osh6p-GFP was similar to that of Tlg1p and Pep12p. Osh7p-GFP was found largely in S100 with a minute pool in P100. Same results were observed for native Osh6p and Osh7p, suggesting that the GFP tag had little effect on distribution. To better understand the subcellular localization of Osh6p and Osh7p, cell extracts were subjected to continuous sucrose density gradient analysis. A total of 13 fractions were collected from top to bottom (1–13) and probed for the presence of GFP, hexokinase, Tlg1p and Pep12p, Vph1p (vacuolar membrane), Gas1p, Vps10 (Golgi) and Dpm1p by immunoblotting. As shown in Figure 1C, Osh6p-GFP distribution was rather broad and three peaks were resolved: the F2 peak representing cytosolic protein; the F7 peak representing dense membranes like early endosome (Tlg1p) and ER (Dpm1p); and the third minor peak (F13) representing PM. Osh7p-GFP was distributed predominantly in F1–F3, which indicates that Osh7p-GFP is a cytosolic protein. The native Osh6p and Osh7p showed similar distribution patterns as the GFP fusion proteins. Combined with data obtained from microscopy and differential centrifugation, it appears that Osh6p-GFP is mainly cytosolic with a small endosomal or PM pool whereas Osh7p-GFP is predominantly cytosolic with a hardly detectable membrane-associated pool in WT cells. These techniques do not allow us to pinpoint the subcellular localization of Osh6p and Osh7p and it is not surprising to find Osh6p or Osh7p in multiple locations since they may function to shuttle lipids between various subcellular membranes.
To examine the nature of Osh6p-GFP association with the pellet fraction (P13 and P100), the combined pellet fraction (P13+P100) was subjected to a number of treatments. As shown in Figure 1D, treating the pellets with 6 M urea, 1% Triton, 1 M NaCl and 0.1 M Na2CO3 (pH 11) solubilized Osh6p-GFP almost completely. Pep12p (transmembrane protein) was completely solubilized by 1% Triton and partially solubilized by 6 M urea; however, it was not affected by treatments with 1 M NaCl and 0.1 M Na2CO3, which dissociate peripheral membrane proteins only. Thus, Osh6p-GFP is a peripheral membrane protein with a major cytoplasmic pool.
Osh6p and Osh7p interact with Vps4p
In a large-scale screen for Vps4p interacting proteins using the yeast two-hybrid system (Yeo et al, 2003), Osh7p was isolated but first reported here. The original yeast two-hybrid clone encodes amino-acid residues 366–437 of Osh7p, within which a putative coiled-coil domain exists (Beh et al, 2001). The interaction between full-length Osh7p and Vps4p was confirmed by yeast two-hybrid assays (Figure 2A and B). The interaction between Vps4p and full-length Osh7p was rather weak whereas the coiled-coil domain of Osh7p (Osh7pCC, amino acid residues 366–437 of Osh7p) showed much higher affinity for Vps4p, suggesting that the coiled-coil motif may mediate this interaction. To further characterize the interaction between Vps4p and Osh6/7p, an in vitro GST pull-down assay was performed. GST fusions were expressed in Escherichia coli and purified using affinity chromatography. Same amount of purified protein was used to pull down Myc-tagged Vps4p from yeast lysates. As shown in Figure 2C, GST-Osh7p or Osh7pCC pulled down Vps4p-Myc (bound fraction, lanes 3 and 4 are duplicates) from yeast lysates. As a control, Osh5p did not pull down Vps4p whereas Osh6p did. A weak interaction between Osh6p and Vps4p was also detected by the yeast two-hybrid assay (data not shown). This is not surprising since Osh6p and Osh7p share ∼80% sequence identity. Interestingly, GST-Osh7pCC seemed to pull down a little more Vps4p-Myc than the full-length Osh7p protein, but the difference was not as great in the protein binding assay as was seen in the two-hybrid assays (Figure 2A and B). This could be due to the fact that the Osh proteins, like OSBP, may bind sterols (Wang et al, 2005). When produced in sterol-free bacteria, Osh7p may assume a conformation that allows stronger interaction with Vps4p. To test this hypothesis, bacterially expressed Osh7p and Osh7pCC were incubated with or without ergosterol prior to the pull-down assay. As shown in Figure 2D, Osh7p pulled down little Vps4p after incubation with ergosterol, whereas the interaction between Osh7pCC and Vps4 was not affected by ergosterol. In summary, these results indicate that Vps4p specifically interacts with a subset of Osh proteins and that this interaction is probably regulated by ergosterol in vivo.
Figure 2.
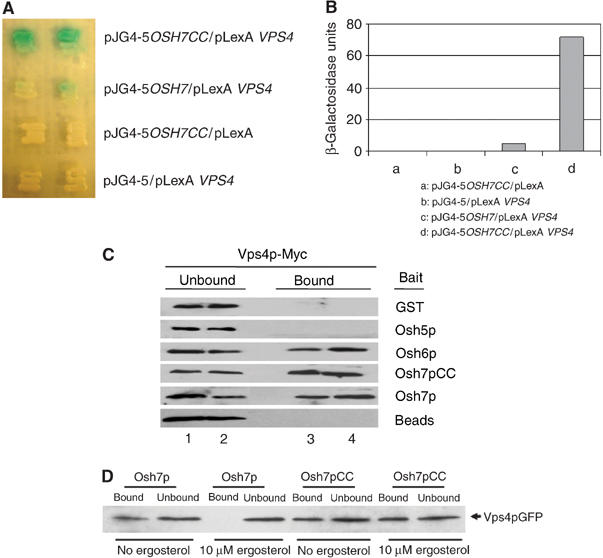
Osh6p and Osh7p interact with Vps4p in vivo and in vitro. (A) Two-hybrid interaction. The LexA-based bait plasmids pLexA and pLexA-VPS4 and the pB42AD-based prey plasmids pJG4-5, pJGOSH7CC and pJGOSH7 were introduced into EGY48 together with the reporter plasmid PSH18-34 and grown in synthetic galactose/raffinose medium containing X-gal at 30°C for 2 days. (B) Quantification of yeast two-hybrid interactions. Cells were lysed by freeze–thaw cycles using liquid nitrogen and β-galactosidase activity was assayed using ONPG as a substrate. (C) GST pull-down. GST fusion proteins were purified from E. coli using glutathione (GSH) agarose beads. The fusion proteins were incubated with yeast lysates prepared from Vps4p-Myc expressing Y10000 cells. After stringency washes, bound proteins were eluted and separated on SDS–PAGE. Vps4p-Myc was detected using an anti-Myc monoclonal antibody. Lanes 1 and 2: unbound fraction; lanes 3 and 4: bound fraction. (D) Incubation with ergosterol affects the interaction between Osh7p and Vps4p in vitro. GST fusion proteins were purified from E. coli using GSH agarose beads. Fusion proteins containing full-length Osh7p or Osh7pCC were incubated in the presence or absence of 10 μM ergosterol for 2 h at 30°C, and then mixed with yeast lysates prepared from Vps4p-GFP-expressing Y10000 cells. After stringency washes, bound proteins were eluted and separated on SDS–PAGE.
