Abstract
[URE3] is a prion (infectious protein) of the Ure2 protein of yeast. In vitro, Ure2p can form amyloid filaments, but direct evidence that these filaments constitute the infectious form is still missing. Here we demonstrate that recombinant Ure2p converted into amyloid can infect yeast cells lacking the prion. Infection produced a variety of [URE3] variants. Extracts of [URE3] strains, as well as amyloid of Ure2p formed in an extract-primed reaction could transmit to uninfected cells the [URE3] variant present in the cells from which the extracts were made. Infectivity and determinant of [URE3] variants resided within the N-terminal 65 amino acids of Ure2p. Notably, we could show that amyloid filaments of recombinant Ure2p are nearly as infectious per mass of Ure2p as extracts of [URE3] strains. Sizing experiments indicated that infectious particles in vitro and in vivo were >20 nm in diameter, suggesting that they were amyloid filaments and not smaller oligomeric structures. Our data indicate that there is no substantial difference between filaments formed in vivo and in vitro.
Keywords: amyloid, prion transformation, prion variants, [URE3]
Introduction
Amyloid is a filamentous protein form characterized by high β-sheet content in a cross-β conformation, resistance to protease digestion, and birefringence upon staining with the dye Congo Red (Kirschner et al, 2000). Amyloid in humans is generally a pathologic finding, but amyloid serves a protective function on the surface of Escherichia coli (Chapman et al, 2002), fungi (Gibbink et al, 2005), and fish eggs (Podrabsky et al, 2001). Amyloid diseases are characterized by a spontaneous form and an inherited form. The spontaneous form is believed to be due to the stochastic initiation of amyloid formation, with spread of amyloid through the patient's tissues, similar to the autocatalytic formation of amyloid in vitro. The inherited form of the disease is generally due to alteration of the amyloid-forming protein increasing its tendency to form amyloid, change of a protein needed for amyloid formation or degradation, or elevated production of the amyloid-forming protein.
Among amyloid diseases, only the prion diseases are infectious, and the word ‘prion' means simply ‘infectious protein' (Prusiner, 1982). The transmissible spongiform encephalopathies (TSEs) are naturally infectious by the oral route (as in scrapie of sheep and chronic wasting disease of elk and deer) and are experimentally transmitted by inoculation, but their long incubation periods (months to years) slow the progress of their study. Considerable evidence suggests that the TSEs are infectious forms of the PrP protein, and lack the requirement of a nucleic acid in the infectious material, but definitive proof has been difficult to demonstrate. Progress toward the long-sought goal of showing infectivity of recombinant PrP may have been achieved (Legname et al, 2004), but debate continues even to the present time. Variants (or ‘strains') of the TSEs have been well defined, with different isolates showing different incubation times, disease symptoms, and brain regions affected in genetically identical hosts (reviewed in Bruce, 2003). So far, the molecular nature of these variant differences is not known.
Infectious elements of yeast and filamentous fungi, such as viruses and plasmids, are naturally transmitted by cytoplasmic mixing during mating or experimentally by cytoduction or heterokaryon formation. The Saccharomyces cerevisiae nonchromosomal genetic elements [PSI+] (Cox, 1965) and [URE3] (Lacroute, 1971) were shown to be prions of Sup35p and Ure2p, respectively, based on their unique genetic properties (Wickner, 1994). Both prions exist in different variants (Derkatch et al, 1996; Schlumpberger et al, 2001). Likewise, [Het-s] was discovered as a nonchromosomal genetic element of Podospora anserina (Rizet, 1952), and identified as a prion based on its genetic characteristics (Coustou et al, 1997).
The ease of yeast or fungal prion transmission by cytoplasmic mixing during mating or cytoduction has greatly facilitated study in these systems. Efficient infection of P. anserina with the [Het-s] prion by introduction of amyloid formed in vitro from recombinant HET-s protein showed convincingly that amyloid is not just a side-product, but the infectious material (Maddelein et al, 2002). It provided biochemical proof that [Het-s] is a prion. Similarly, amyloid formed in vitro from recombinant Sup35p could efficiently convert S. cerevisiae to [PSI+] (King and Diaz-Avalos, 2004; Tanaka et al, 2004). This method has been applied to study the structural differences that are presumed to account for different prion variants.
The [URE3] prion is a self-propagating inactive amyloid form of Ure2p (Wickner, 1994), a negative transcriptional regulator of genes encoding enzymes and transporters (such as the allantoate permease Dal5p) needed for catabolism of poor nitrogen sources (Cooper, 2002; Magasanik and Kaiser, 2002). In cells with a good nitrogen source, such as ammonia or glutamine, Ure2p binds to the transcription factor Gln3p, keeping it in the cytoplasm and thereby preventing expression of DAL5 (among many other genes). Uptake by Dal5p of ureidosuccinate (which resembles allantoate and is an intermediate in uracil biosynthesis) enables growth of ura2 strains on ammonia media without uracil (USA+), and is used as an assay of Ure2p activity (Lacroute, 1971; Turoscy and Cooper, 1987).
The N-terminal 65 residues of Ure2p are necessary and sufficient for prion generation and propagation; this region was designated the prion domain (Masison and Wickner, 1995; Masison et al, 1997). In [URE3] prion-containing cells, Ure2p is protease resistant and aggregated, and filaments of Ure2p can be visualized (Masison and Wickner, 1995; Edskes et al, 1999; Speransky et al, 2001). Just as the Ure2p prion domain induces prion formation in vivo, it can induce amyloid formation by the full-length protein in vitro (Taylor et al, 1999). Although substantial correlative evidence indicates that amyloid of Ure2p is the infectious prion form, no direct evidence exists for this conclusion. It has even been proposed that the [URE3] prion might be structurally different from in vitro-formed amyloid filaments of Ure2p (Ripaud et al, 2004). Another group has proposed that prion formation involves filaments formed by a repeated head-to-tail interaction between N-terminus and C-terminus (Bousset et al, 2002, 2004).
Using the earlier work on [PSI+] as a starting point, we sought to establish a transformation procedure with recombinant Ure2p in order to confirm that amyloid is the molecular basis of [URE3], the N-terminal prion domain as the mediator of amyloid and prion structure, and to initiate the study of the structural basis of prion variants and infectivity of this prion. An ADE2-based reporter system enabled us to demonstrate and quantify infectivity in yeast for recombinant amyloid as well as cellular extracts. It showed that in both cases, infectious particles are >20 nm. Interestingly, some in vitro filament fractions were nearly as infectious as in vivo filaments.
Results
Construction of reporters to monitor and select [URE3] in yeast cells
The assay for [URE3] measures deficiency of active Ure2p resulting in inappropriate expression of DAL5 (Lacroute, 1971; Turoscy and Cooper, 1987), making cells USA+ (Figure 1B). Initial attempts to transform Ure2p filaments and extracts from [URE3] cells into protoplasts from ura2Δ strains selecting USA+ cells were unsuccessful (data not shown).
Figure 1.
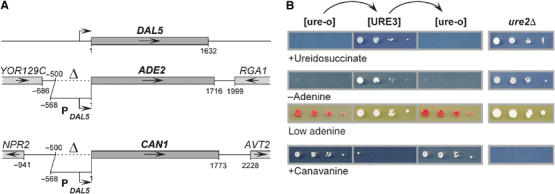
Assays for [URE3]. Ure2p negatively regulates the DAL5 promoter in the presence of a rich nitrogen source. Inactivation of Ure2p, either by prion formation, or in ure2Δ strains, leads to transcription from the DAL5 promoter. DAL5 encodes the allantoate transporter, which also takes up ureidosuccinate, the product of Ura2p (aspartate transcarbamylase). In this study, the DAL5 promoter was placed upstream of the ADE2 and CAN1 open reading frames. In [URE3] or ure2Δ strains containing these constructs, a derepressed DAL5 promoter allowed growth of a ura2 mutant when ureidosuccinate was substituted for uracil. Adenine prototrophy, white colony color on limiting adenine, and sensitivity to the arginine analog canavanine taken up by Can1p can be observed. (A) Schematic representation of the genomic DAL5, ADE2, and CAN1 loci in the reporter strains. Dotted lines indicate the deleted promoter regions. (B) BY334 cells lacking [URE3] (‘[ure-o]') were converted to [URE3] by cytoduction from strain 3310 (‘[URE3]'). Those cytoductants were cured of [URE3] by growth for 3 days on 1/2 YPD plates containing 3 mM guanidine (‘[ure-o]'). ure2Δ control strains were 4132 and BY256. Serial 10-fold dilutions were spotted on SD+Ade,HLW plates containing 33 mg/l ureidosuccinate (‘+Ureidosuccinate'), on SC−Ade plates (‘−Adenine'), on 1/2 YPD plates (‘Low adenine'), and on HC−R plates containing 200 mg/l canavanine (‘+Canavanine').
The usefulness of the red ade2 marker in work on the [PSI+] prion (Cox, 1965) and its previous successful adaptation to the [URE3] system (Schlumpberger et al, 2001) led us to construct a variety of new reporters of [URE3]. We replaced 500 bp of the chromosomal ADE2 promoter with 568 bp of the DAL5 promoter (Figure 1A), placing the ADE2 gene under Ure2p control. An active Ure2p makes such a strain Ade− and red on adenine-limiting medium (1/2 YPD) because of the accumulation of a red pigment in ade2 mutants. The intensity of the red color is proportional to the block of ADE2 transcription, with [URE3] clones being white and [ure-o] (the absence of [URE3]) clones being red. Thus, [URE3] can be directly assessed and variants of [URE3] can be distinguished by color intensity and stability, as has been carried out for [PSI+] (Derkatch et al, 1996).
Similarly, we constructed PDAL5:CAN1 by replacing 500 bp of the chromosomal CAN1 gene with 568 bp of the DAL5 promoter (Figure 1A). CAN1 encodes the arginine permease, the only permease that takes up the toxic arginine analog canavanine. Therefore, as PDAL5:ADE2 allows selection for [URE3] cells, PDAL5:CAN1 allows selection against [URE3] cells, since canavanine kills cells with an active Can1p.