Vps4p regulates the membrane association of Osh6p and Osh7p
Since Osh6p and Osh7p physically interact with Vps4p, we were interested to know the functional relevance of these interactions. Vps4p is a well-established AAA ATPase required for efficient transport in the multivesicular body (MVB) sorting pathway, where it functions to disassemble the ESCRT (endosomal sorting complex required for transport) complexes (Babst et al, 1997, 1998, 2002a, 2002b; Katzmann et al, 2001). It is possible that Vps4p interacts with Osh6p/Osh7p transiently and help them dissociate from membranes so that they can be recycled into the cytoplasm. To test this, we expressed Osh6p-GFP and Osh7p-GFP in WT and vps4Δ cells and investigated their cellular localization. The intracellular distribution of Osh6p-GFP was altered significantly in vps4Δ cells (Figure 3A). In WT cells, few internal fluorescent structures containing Osh6p-GFP were observed, whereas there were bright, punctate structures in vps4Δ cells. We also used Osh4p-GFP as a negative control, whose cellular distribution remained largely unchanged (not shown). The fluorescence from Osh7p-GFP was rather weak and appeared to be cytoplasmic in vps4Δ (Figure 3B), but we could not exclude the possibility that a membrane-associated pool was masked by the cytosolic pool. To find out the nature of structures containing Osh6p, we performed colocalization experiment using FM4-64, which is endocytosed and delivered to the vacuole membrane via the endocytic pathway (Vida and Emr, 1995). Osh6p-GFP was found to partially colocalize with FM4-64-stained endosomal compartments, implying that a small portion of Osh6p-GFP associates with the endosomal membrane (Figure 3C).
Figure 3.
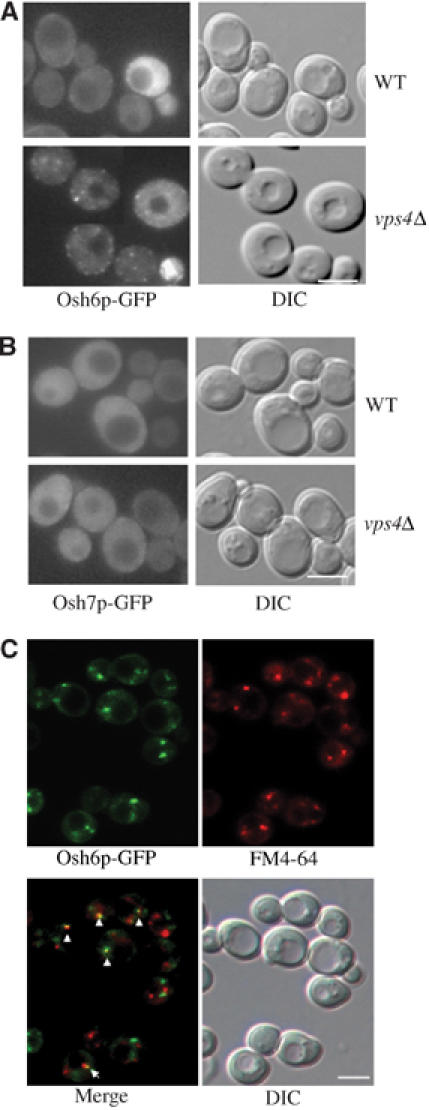
Vps4p mediates the membrane dissociation of Osh6p and Osh7p. (A, B) Fluorescence images of Osh6p-GFP and Osh7p-GFP in WT (Y10000) and vps4Δ (Y15588) strains. (C) Colocalization of Osh6p-GFP with FM4-64-positive compartments. Exponentially growing vps4Δ (Y15588) cells expressing Osh6p-GFP were labeled with FM4-64 on ice for 30 min and then shifted to 15°C for 20 min allowing FM4-64 to be internalized. Cells were immediately put back on ice and washed thoroughly with 1 × phosphate-buffered saline (PBS, pH 7.4, 20 mM NaN3/NaF). GFP and FM4-64 images were acquired via a GFP filter and Texas red filter, respectively. Arrows point to points of colocalizations. DIC: differential interference contrast. Scale bar: 5 μm.
To further analyze the impact of VPS4 deletion on the distributions of Osh6p and Osh7p, we performed subcellular fractionation experiments. Cleared cell lysate was centrifuged at 100 000 g, resulting in fractions S (supernatant) and P (pellet). As shown in Figure 4A, more Osh6p-GFP was found to be associated with the pellet fraction in vps4Δ cells than that in WT cells, whereas a significantly large amount of Osh7p-GFP was detected in the pellet fraction only in vps4Δ cells. The relocation of Osh6p/Osh7p-GFP in vps4Δ cells was specific because Osh4p-GFP and Osh1p (615–1188)-GFP did not relocate. We also performed sucrose density gradient analysis on Osh7p-GFP in WT and vps4Δ strains. As shown in Figure 4B, a second peak of Osh7p-GFP, which overlaps with markers of internal membranes (data not shown), was observed only in vps4Δ strain. Slight change of Osh6p-GFP on sucrose gradient was also observed in vps4Δ cells but it was not as striking as that of Osh7p-GFP (data not shown). Next, we tested whether this regulation process needs the ATPase activity of Vps4p. Thus, we reintroduced WT VPS4 allele and vps4 E233Q defective in ATP hydrolysis (Babst et al, 1997) on a CEN plasmid into vps4Δ cells, and examined the cellular localization of Osh6p-GFP and Osh7p-GFP. As shown in Figure 4C, WT VPS4 corrected mislocalization of Osh6p-GFP and Osh7p-GFP in vps4 cells, but E233Q mutant allele failed to do so. This suggests that Vps4p ATPase activity is required for regulating membrane association of Osh6p and Osh7p. To investigate the biochemical nature of pellets containing Osh6p-GFP or Osh7p-GFP, the pellet fractions from vps4Δ cells were treated with 1% Triton X-100 on ice for 30 min, and fractionated again by centrifugation into S (supernatant) and P (pellet). As shown in Figure 4D, Osh6p-GFP was almost completely solubilized, indicating that the pellet fraction of Osh6p-GFP was membrane associated. However, Osh7p-GFP could not be solubilized, indicating that, similar to certain members of the class E Vps proteins, it could be associated with large protein aggregates in vps4Δ cells (Babst et al, 1998). To further determine whether the Osh7p aggregates associate with membranes, a floatation experiment was performed (Babst et al, 1998). When subjected to ultracentrifugation, membranes migrate out of 60% sucrose and into 35% sucrose due to their intrinsic buoyant density (Walworth et al, 1989). The pellet fraction from vps4Δ cells was loaded at the bottom of a sucrose step gradient. After centrifugation, three fractions were collected: the top fraction containing membrane-associated materials (F), the middle fraction containing the nonfloat material (N) and the pellet containing large nonmembrane-associated material. As shown in Figure 4E, Osh7p-GFP floated up to the top fraction, suggesting that the pellet fraction of Osh7p-GFP in vps4Δ cells was indeed membrane associated. Lastly, the effects of other class E vps mutants on cellular distribution of Osh6p-GFP and Osh7p-GFP were examined. As shown in Figure 4F, deletion of VPS2, VPS24, VPS23, VPS27 or VPS36 did not affect Osh6/7p-GFP localization.
Figure 4.