A wild-type ([ure-o]) strain has active Ure2p, so transcription of PDAL5:ADE2 and PDAL5:CAN1 is shut off and cells are Ade−, dark red, and resistant to canavanine (Figure 1B). Introduction of [URE3] by cytoduction inactivates Ure2p, thereby derepressing transcription and makes cells Ade+, white, and canavanine sensitive, like a ure2Δ strain (Figure 1B). Curing [URE3] by growth on 3 mM guanidine (M Aigle and F Lacroute, cited in Cox, 1993) restores the normal phenotype (Figure 1B). Different strains carrying the same [URE3-1] variant (Lacroute, 1971) can show differences in redness and in frequency of [URE3] loss (Supplementary Figure 1).
Establishment of the transformation procedure
Following and adapting the procedure of Tanaka et al (2004), we introduced filaments or extracts into spheroplasts, cotransforming with the LEU2 2μ plasmid pH125 to select cells that had taken up materials from the transformation mixture. Plating on complete medium with 1 M sorbitol and lacking adenine and leucine, and plates lacking only leucine, we found that transformation of Ure2p filaments (Figure 2A) indeed gave Ade+ colonies, although with very low efficiency (not shown). Sonication of filaments (Figure 2B) increased conversion to [URE3] to about 5% of total Leu+ pH125 transformants (not shown). All Ade+ clones were curable by guanidine, and sample clones were shown transmissible by cytoduction, indicating that they were [URE3]. However, among the clones selected only for Leu+, the number of Ade+ colonies was lower than when both prototrophies were simultaneously selected (not shown), pointing to an effect of the adenine concentration on the establishment of [URE3].
Figure 2.
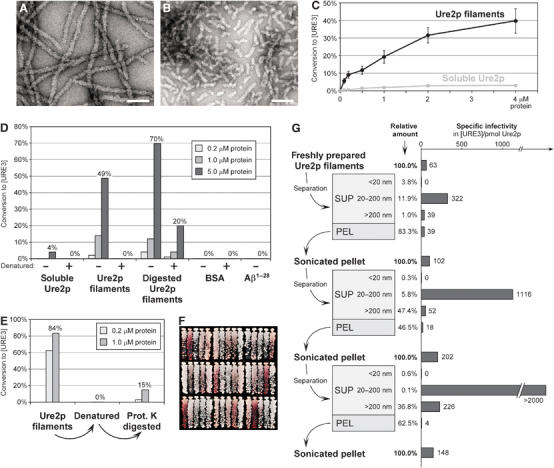
Transformation of in vitro-formed Ure2p filaments into yeast spheroplasts. (A) Electron micrograph of negatively stained Ure2p filaments. Bar, 100 nm. (B) Negatively stained Ure2p filaments after sonication two times for 15 s at 60 W. Bar, 100 nm. (C) Ure2p concentration dependence of conversion to [URE3]. Indicated concentrations of Ure2p filaments and soluble Ure2p were transformed into BY241. Values of [URE3] colonies relative to all Leu+ transformants are the mean of at least three independent experiments (±standard error). (D) Infectivity of proteinase K-digested Ure2p filaments (see Materials and methods) and effect of heat denaturation. Heat denaturation of protein samples was achieved by incubation at 95°C for 5 min. Indicated protein concentrations were transformed into BY241. (E) Recovery of infectivity from heat-denatured Ure2p filaments by proteinase K digestion. Heat denaturation, proteinase K treatment, and transformation were performed as in panel D. (F) Spectrum of [URE3] variants in randomly chosen colonies after transformation of BY241 with Ure2p filaments. (G) Fractionation of freshly prepared Ure2p filaments (see Materials and methods) and specific infectivity following transformation into BY241. The starting concentration of Ure2p filaments was 2 μM; concentrations of each fraction are presented as relative values. Electron micrographs of filament fractions and [URE3] variant spectra of the respective transformants are provided in Supplementary Figure 3.
To establish the optimal adenine concentration for recovery of [URE3] clones, sonicated filaments were transformed into cells and plated at different adenine concentrations. With increasing adenine concentration in the medium, the number of white ([URE3]) colonies increased, but at still higher adenine concentrations, abundant red Ade− [ure-o] colonies appeared, and at even higher concentrations, all colonies were white because adenine was no longer limiting (Supplementary Figure 2A). Thus, there is an optimum adenine concentration of approximately 0.5 mg/l in the top agar (equals 0.17 mg/l adenine in the whole plate) for recovery of [URE3] clones after transformation (Supplementary Figure 2B). The tested strains differed slightly in their optimum concentration, but the standardized procedure described in Materials and methods gave reproducible transformation efficiencies with most strains without allowing growth of many Ade− clones (Supplementary Figure 2A and B). Among the strains tested, BY241 was the best, and was used in subsequent experiments. Similar optimization of selection for USA+ colonies (adding low amounts of uracil to the selection plates) improved yields of [URE3] transformants, but transformation efficiencies remained <1% (not shown).
Characterization of infectious material
Ure2p filament transformation showed concentration dependence (Figure 2C). Even soluble Ure2p gave rise to some [URE3] transformants at higher concentrations. Infectivity of soluble Ure2p went up with length of storage at −80°C, suggesting that this infectivity was due to spontaneous filament formation. Proteinase K digestion of Ure2p filaments destroys the C-terminal part of the molecule and leaves only a highly resistant amyloid core fiber consisting of residues 1 to ∼70, essentially the prion domain (Baxa et al, 2003). Proteinase K-digested filaments were as infectious as undigested filaments (Figure 2D), pointing to the N-terminus or the core fiber being the infectious element. BSA and Aβ amyloid did not induce the appearance of [URE3] (Figure 2D). Heat denaturation led to complete loss of infectivity, but not in the case of filaments already digested with proteinase K (Figure 2D). For denatured filaments of full-length Ure2p, the activity was partially restored by proteinase K digestion (Figure 2E), suggesting that aggregation of the heat-denatured filaments prevented them either from acting as seeds or from entering the spheroplasts.
Transformation with either Ure2p filaments (Figure 2F), protease-digested filaments, or soluble Ure2p (not shown) gave rise to a number of different [URE3] variants, as shown by different color on 1/2 YPD medium. The spectrum of colors on 1/2 YPD was somewhat dependent on recipient strain, but was independent of the amount of adenine in the selection plates (Supplementary Figure 2C).
Ure2p filaments were fractionated by sedimentation and filtration to determine the size of the infectious material (Figure 2G). No infectivity was found in samples passing a 20 nm filter, and only low infectivity was found in the 13 000 g pellet or in material retained by a 200 nm filter. The highest infectivity was found for filaments between 20 and 200 nm, exactly the size of the filaments in Figure 2B. Sonication of the pellet fraction released new infectious particles from the larger filaments, and again most of this infectivity was in the range of 20–200 nm. Again, no infectivity was present in particles less than 20 nm. Repeating this procedure on the second pellet fraction again generated increased infectivity (Figure 2G), indicating that the large material carried latent infectivity. Electron micrographs of the filament fractions revealed that infectivity correlated with the presence of small filament fragments, especially obvious in the supernatant fractions <200 nm (Supplementary Figure 3). Pellet fractions contained larger aggregates from which smaller filaments were liberated through sonication (Supplementary Figure 3). The noninfectious fractions <20 nm did not contain any visible material, although they contained up to 4% of the total protein (not shown). Size fractionation had no apparent influence on the spectrum of variants obtained (Supplementary Figure 3).
Transformation of filaments from N-terminal fragments of Ure2p
Filaments of Ure2p, Ure2p1−89, Ure2p1−65, and Ure2p10−39, along with protease-digested filaments of Ure2p were prepared and vigorously sonicated prior to transformation (Figure 3A). All except Ure2p10−39 were infectious, but Ure2p1−89 and Ure2p1−65 were only very weakly so (Figure 3B). As fragmentation of filaments of Ure2p increased their infectivity, the reduced infectivity of the filaments composed of shorter Ure2p fragments (and even the apparent lack of infectivity of Ure2p10−39) may be due to clumping, limiting their ability to enter the cells. A similar spectrum of variants was observed for each of the fragments (Figure 3C). For Ure2p1−65, this was unexpected, since Schlumpberger et al (2001) observed only a single variant when inducing [URE3] by overexpression of this fragment. To explore this discrepancy further, we induced [URE3] appearance by overexpression of Ure2p1−65 in several strains (Supplementary Figure 4). We observed distinguishable [URE3] variants in each host strain tested, but the degree of variation depended on the strain, suggesting that yeast strain differences might be responsible for the disagreement.
Figure 3.
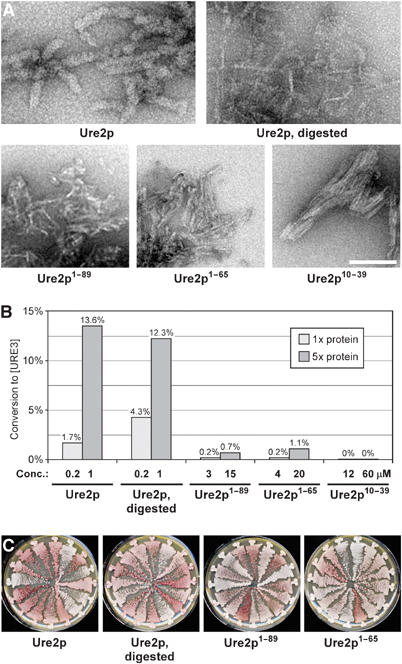
Transformation of filaments from Ure2p N-terminal fragments. (A) Negatively stained filaments of the indicated proteins. Bar, 100 nm. (B) Infectivity of filaments from Ure2p N-terminal fragments in comparison to full-length Ure2p filaments and proteinase K-digested Ure2p filaments. Indicated protein concentrations were transformed into BY241. (C) Spectrum of [URE3] variants in randomly chosen transformants obtained from the successful transformations in panel B.