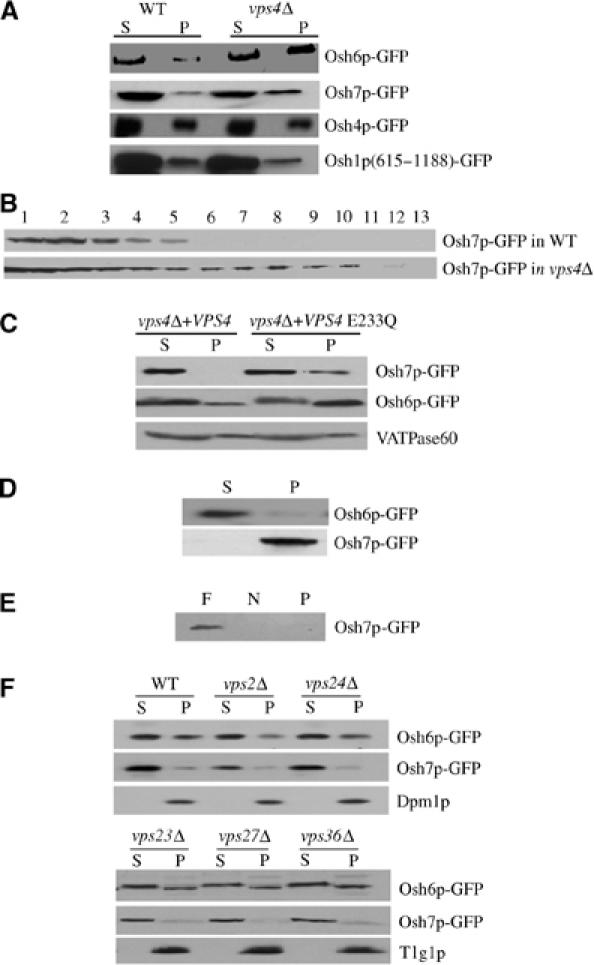
Vps4p but not other class E Vps proteins regulates the membrane dissociation of Osh6p and Osh7p. (A) Subcellular fractionation by differential centrifugation. WT (Y10000) or vps4Δ (Y15588) cells expressing various GFP fusion proteins were spheroplasted and lysed and cell lysates were fractionated into S (supernatant) and P (pellet) by centrifugation at 100 000 g. (B) Fractionation of Osh7p-GFP by density gradient centrifugation. WT (Y10000) and vps4Δ (Y15588) cells expressing Osh7p-GFP were lysed using a bead beater and then cleared. The cleared lysate were resolved on a 20–50% linear sucrose gradient by centrifugation at 100 000 g. (C) The ATPase activity of Vps4p is required for dissociation of Osh6p/Osh7p-GFP from membranes. Differential centrifugation was performed as in panel A. vps4Δ+vps4 E233Q denotes vps4Δ cells carrying pRS316vps4-E233Q plasmid encoding the ATPase-defective Vps4p-E233Q mutant protein. vps4Δ+VPS4 indicates vps4Δ cells carrying pRS316VPS4 plasmid encoding WT Vps4p. (D) Membrane extraction. The 100 000 g pellets prepared from vps4Δ cells were treated with cold 1% Triton X-100 and centrifuged at 100 000 g again resulting in supernatant (S) and pellet (P) fractions. (E) Membrane floatation. The P100 fraction of vps4Δ cells expressing Osh7p-GFP was subjected to density gradient flotation (see Materials and methods for details). F: float; N: nonfloat; P: pellet. (F) Fractionation of Osh6p/Osh7p-GFP from WT (Y10000), vps2Δ (Y14850), vps24Δ (Y14890), vps23Δ (Y13426), vps27Δ (Y15381) and vps36Δ (Y15325) was performed as described in panel A.
Vps4p regulates cellular sterol metabolism
Deletion of all seven OSH genes caused specific changes in cellular sterol homeostasis, such as a three-fold increase in total cellular sterol level and accumulation of free sterol in internal organelles (Beh et al, 2001; Beh and Rine, 2004). Since Vps4p regulates membrane association of Osh proteins and sterol accumulation was observed in mammalian cells overexpressing an ATPase defective VPS4 (Bishop and Woodman, 2000), Vps4p in yeast may therefore play a role in sterol metabolism. We investigated sterol esterification in osh6Δosh7Δ and vps4Δ strains. Double deletions of OSH6 and OSH7 resulted in an ∼30% decrease in oleic acid incorporation into sterol esters whereas incorporation into TAG was not affected (Figure 5A), indicating that fatty acid uptake was normal. Interestingly, the rate of oleate incorporation into sterol esters was also significantly reduced in vps4Δ (Figure 5B). As a positive control, deletion of the major acyl-coenzyme A: sterol acyl transferase gene, ARE2, drastically reduced sterol esterification (Yang et al, 1996). The same reduction was also observed in the vps4ts mutant after incubating at 37°C for 40 min, implying that the defect is an immediate and direct effect of loss of Vps4p function (Figure 5C). Most importantly, overexpression of OSH7 resulted in a significant increase in sterol esterification in the vps4Δ strain, suggesting a functional relationship between Vps4p and Osh7p. Overexpression of OSH6 also partially suppressed the sterol esterification defect in the vps4Δ strain (Supplementary Figure S2). Lastly, no decrease in sterol esterification was observed in vps23Δ, vps27Δ or vps36Δ mutants (data not shown). Oleate incorporation into TAG remained constant in all experiments (data not shown except Figure 5A).
Figure 5.