Transformation of filaments from fusion proteins
To test the influence of C-terminal domains on prion transmission and variant determination, we prepared and tested the infectivity of proteins in which different N-terminal fragments of Ure2p were fused to various enzymes, aspartate aminotransferase (AAT), green fluorescent protein (GFP), glutathione S transferase (GST), and carbonic anhydrase (CA). Ure2p1−90-AAT, Ure2p1−80-GFP, Ure2p1−65-GFP, Ure2p1−65-GST, and Ure2p1−65-CA all formed filaments readily during agitated incubation (Figure 4A), although with different kinetics. After sonication, Ure2p1−90-AAT filaments remained very soluble and showed a gel-like appearance upon sedimentation, whereas Ure2p1−65-CA filaments remained very long, which can be seen by electron microscopy (EM) (Figure 4A). All fusion protein filaments were infectious upon transformation into yeast, but the infectivity varied greatly and correlated with the solubility of the respective filaments, with Ure2p1−90-AAT filaments being at least 10 times more infectious than Ure2p filaments (Figure 4C). Ure2p1−65-CA filaments showed only very low infectivity, probably due to their large size (Figure 4A and C). The different fusion proteins differed also in the infectivity of the soluble protein (Figure 4C). This correlated with the rate of spontaneous filament formation, with Ure2p1−80-GFP being the fastest and Ure2p1−90-AAT being the slowest. Indeed, filaments similar to those previously described (Baxa et al, 2002) could be visualized by EM in ‘soluble' Ure2p1−80-GFP (data not shown). Each of the fusion protein filaments gave rise to a similar spectrum of [URE3] variants (Figure 4B).
Figure 4.
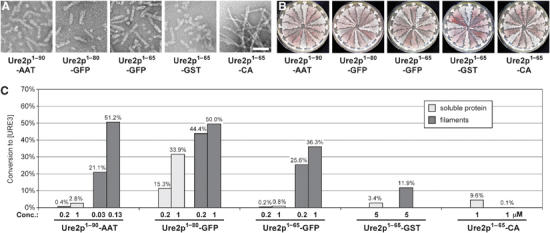
Transformation of fusion protein filaments. (A) Negatively stained filaments of the indicated proteins. Bar, 100 nm. (B) Spectrum of [URE3] variants resulting from transformation of BY241 with fusion protein filaments. In the case of Ure2p1−65-CA, transformants were obtained from transformation of soluble protein. (C) Infectivity of soluble protein and filaments from fusion proteins, as determined by transformation of strain BY241 with the indicated concentrations of protein.
Transformation of [URE3] variants
We selected three different [URE3] variants that were easily distinguishable. Variant 1 is the original [URE3-1] isolate (Lacroute, 1971), while variants 2 and 3 arose from the induction experiment in Supplementary Figure 4. Variants could be distinguished in BY241 by their color on 1/2 YPD plates (1 and 2 are white, 3 is pink), their stability (1 is very stable and almost never reverts, 2 and 3 are unstable, but revert with different frequencies) and their canavanine sensitivity (only 1 is sensitive, 2 and 3 are not) (Figure 5A). These characteristics were maintained after transfer through a different strain. Although the variants behaved slightly differently in these strains, they were distinguishable in either host (Figure 5A and Supplementary Figure 5). When Ure2p1−80-GFP fusion proteins were expressed in the different [URE3] strains, fluorescent foci within the cells could be observed by fluorescence microscopy, whereas in [ure-o] strains the fluorescence was evenly distributed within the cytoplasm (data not shown). No obvious differences between the three [URE3] variants could be detected.
Figure 5.
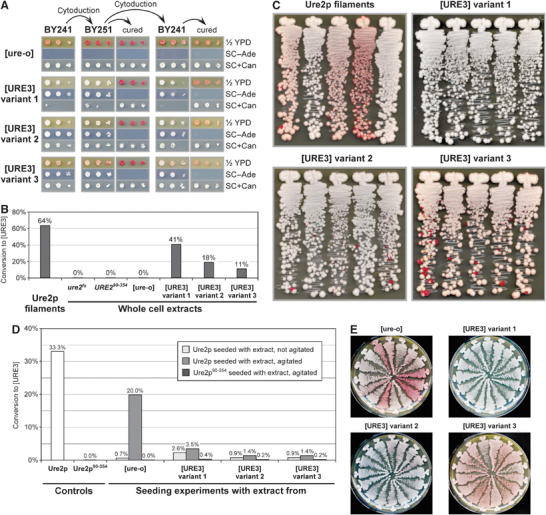
Transformation of [URE3] variants. (A) Conservation of variant characteristics upon transmission of [URE3] variants. The cytoplasm of BY241 strains that were [ure-o] or one of the three different [URE3] variants was transferred to BY251 by cytoduction, and then cytoduced back into BY241. Recipient clones were cured of [URE3] by growth on 1/2 YPD plates containing 3 mM guanidine, and demonstrated reversion to [ure-o] in all cases. Serial 10-fold dilutions were spotted on the indicated media and incubated at 30°C for 3 days (1/2 YPD and SC+Can) or 5 days (SC−Ade). Note the differences in color on 1/2 YPD plates and sensitivity to canavanine between the three [URE3] variants and between the two strains. Data from a parallel experiment performed with BY252 instead of BY251 are provided in Supplementary Figure 5. (B) Infectivity of whole cell extracts from strains BY259 (ure2fs), BY277 (URE290−354), and BY241 with [ure-o] or one of the three [URE3] variants, as determined by transformation into BY241. Protein extracts were transformed at the following concentrations (in mg/ml): ure2fs, 0.5; URE290−354, 0.8; [ure-o], 1.3; [URE3] variant 1, 1.1; [URE3] variant 2, 1.3; [URE3] variant 3, 1.3. A control transformation was performed with 10 μM Ure2p filaments. Control transformations to ensure that the obtained [URE3] clones are indeed transformants and not cells remaining in the extracts are described in Supplementary data. (C) Spectrum of [URE3] variants following successful transformations in panel B. Five randomly chosen clones were streaked on 1/2 YPD plates and incubated at 30°C for 5 days. (D) Seeding experiment with whole cell extracts. Solutions of Ure2p and Ure2p90−354 were incubated with or without agitation (on a vertical roller at 10 r.p.m.) at 4°C for 12 h with whole cell extracts from BY241 strains that were [ure-o] or contained one of the three [URE3] variants. The Ure2p to extract ratio was roughly 1/150 and corresponds to a Ure2p ratio of about 1/2 × 106. The solutions were sonicated and transformed into BY241 at a concentration of 2 μM. For comparison, soluble Ure2p and Ure2p90−354 were incubated in the same way and also transformed into BY241. (E) Spectrum of [URE3] variants following transformation of agitated seeding solutions in panel D.