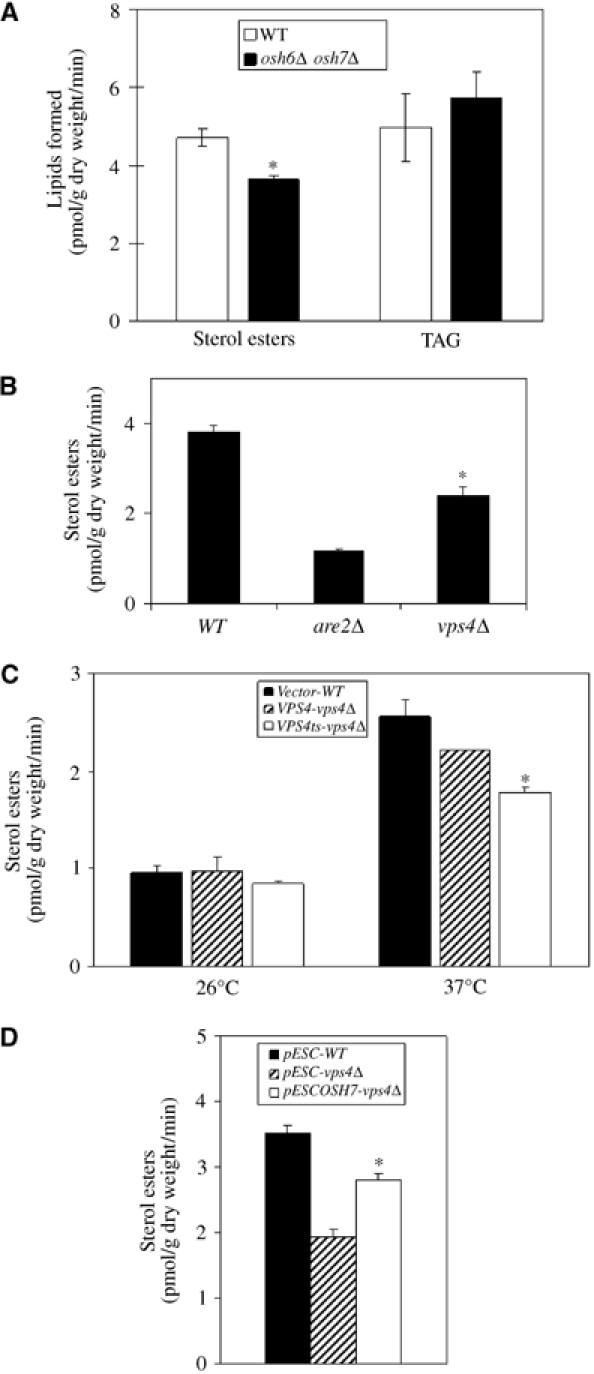
Oleate incorporation into sterol esters. Cells were pulsed with [3H]oleic acid and dried. Lipids were extracted with hexane and separated by thin-layer chromatography (TLC). Oleate incorporation into different lipid species was quantified using a scintillation counter. Data were obtained from three independent experiments (n=3). *P<0.05. (A) Oleate incorporation is impaired in the osh6Δosh7Δ strain. WT (SEY6210) and osh6Δosh7Δ (JRY6207) were studied. (B) Oleate incorporation is impaired in the vps4Δ strain. WT (Y10000), vps4Δ (Y15588) and are2Δ (Y15394) were studied. (C) vps4ts mutant has defects in oleate incorporation. Exponentially growing yeast strains were preincubated at 26 or 37°C for 10 min and then radiolabeled for 30 min at the same temperature. Lipids were extracted as described above. Vector-WT: WT (RH448) cells carrying YCplac111; VPS4-vps4Δ: RH2906 (vps4Δ) cells expressing WT Vps4p from YCpVPS4; vps4ts-vps4Δ: RH2906 cells expressing the temperature-sensitive Vps4p from YCpvps4ts. (D) Overexpression of OSH7 suppresses defects in oleate incorporation in vps4Δ. Cells were grown in glucose medium to mid-log phase and transferred to galactose/raffinose medium for 4 h. Cells were then labeled with [3H]oleate for 30 min and lipids were extracted as described above. pESC-WT: WT (Y10000) cells carrying pESC; pESC-vps4Δ: vps4Δ (Y15588) cells carrying pESC; pESCOSH7-vps4Δ: vps4Δ (Y15588) cells carrying pESCOSH7.
Overexpression of the coiled-coil domain of Osh7p causes an MVB sorting defect
Since a strong interaction between the coiled-coil domain of Osh7p (Osh7pCC) and Vps4p was detected, we wonder whether Osh7pCC could act as a dominant negative fragment in vivo. The vacuolar enzyme carboxypeptidase S (CPS) is synthesized as a membrane-bound precursor (pro-CPS), and is ubiquitylated before being sorted into the lumenal vesicles of MVBs. Once delivered to the vacuole lumen, pro-CPS is proteolytically clipped from its transmembrane domain, resulting in a soluble enzyme within the vacuole lumen (Katzmann et al, 2001). In the vps4Δ mutant strain, MVB is not formed and a GFP-tagged version of CPS (GFP-CPS) was found on the limiting membrane of vacuole and an abnormal endosomal compartment (Babst et al, 2002b; Figure 6A). Overproduction of Osh7pCC, but not the full-length Osh7p, caused a similar GFP-CPS distribution pattern as in vps4Δ (Figure 6A). Since no carboxypeptidase Y (CPY) sorting defect was observed even when all OSH genes are deleted, this MVB sorting defect possibly results from inhibition of normal Vps4p function as a result of the strong interaction between Osh7pCC and Vps4p. To further evaluate the MVB sorting defect in these strains, GFP-CPS was subjected to Western blot analysis. As shown in Figure 6B, free GFP was detected in WT cells and in cells overexpressing OSH7 or OSH7CC while a GFP product that is 4 kDa larger was found in vps4Δ cells and interestingly, also in cells overexpressing OSH7CC. As reported previously, the larger GFP product corresponds to cleavage immediately beyond the transmembrane domain of CPS since an MVB sorting defect prevents the GFP tag from entering the vacuole (Reggiori and Pelham, 2001; Shiflett et al, 2004).
Figure 6.
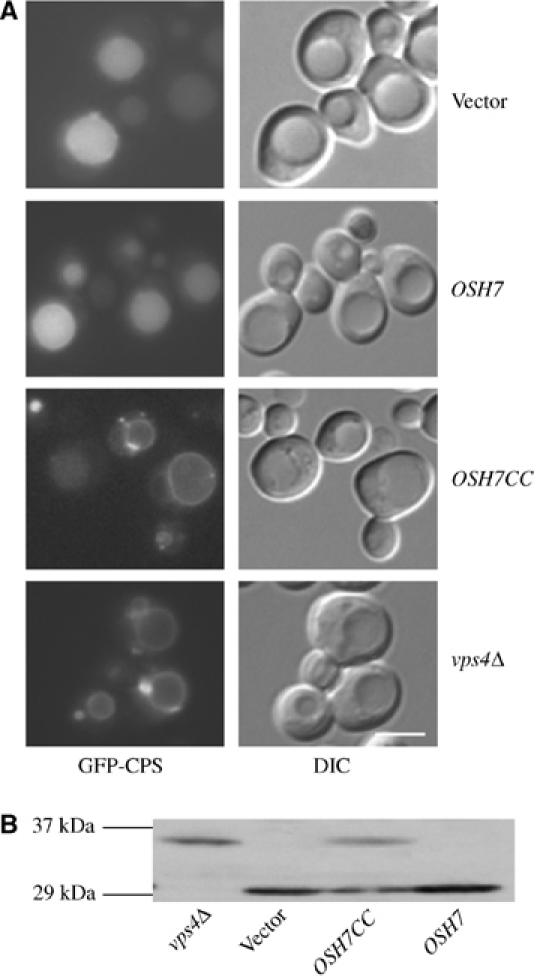
Overexpression of OSH7CC causes an MVB sorting defect. WT cells (Y10000) expressing Osh7CC/Osh7p from pESCOSH7CC/pESCOSH7 were grown in glucose medium to mid-log phase and transferred to galactose/raffinose medium for 4 h. (A) GFP-CPS was visualized using a Leica fluorescence microscopy via a GFP filter. DIC: differential interference contrast. Scale bar: 5 μm. (B) Western blot analysis of total cell extracts from indicated strains with anti-GFP antisera. Equivalent amounts of total protein were loaded in each lane.
Discussion
Lipids are essential components of all cell membranes, but each subcellular organelle has a unique lipid profile. In order to maintain a nonhomogenous distribution of lipids in different membranes, cells must selectively transport certain lipids in a desired direction. The molecular mechanisms for subcellular lipid transport are poorly understood; however, recent studies point to at least two possible mechanisms: vesicular and nonvesicular lipid transport (Funato and Riezman, 2001; Hanada et al, 2003). Nonvesicular lipid transport is likely to take place in MCS with the aid of a wide range of lipid transfer proteins (Levine, 2004). To meet the criteria as cytosolic lipid carriers between membranes, the lipid transfer proteins should ideally be able to interact with different membranes, be able to bind and extract lipids and lastly, be able to associate with and dissociate from membranes in a regulated manner for efficient lipid transport. A large family of proteins in eukaryotic cells, the OSBP and certain ORPs possess a well-conserved lipid binding motif (ORD) and can target multiple membranes; therefore, they have been proposed to mediate nonvesicular lipid transport (Levine, 2004; Olkkonen and Levine, 2004). Our results here support such a hypothesis by providing a mechanism whereby the membrane association of putative lipid transfer proteins, such as ORPs, is regulated.
AAA proteins have been implicated in functions as diverse as peroxisome biogenesis (Pex1p and Pex6p), vesicle transport (Sec18p/NSF), cell division (Cdc48p/p97) and endosomal function (Vps4p/SKD1) (Lupas and Martin, 2002). They may also play a role in lipid trafficking: it has been shown that an ATPase-defective VPS4/SKD1 caused a delay in cholesterol transport out of endosomes (Bishop and Woodman, 2000). We also demonstrated here that a vps4 mutant exhibits specific defects in sterol metabolism. These defects might be secondary effects since certain AAA proteins have defined functions, such as in multivesicular body sorting for Vps4p. However, AAA proteins could have a much more direct role in lipid metabolism. Here, we provided evidence suggesting that AAA proteins may regulate lipid transport by controlling membrane association/dissociation of soluble lipid carriers. We show that members of the Osh protein families interact with AAA proteins and that the Osh proteins become trapped on membranes when AAA proteins' function is compromised.
Osh6p and Osh7p both interact with Vps4p, an AAA ATPase that plays a vital role in the MVB sorting pathway by catalyzing the dissociation and disassembly of all three ESCRT complexes from endosomal membranes (Katzmann et al, 2002). We show that Osh6p and Osh7p interact with Vps4p by the yeast two-hybrid assay and by GST pull-down. Deletion of VPS4 resulted in significant redistribution of Osh6p and Osh7p to the membrane fraction, a phenotype that is shared with components of the ESCRT complexes. We further demonstrate that the accumulation of Osh6p and Osh7p in the membrane fraction is a specific and direct consequence of loss of Vps4p ATPase activity (Figure 4C). These results suggest that Osh6p and Osh7p may transiently associate with cell membranes and that their dissociation requires the ATPase activity of Vps4p. It is interesting to note that the putative coiled-coil motif of Osh7p interacts much more strongly with Vps4p than the full-length protein and exhibits dominant negative effects upon overexpression. One possibility is that the coiled-coil motif is partially masked by a lipid ligand, such as ergosterol (Figure 2D), and only upon removal of this lipid ligand is the coiled-coil domain available for interaction with Vps4p. Results from a recent study also lend some support to this hypothesis. Wang et al (2005) showed that OSBP is a cholesterol-regulated scaffolding protein that oligomerizes with a member of the PTPBS family of tyrosine phosphatases and the serine/threonine phosphatase PP2A. Cholesterol depletion leads to a conformation change in OSBP, resulting in the disassembly of a high molecular weight complex. However, it is not clear how cholesterol depletion breaks protein–protein interactions. Based on results from this study, an interesting possibility is that cholesterol depletion causes exposure of a domain on OSBP for interaction with an AAA ATPase, which provides forces to break protein–protein interactions. This possibility remains to be further tested in both yeast and mammalian systems.
Our results also suggest that Osh6p and Osh7p may not exist in the same membrane-associated protein complex. Although both of them relocate to the membrane fraction in the absence of Vps4p, Osh6p was soluble in Triton X-100 whereas Osh7p was resistant to detergent extraction. Osh7p could therefore oligomerize into a very high molecular weight protein complex with other proteins, a common feature of class E VPS proteins (Babst et al, 1998). Although Osh7p behaves in a strikingly similar manner to the core components of the ESCRT complex, such as Snf7p or Vps24p, we argue that the interaction between Osh7p and Vps4p likely represents a novel pathway that is separate from the MVB pathway and does not involve other class E VPS proteins. First, deletion of OSH6, OSH7 or both has no effect on the VPS pathway (P Wang and H Yang, unpublished observations). In fact, when all seven OSH genes are deleted, no CPY sorting defect was detected (Beh and Rine, 2004). Second, deletion of VPS2 or VPS24 has little effect on the membrane association of Osh7p. The Vps2p–Vps24p subcomplex of ESCRT III recruits Vps4p to the endosomes to catalyze the release of class E VPS proteins (Babst et al, 2002a). If Osh7p oligomerizes with class E proteins, it should remain oligomerized and trapped on the membranes when either VPS2 or VPS24 is deleted. Third, the decrease in sterol esterification was only detected in vps4Δ cells but not in other class E vps mutants tested, including vps23Δ, vps27Δ and vps36Δ strains. Current ongoing effort in our laboratory is to identify the proteins that oligomerize with Osh6p and Osh7p.
As an extension to our studies on Vps4p and Osh6/7p, we found that Afg2p, an essential AAA protein that has strong homology to Cdc48p (Patel and Latterich, 1998), regulates the membrane association of Osh1p and sterol metabolism (P Wang and H Yang, unpublished results). A large-scale characterization of multiprotein complexes in yeast has previously identified Afg2p and Osh1p in the same protein complex (Gavin et al, 2002). Therefore, it appears that these two families (OSBP and AAA) of proteins may function together to control subcellular lipid transport. Furthermore, it is conceivable that Afg2p could also regulate the membrane association/oligomerization of other lipid binding proteins, especially that of CERT, a protein with high structural and sequence similarity to that of OSBP. One of the outstanding issues in CERT-mediated ceramide transport is the requirement for ATP. ATP might be needed to produce phosphatidylinositol-4-phosphate or it might be required for phosphorylation as proposed by Hanada et al (2003) and Munro (2003). Our data, however, suggest that ATP might be required for AAA proteins to mediate the membrane dissociation and recycling of CERT into cytoplasm.
In summary, the data presented here represent the first physical and functional link between two important families of proteins: the OSBP family and the AAA ATPases. Our results offer exciting new avenues for future research. For instance, is there an AAA protein that interacts with Osh4p/Osh5p and regulates their membrane association? If so, how would it affect the SEC14 pathway? What about AAA proteins and mammalian OSBP and ORPs? Do AAA proteins play any role in the regulation of CERT, whose function requires ATP? Studies along these lines may not only provide mechanistic insights into the function of OSBP and ORPs, but may also further our understanding on the mechanism(s) of cellular lipid transport in general.
Materials and methods
Materials
Mouse anti-Pep12p, Dpm1p, ALP, Vph1p, V-ATPase60kD subunit, Vps10p, Porin and rabbit anti-GFP, hexokinase antibodies were purchased from Molecular Probes (Eugene, OR, USA). Rabbit anti-Tlg1p was a gift from Professor W Hong (IMCB, Singapore) (Coe et al, 1999). Rabbit anti-Gas1p was a gift from Dr Howard Riezman (University of Geneva, Switzerland). [3H]oleic acid (5.0 mCi/ml) and [14C]cholesterol (0.1 mCi/ml) were obtained from Amersham Biosciences (Uppsala, Sweden). Rabbit anti-Osh6p/Osh7p antibodies were made as part of this study.