Protein extracts were made from the same host carrying each of these variants, as well as from URE2 [ure-o], ure2fs, and URE290−354 (expressing only Ure2p90−354) strains. Extracts were transformed into strain BY241 and, as expected, only extracts from [URE3] strains were infectious (Figure 5B). Variant 1 was modestly more infectious than variant 2 or 3, possibly because the former was more stable while about 50% of cells in the cultures of variants 2 and 3 were already [ure-o] at the point when extracts were made (data not shown). Transformants displayed characteristics of the variants from which the extracts were made (Figure 5C), with faithful transmission of ADE2 expression level, stability, and canavanine phenotype of the donor.
In vitro seeding experiments were carried out in which 1/150 part (w/w) of these extracts by protein (approximately 1/2 000 000 part by weight of Ure2p) was added to purified full-length Ure2p, filament formation was allowed to occur, and the products were sonicated and used to transform. Compared to a control lacking the prion domain (Ure2p90−354), or a control with extract alone (data not shown), a 5- to 10-fold increase in infectivity was obtained (Figure 5D). At least 90% of transformants showed variant characteristics of the donor, whereas the transformants from the [ure-o] seeding experiment, or Ure2p without any seeding, included a range of variants (Figure 5E). Surprisingly, incubation of Ure2p with the [URE3] extracts led to a substantial reduction in infectivity compared to incubation of Ure2p alone or with [ure-o] extract, probably by very effective sequestration of soluble Ure2p into the existing filaments before spontaneous filament formation could occur (Figure 5D). Cotransformation of preformed Ure2p filaments with extracts from URE2 [ure-o], ure2fs, or URE290−354 strains had no influence on infectivity (data not shown). Seeding experiments were complicated by the rapid filament formation of Ure2p protein alone. This was evident in unsuccessful attempts to seed with higher dilutions of extracts and in attempts at reseeding (Supplementary Figure 6). We suggest that the seeded reactions quickly form large filaments with a low specific infectivity, but the unseeded reactions spontaneously form many smaller, more infectious particles.
Extracts of [URE3] strains can also seed filament formation by some fusion proteins. Ure2p1−90-AAT gives particularly clear results with fidelity of variant propagation (Supplementary Figure 6B), although reseeding was unsuccessful in this case as well.
Separation of [URE3] extracts prior to transformation was performed as described above for Ure2p filaments. Once again, no infectivity was observed in fractions <20 nm (Figure 6A). No Ure2p protein was found in these fractions from different [URE3] strains, whereas fractions <20 nm from [ure-o] strains contained up to 15% of total Ure2p (data not shown). The pellet fractions were significantly less infectious, but infectivity was enhanced after sonication (Figure 6A). In contrast to in vitro-formed filaments (Figure 2), [URE3] extracts showed more infectivity in larger particles (Figure 6A), suggesting that their form or association with other proteins in the extracts might facilitate transformation. Overall, infectivity from [URE3] extracts was one to two orders of magnitude higher than that of recombinant in vitro filaments. As noted before, simple addition of [ure-o] cell extracts to transformations of Ure2p filaments did not increase infectivity (data not shown). Fractionation had no influence on the faithful transmission of variants (Figure 6B).
Figure 6.