Construction of plasmids and strains
Subcloning was carried out as described (Sambrook et al, 1989). Yeast plasmid transformation was performed using a lithium acetate/single-stranded carrier DNA/polyethylene glycol method (Gietz and Woods, 2002). To disrupt a gene of interest, PCR-based method was applied (Baudin et al, 1993). See Tables I and II for plasmids and strains used in this study.
Table 1.
Plasmids used in this study
| Plasmids | Description | References |
|---|---|---|
| pGEXOSH5 | pGEX4T-1, GST, OSH5 | |
| pGEXOSH6 | pGEX4T-1, GST, OSH6 | |
| pGEXOSH7 | pGEX4T-1, GST, OSH7(1–437/end) | |
| pGEXOSH7CC | pGEX4T-1, GST, OSH7CC(366–437/end) | |
| YCpOSH6-GFP | YCplac111-scGFP, OSH6 promoter, OSH6-GFP, LEU2 | |
| YCpOSH7-GFP | YCplac111-scGFP, OSH7 promoter, OSH7-GFP, LEU2 | |
| pRS316-OSH6-GFP | pRS316, OSH6 promoter, OSH6-GFP, URA3 | |
| pASOSH6 | pAS2-1, GAL4opBD, OSH6, TRP1 | |
| pACTVPS4 | pACT2, GAL4opAD, VPS4, LEU2 | |
| YCpVPS4MYC | YCplac111, VPS4 promoter, VPS4-MYC, LEU2 | Zahn et al (2001) |
| YCpOSH4-GFP | YCplac111-scGFP, OSH4 promoter, OSH4, LEU2 | |
| pRS316-OSH7-GFP | pRS316, OSH7 promoter, OSH7-GFP, URA3 | |
| pJGOSH7 | pJG4-5 LexAop AD, OSH7, TRP1 | |
| pJGOSH7CC | pJG4-5 LexAop AD, OSH7CC (366–437/end), TRP1 | |
| pLexAVPS4 | pLexA, LexAop BD, VPS4, HIS3 | Yeo et al (2003) |
| pRS316VPS4 | pRS316, VPS4 promoter, VPS4, URA3 | |
| pRS316VPS4m | pRS316, VPS4 promoter, vps4E233Q, URA3 | |
| PSH18-34 | PSH18-34, pGAL1, LexA op, LacZ, URA3 | BD Biosciences |
| pGEX4T-1 | GST, copper-inducible promoter | Amersham |
| pYEX4T-1 | Pcup1, GST, URA3, leu-2d, 2μ | Clontech |
| YCplac111 | CEN, LEU2 | Yeo et al (2003) |
| YCplac111-scGFP | CEN, LEU2, GFP | Yeo et al (2003) |
| YEplac181-scGFP | 2μ, LEU2, GFP | Yeo et al (2003) |
| pAS2-1 | GAL4opBD, TRP1 | BD Biosciences |
| pACT2 | GAL4opAD, LEU2 | BD Biosciences |
| pLexA | LexAop BD, HIS3 | BD Biosciences |
| pJG4-5 | LexAop AD, TRP1 | BD Biosciences |
| pESC | GAL1, GAL10 promoter, LEU2 | Stratagene |
| pESCOSH7CC | pESC, GAL1 promoter, OSH7CC (366–437/end), LEU2 | |
| pESCOSH7 | pESC, GAL1 promoter, OSH7 (1–437/end), LEU2 | |
| pGO45 | pRS426, GFP-CPS, URA3 | Odorizzi et al (1998) |
| YCpVPS4 | YCplac111, VPS4 | Zahn et al (2001) |
| YCpvps4-ts | YCplac111, vps4 M307T and L327S | Babst et al (1997) |
| YEpOSH1s-GFP | YEplac181-scGFP, OSH1 promoter, OSH1(615–1188/end) | |
| YCpVPS4GFP | YCplac111, VPS4 promoter, VPS4GFP, LEU2 | |
| Unless specified, all the plasmids were constructed as part of this study. | ||
Table 2.
Strains used in this study
| Strain | Genotype | Sources |
|---|---|---|
| Y10000 | BY4742: MAT α; his31; leu20; lys20; ura30 | EUROSCARF |
| pJ69-4A | MATa; trpl-901; leu2-3,112; ura3-52; his3-200; ga14Δ; ga180Δ; LYS2∷GAL1-HIS3; GAL2-ADE2; met2∷GAL7-lacZ | James et al (1996) |
| Y15394 | BY4742: are2∷kanMX4 | EUROSCARF |
| EGY48 | MAT α; his3; trp1; ura3; 6LexAop-LEU2 | BD Biosciences |
| Y15588 | BY4742: vps4∷kanMX4 | EUROSCARF |
| Y00000 | BY4741: MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | EUROSCARF |
| JRY6326 | SEY6210: TRP1∷PMET3-OSH2; osh1Δ∷kan-MX4; osh2Δ∷kan-MX4; osh3Δ∷LYS2; osh4Δ∷HIS3; osh5Δ∷LEU2; osh6Δ∷LEU2; osh7Δ∷HIS3 | Beh et al (2001) |
| RH448 | MATa his4; leu2; ura3; bar1; lys2 | H Riezman |
| RH2906 | MATa his4; leu2; ura3; bar1; lys2; vps4Δ∷ URA3 | Zahn et al (2001) |
| SEY6210 | MATα, ura3-52, his3Δ200, trp1Δ901, leu2-3, 112; suc2Δ9, lys2-801 | Beh et al (2001) |
| Y14850 | BY4742: vps2Δ∷kanMX4 | EUROSCARF |
| Y14890 | BY4742: vps24Δ∷kanMX4 | EUROSCARF |
| Y13416 | BY4742: vps23Δ∷kanMX4 | EUROSCARF |
| Y15381 | BY4742: vps27Δ∷kanMX4 | EUROSCARF |
| Y15325 | BY4742: vps36Δ∷kanMX4 | EUROSCARF |
| JRY6207 | SEY6210: osh6Δ∷LEU2; osh7Δ∷HIS3 | Beh et al (2001) |
Biochemical assays
Differential centrifugation was performed as described (Gaynor et al, 1994). Briefly, 10 OD600 units of cells were pretreated with 1 ml of suspension buffer (50 mM Tris–HCl pH 8.0, 1% β-mercaptoethanol) at 30°C for 10 min. Cells were then spheroplasted with 1 ml of lysis buffer I (1.2 M sorbitol, 50 mM KH2PO4 pH 7.5, 1 mM MgCl2, 0.15 mg/ml 20T zymolase) at 30°C for 40 min. Spheroplasts were then washed once with 1.2 M sorbitol and then lysed in 1 ml of ice-cold lysis buffer II (50 mM Tris–HCl pH 7.5, 1 mM EDTA, 0.2 M sorbitol) containing complete protease inhibitor. The lysate was cleared at 500 g for 5 min resulting in supernatant S5. S5 was then subjected to centrifugation at 13 000 g for 15 min resulting in supernatant S13 and pellet P13. S13 was centrifuged at 100 000 g for 45 min to produce pellet P100 and supernatant S100. Proteins were trichloroacetic acid (TCA) precipitated from S100. All pellets were dissolved in an appropriate volume of SDS–PAGE sample buffer (5% SDS, 40 mM Tris–HCl pH 6.8, 0.1 mM EDTA, 0.4 mg/ml bromphenol blue, 10% β-mercaptoethanol) containing 8 M urea and resolved by SDS–PAGE.