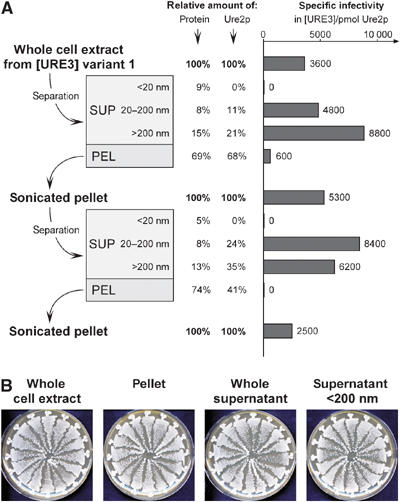
Fractionation of whole cell extracts. (A) Cellular extract of strain BY251 [URE3] variant 1 was fractionated (see Materials and methods) and transformed into BY241. The starting concentration was 25 mg/ml; relative amounts of Ure2p were estimated by determining whole protein concentration and relative Ure2p content in each fraction (see Materials and methods). Similar results were obtained with cell extracts from different strains and [URE3] variants (data not shown). (B) Fractionation does not interfere with faithful transmission of [URE3] characteristics to the transformants. All randomly chosen transformants resulting from a transformation with BY241 [URE3] variant 1 extract fractions show the characteristic white appearance on 1/2 YPD plates. Extract fractionation was performed as in previous experiments, except that pelleting was performed at 45 000 g for 45 min.
Discussion
Using the PDAL5:ADE2 reporter system, we were able to demonstrate the infectivity of amyloid Ure2p filaments. Infectivity required both the ability to enter cells and the capacity to prime amyloid formation of Ure2p once inside the cell. Thus, particles larger than 200 nm consisting of very long filaments and filament aggregates were poorly infectious, although they could readily prime amyloid formation in vitro and could be made infectious by sonication. Filaments that avidly adhered to each other had lower infectivity than they might otherwise, probably because they could not readily enter cells. The average filament length in the most infectious fractions was 50–200 nm as judged by electron microscopy, equivalent to 100–400 monomers of Ure2p (Baxa et al, 2003). Enhancement of infectivity by sonication of larger Ure2p filaments is also consistent with the filament ends being the initiator of polymerization of cellular Ure2p. Significantly, we never observed infectious particles <20 nm either from amyloid formed in vitro or in extracts of [URE3] cells. This suggests that small oligomers, if they exist, as in other amyloids (Caughey and Lansbury, 2003), are not infectious.
Sonicated amyloid filaments of recombinant Ure2p produced about 100 [URE3] colonies per pmol of protein monomer, but some fractions had a higher specific infectivity of more than 1000 [URE3] per pmol. Under the same conditions, extracts of [URE3] strains produced about 100 [URE3] colonies per μg of total protein. This would be equivalent to only about 6.7 ng of Ure2p, or about 5000 [URE3] colonies per pmol of Ure2p monomer. Thus, based on the weight of Ure2p, infectivity of Ure2p in whole cell extracts is about five-fold higher than infectivity of sonicated filament fractions generated in vitro from recombinant Ure2p. Enhanced infectivity of [URE3] cell extracts could reflect the presence of other cellular components, such as chaperones, that might facilitate the addition of new Ure2p to filaments or prevent aggregation of infectious particles. However, addition of the same proportion of an extract from [ure-o] cells did not enhance infectivity of amyloid filaments made in vitro (data not shown). Alternatively, Ure2p amyloid in extracts may be optimal in size for entry into cells, or associated with membranes or other cellular components that facilitate entry into the spheroplasts. Fractionation of infectious material, using the assay we report here, is one route to examine this issue. Yet another possibility is that a portion of in vitro-formed filaments might adopt amyloid structures that cannot seed prions inside yeast cells. Such filament populations might be either too stable or too unstable and thus cannot propagate to daughter cells.
In experiments with PrP, the difference between in vitro and ex vivo material is many orders of magnitude more pronounced. Amyloid of a fragment of recombinant PrP produces a TSE disease in mice, but only after inoculation of a large amount of material and a prolonged incubation time (Legname et al, 2004). Briefly, inoculation of 15 μg of PrP amyloid produced disease after 520 days, while inoculation of 30 μl of a 1% brain extract infected with the RML strain (about 50 ng of PrP; G Raymond, personal communication) produced disease after 160 days. This 360 days difference in incubation time implies an astronomical difference in titer of >108 (Prusiner et al, 2004; B Chesebro, personal communication), for a total titer/PrP difference of >1010. Although earlier studies showed abundant infectivity of recombinant Sup35p amyloid filaments and of [PSI+] extracts, those data do not allow a quantitative comparison (King and Diaz-Avalos, 2004; Tanaka et al, 2004).
Infectivity clearly resides within the N-terminal domain (the amyloid core), as was shown by infection with protease-digested filaments, N-terminal fragments, and prion domain fusion proteins, all of which have been shown to form amyloid (Baxa et al, 2002, 2005). Furthermore, since particles smaller than 20 nm showed no infectivity and large amyloid filaments could be made more infectious by sonication, it appears to be amyloid that carries infectivity, not a soluble form of Ure2p. The low level of infectivity of soluble Ure2p is probably due to the unavoidable formation of amyloid during the incubations.