Sucrose density gradient centrifugation was performed according to a previously described method (Kolling and Hollenberg, 1994). Briefly, cells were lysed using a bead beater in STED10 (10% sucrose w/w in 10 mM Tris–HCl pH 7.6, 1 mM EDTA, 1 mM DTT, protease inhibitor). The resulting lysate was cleared at 300 g for 10 min and loaded onto the top of a linear sucrose gradient (20–50% w/w) and then centrifuged at 100 000 g for 16 h. A total of 13 equal volumes of fractions were collected from the top to bottom and analyzed by Western blotting.
To determine the nature of protein association with P13/P100, membrane floatation and extraction were performed as described (Horazdovsky and Emr, 1993) with modification. The pellets were treated with 1% Triton X-100, 6 M urea, 0.1 M Na2CO3 pH 11, 1 M NaCl, or lysis buffer (50 mM Tris–HCl pH 7.5, 1 mM EDTA, 0.2 M sorbitol) (mock) respectively on ice or room temperature for 30 min, and then centrifuged at 100 000 g for 45 min resulting in S (supernatants) and P (pellets).
Membrane floatation or Western blotting analysis of GFP-CPS was performed essentially according to previously described methods (Babst et al, 1998; Shiflett et al, 2004).
In vitro GST pull-down was performed according to a method of Yeo et al (2003) with modification. In brief, GST fusion proteins were expressed in E. coli BL21 and purified by affinity chromatography using GSH agarose beads. Cells expressing Vps4p-Myc from a YCpVPS4-Myc were lysed with a bead beater in yeast extraction buffer (0.2 M sorbitol, 20 mM HEPES pH 7.5, 1 mM EDTA, 10 mM MgCl2, 0.1 M K-acetate, 1 mM DTT, plus complete protease inhibitor). Triton X-100 (10%) was added to the lysate to a final concentration of 1%. The lysate was incubated on ice for 10 min and then cleared at 100 000 g for 0.5 h. The GST fusion protein-bound GSH beads (for mock, GSH beads without GST fusion proteins were used) were mixed with the cleared yeast lysate. After incubation for 2 h at room temperature, the beads (bound fraction) were spun down and washed thrice, respectively, with ice-cold yeast extraction buffer, yeast extraction buffer containing 0.3 M NaCl and 1 × PBS containing 1% Triton X-100. Proteins in the supernatant (unbound) were TCA precipitated and dissolved in SDS–PAGE sample buffers with 8 M urea. The bound proteins were then eluted from beads with SDS–PAGE sample buffer by heating at 95°C for 3 min.
Fluorescence microscopy
Imaging of GFP chimeras in live cells by fluorescence microscopy was performed as described (Levine and Munro, 2001). FM4-64 short chase was carried out according to a method of Vida and Emr (1995).
Two-hybrid analysis
Yeast two-hybrid screen was performed as described by Yeo et al (2003). The full-length Vps4p was used in the original screening described by Yeo et al. For assays carried out in this study, the full-length OSH7 (1–437/end) or OSH7CC (366–437/end) was cloned into pJG4-5 vector and VPS4 was cloned into pLexA and both plasmids were then transformed into EGY48 together with a LacZ reporter plasmid PSH18-34. For Osh6p, the full-length OSH6 (1–448/end) was cloned into pAS2-1 and VPS4 into pACT-2 for direct two-hybrid assay; both plasmids were then transformed into pJ69-4A (James et al, 1996) with the integrated LacZ reporter gene. β-Galactosidase unit was calculated according to the formula β-galactosidase units=1000A420/(TV OD600), where T is the elapsed time (in minutes) of incubation, V=0.5 ml and OD600=A600 of 1 ml of culture.
Sterol esterification assay
Sterol esterification was assessed by incorporation of [3H]oleic acid into sterol esters as described (Yang et al, 1996). Exponentially growing yeast cells were pulsed with [3H]oleic acid for 30 min at 30°C. For temperature-sensitive strains, cells were preincubated at 37°C for 10 min and then pulsed for 30 min. Cells were harvested and washed twice with ice-cold wash buffer (0.5% NP-40, 20 mM NaN3) and once with distilled water. Cells were then lyophilized overnight. The dried cells were lysed in lysis buffer (1700 U/ml lyticase, 10% glycerol, 0.02% sodium azide) at 37°C for 15 min. The cell suspension was then frozen at −70°C for 1 h and then thawed at 37°C for 15 min. Lipids were extracted with hexane and resolved by TLC.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Acknowledgments
This work was supported by a Young Investigator Award from the National University of Singapore, a grant from the National Medical Research Council, Singapore (to HY) and by A*STAR (Singapore) and the National Health and Medical Research Council (Australia) Project Grant 252750 (to ALM). We are indebted to Mahendra Wagle for his contribution in the early part of this project. We thank Dr ND Ridgway for pointing out the interaction between Afg2p and Osh1p and Drs Ta-Yuan Chang, Peter Pentchev and Richard Deckelbaum for continuous support and encouragement. We are grateful to Dr Victoria Boulton for advice on yeast subcellular fractionation techniques. We also thank Drs H Bergler, H Riezman, S Emr and W Hong for yeast strains, plasmids and antisera.
References
- Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD (2002a) Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell 3: 271–282 [DOI] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD (2002b) Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell 3: 283–289 [DOI] [PubMed] [Google Scholar]
- Babst M, Sato TK, Banta LM, Emr SD (1997) Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J 16: 1820–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Wendland B, Estepa EJ, Emr SD (1998) The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J 17: 2982–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C (1993) A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res 21: 3329–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beh CT, Cool L, Phillips J, Rine J (2001) Overlapping functions of the yeast oxysterol-binding protein homologues. Genetics 157: 1117–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beh CT, Rine J (2004) A role for yeast oxysterol-binding protein homologs in endocytosis and in the maintenance of intracellular sterol-lipid distribution. J Cell Sci 117: 2983–2996 [DOI] [PubMed] [Google Scholar]
- Bishop N, Woodman P (2000) ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol Biol Cell 11: 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe JGS, Lim ACB, Xu J, Hong W (1999) A role for Tlg1p in the transport of proteins within the Golgi apparatus of Saccharomyces cerevisiae. Mol Biol Cell 7: 2407–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson PA, Ridgway ND, Slaughter CA, Brown MS, Goldstein JL (1989a) cDNA cloning and expression of oxysterol-binding protein, an oligomer with a potential leucine zipper. J Biol Chem 264: 16798–16803 [PubMed] [Google Scholar]
- Dawson PA, Van der Westhuyzen DR, Goldstein JL, Brown MS (1989b) Purification of oxysterol binding protein from hamster liver cytosol. J Biol Chem 264: 9046–9052 [PubMed] [Google Scholar]
- Fang M, Kearns BG, Gedvilaite A, Kagiwada S, Kearns M, Fung MK, Bankaitis VA (1996) Kes1p shares homology with human oxysterol binding protein and participates in a novel regulatory pathway for yeast Golgi-derived transport vesicle biogenesis. EMBO J 15: 6447–6459 [PMC free article] [PubMed] [Google Scholar]
- Funato K, Riezman H (2001) Vesicular and nonvesicular transport of ceramide from ER to the Golgi apparatus in yeast. J Cell Biol 155: 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147 [DOI] [PubMed] [Google Scholar]
- Gaynor EC, te Heesen S, Graham TR, Aebi M, Emr SD (1994) Signal-mediated retrieval of a membrane protein from the Golgi to the ER in yeast. J Cell Biol 127: 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350: 87–96 [DOI] [PubMed] [Google Scholar]
- Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M (2003) Molecular machinery for non-vesicular trafficking of ceramide. Nature 426: 803–809 [DOI] [PubMed] [Google Scholar]
- Horazdovsky BF, Emr SD (1993) The VPS16 gene product associates with a sedimentable protein complex and is essential for vacuolar protein sorting in yeast. J Biol Chem 268: 4953–4962 [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandutsch AA, Taylor FR, Shown EP (1984) Different forms of the oxysterol-binding protein. Binding kinetics and stability. J Biol Chem 259: 12388–12397 [PubMed] [Google Scholar]
- Katzmann DJ, Babst M, Emr SD (2001) Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106: 145–155 [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Odorizzi G, Emr SD (2002) Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol 3: 893–905 [DOI] [PubMed] [Google Scholar]
- Kolling R, Hollenberg CP (1994) The ABC-transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO J 13: 3261–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagace TA, Byers DM, Cook HW, Ridgway ND (1997) Altered regulation of cholesterol and cholesteryl ester synthesis in Chinese-hamster ovary cells overexpressing the oxysterol-binding protein is dependent on the pleckstrin homology domain. Biochem J 326: 205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagace TA, Byers DM, Cook HW, Ridgway ND (1999) Chinese hamster ovary cells overexpressing the oxysterol binding protein (OSBP) display enhanced synthesis of sphingomyelin in response to 25-hydroxycholesterol. J Lipid Res 40: 109–116 [PubMed] [Google Scholar]
- Lehto M, Laitinen S, Chinetti G, Johansson M, Ehnholm C, Staels B, Ikonen E, Olkkonen VM (2001) The OSBP-related protein family in humans. J Lipid Res 42: 1203–1213 [PubMed] [Google Scholar]
- Levine TP (2004) Short-range intracellular trafficking of small molecules across endoplasmic reticulum junctions. Trends Cell Biol 14: 483–490 [DOI] [PubMed] [Google Scholar]
- Levine TP, Munro S (1998) The pleckstrin homology domain of oxysterol-binding protein recognises a determinant specific to Golgi membranes. Curr Biol 8: 729–739 [DOI] [PubMed] [Google Scholar]
- Levine TP, Munro S (2001) Dual targeting of Osh1p, a yeast homologue of oxysterol-binding protein, to both the Golgi and the nucleus–vacuole junction. Mol Biol Cell 12: 1633–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Rivas MP, Fang M, Marchena J, Mehrotra B, Chaudhary A, Feng L, Prestwich GD, Bankaitis VA (2002) Analysis of oxysterol binding protein homologue Kes1p function in regulation of Sec14p-dependent protein transport from the yeast Golgi complex. J Cell Biol 157: 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen CJ, Roy A, Levine TP (2003) A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J 22: 2025–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas AN, Martin J (2002) AAA proteins. Curr Opin Struct Biol 12: 746–753 [DOI] [PubMed] [Google Scholar]
- Munro S (2003) Cell biology: earthworms and lipid couriers. Nature 426: 775–776 [DOI] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD (1998) Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell 95: 847–858 [DOI] [PubMed] [Google Scholar]
- Olkkonen VM, Levine TP (2004) Oxysterol binding proteins: in more than one place at one time? Biochem Cell Biol 82: 87–98 [DOI] [PubMed] [Google Scholar]
- Patel S, Latterich M (1998) The AAA team: related ATPases with diverse functions. Trends Cell Biol 8: 65–71 [PubMed] [Google Scholar]
- Reggiori F, Pelham HR (2001) Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J 20: 5176–5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway ND, Dawson PA, Ho YK, Brown MS, Goldstein JL (1992) Translocation of oxysterol binding protein to Golgi apparatus triggered by ligand binding. J Cell Biol 116: 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Press [Google Scholar]
- Shiflett SL, Ward DM, Huynh D, Vaughn MB, Simmons JC, Kaplan J (2004) Characterization of Vta1p, a class E Vps protein in Saccharomyces cerevisiae. J Biol Chem 279: 10982–10990 [DOI] [PubMed] [Google Scholar]
- Storey MK, Byers DM, Cook HW, Ridgway ND (1998) Cholesterol regulates oxysterol binding protein (OSBP) phosphorylation and Golgi localization in Chinese hamster ovary cells: correlation with stimulation of sphingomyelin synthesis by 25-hydroxycholesterol. Biochem J 336: 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor FR, Saucier SE, Shown EP, Parish EJ, Kandutsch AA (1984) Correlation between oxysterol binding to a cytosolic binding protein and potency in the repression of hydroxymethylglutaryl coenzyme A reductase. J Biol Chem 259: 12382–12387 [PubMed] [Google Scholar]
- Vida TA, Emr SD (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol 128: 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walworth NC, Goud B, Ruohola H, Novick PJ (1989) Fractionation of yeast organelles. Methods Cell Biol 31: 335–356 [DOI] [PubMed] [Google Scholar]
- Wang PY, Weng J, Anderson RG (2005) OSBP is a cholesterol-regulated scaffolding protein in control of ERK1/2 activation. Science 307: 1472–1476 [DOI] [PubMed] [Google Scholar]
- Wyles JP, McMaster CR, Ridgway ND (2002) Vesicle-associated membrane protein-associated protein-A (VAP-A) interacts with the oxysterol-binding protein to modify export from the endoplasmic reticulum. J Biol Chem 277: 29908–29918 [DOI] [PubMed] [Google Scholar]
- Yang H, Bard M, Bruner DA, Gleeson A, Deckelbaum RJ, Aljinovic G, Pohl TM, Rothstein R, Sturley SL (1996) Sterol esterification in yeast: a two-gene process. Science 272: 1353–1356 [DOI] [PubMed] [Google Scholar]
- Yeo SC, Xu L, Ren J, Boulton VJ, Wagle MD, Liu C, Ren G, Wong P, Zahn R, Sasajala P, Yang H, Piper RC, Munn AL (2003) Vps20p and Vta1p interact with Vps4p and function in multivesicular body sorting and endosomal transport in Saccharomyces cerevisiae. J Cell Sci 116: 3957–3970 [DOI] [PubMed] [Google Scholar]
- Zahn R, Stevenson BJ, Schroder-Kohne S, Zanolari B, Riezman H, Munn AL (2001) End13p/Vps4p is required for efficient transport from early to late endosomes in Saccharomyces cerevisiae. J Cell Sci 114: 1935–1947 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2