Transformation with amyloid formed in vitro gave rise to several variants, distinguished by the two reporters, stability and red color, the latter reflecting the proportion of Ure2p inactivated. We can readily distinguish three variants, but it is likely that use of more variables will allow definition of more. At least three strains of [PSI+] may be distinguished by the degree of deficiency of active Sup35p (i.e. the efficiency of nonsense suppression), by the stability of the prion, and by the ability to be transmitted to certain point mutants in the Sup35p prion domain (Derkatch et al, 1996; King, 2001). Different variants could be propagated faithfully in vitro by Sup35p1−61-GFP (King and Diaz-Avalos, 2004) or generated by transformation with amyloid filaments formed from recombinant full-length Sup35p or Sup35NM (Tanaka et al, 2004). In our experiments, Ure2p did not faithfully maintain strain characteristics upon reseeding, probably because of a high rate of spontaneous filament formation. It is likely that in vitro incubation conditions will differentially affect generation of new filaments and propagation of existing filaments, so that conditions strongly favoring propagation can be found. The use of fusion proteins might limit de novo filament formation. Ure2p1−90-AAT with its low rate of spontaneous aggregation and high solubility of filaments is promising in this regard.
In our hands, digested filaments, N-terminal fragments, and fusion proteins all give rise to the same spectrum of [URE3] variants, implying that the N-terminal residues 1–65 are sufficient, at least for the variants we can distinguish. This is the region shown to be sufficient for prion transmission (Masison and Wickner, 1995; Masison et al, 1997), and to constitute the protease-resistant core of Ure2p amyloid (Baxa et al, 2003). Moreover, the prion domain fusion proteins are also sufficient to faithfully propagate the variants. These findings indicate that the C-terminal part of Ure2p is not involved in the determination of the prion variant, and the variant information is completely contained in the prion domain. The fact that amyloid filament fractions formed in vitro from recombinant Ure2p can be nearly as infectious as extracts of [URE3] strains argues that there is no substantial difference between filaments formed in vivo and in vitro. This disagrees with Ripaud et al (2004), who found subtle differences between protease-digestion patterns of Ure2p in filaments formed in vitro and extracts of [URE3] strains and concluded that the in vitro amyloid is not responsible for the [URE3] prion. It is possible that the differences observed by Ripaud et al are due to the binding of chaperones or other proteins to the filaments in extracts. We also observed differences in the size of infectious particles in vitro and in vivo, although this might reflect better uptake of material from cell extracts.
The exact difference between prion strains has not been precisely defined in any system, but current evidence for TSEs and [PSI+] indicates that the amyloid structures of different strains are distinct (Bessen and Marsh, 1992; Caughey et al, 1998; King and Diaz-Avalos, 2004; Tanaka et al, 2004, 2005). Studies of different amyloid variants of Aβ peptide point to parallel in-register structure with differences in the loop region (Petkova et al, 2005). Similar parallel and in-register arrangements of β-sheets have been found for amyloids of different larger peptides (Petkova et al, 2002; Jayasinghe and Langen, 2004). For structural analysis of Ure2p amyloids, it is critical to note that filaments formed in vitro are a mixture of several different variants, and thus not the ideal material. ‘Cloning' of variants by seeding through cell extracts is one possible path to obtain suitable homogenous material that is actually infectious and behaves as a prion.
The reporter system and transformation procedure described here allowed for the first time the direct study of infectious material in [URE3]. The results, that filament fractions from both in vitro and in vivo material could exhibit almost the same infectivity, and that the infectious material was >20 nm in both cases, might be applicable to the other fungal prion systems, as well. Our approach enables monitoring characteristics and stability of in vitro or ex vivo [URE3] variants and will allow purification of infectious material from extracts for the determination of Ure2p structure and associated proteins.
Materials and methods
Construction of plasmids and reporter strains
A fusion protein of the N-terminal 90 amino acids of Ure2p and AAT was amplified by polymerase chain reaction using primers ub15, ub18, ub20, and ub21 (Supplementary Table I), and as templates pKT41-1 (Baxa et al, 2003) and pJO2 (containing a sequence coding for AAT; kind gift of JF Kirsch). This construct was cloned into pET-17b (Novagene) between the NdeI and XhoI sites. The resulting vector pUB13 codes for the expression of MetHis6-Ure2p1−90-SerSerArg-AAT. All other Ure2p constructs have been described previously (Baxa et al, 2002, 2003). Construction of yeast strains (Table I) is described in Supplementary data.
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| S288c | MATα gal2 mal | |
| 1735 | MATα his− ura2 [URE3-1]a | Aigle and Lacroute (1975) |
| 3310 | MATa arg1 kar1-1 [URE3-1]a | Wickner (1994) |
| 3742 | MATa leu2 trp1Δ ura2 kar1 | This study |
| 3920 | MATα leu2 trp1Δ ura2Δ kar1 | This study |
| 4132 | MATα ade2 his3-11,15 leu2-3,112 ura2Δ ure2Δ kar1 | This study |
| 4887-26C | MATa ade2 his3 leu2 trp1 kar1 | This study |
| ∑1287b | MATα | Grenson et al (1966) |
| YMS23 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura2Δ::loxP ura3::pMS87 | Schlumpberger et al (2001) |
| BY103 | MATα ade2 his3-11,15 leu2-3,112 ura2Δ ure2Δ PDAL5:CAN1 kar1 | This study |
| BY178 | MATα ade2 his3 leu2 trp1 ure2Δ::kanMX PDAL5:CAN1 kar1 | This study |
| BY179 | MATα his3 leu2 trp1 ure2Δ::kanMX PDAL5:ADE2 PDAL5:CAN1 kar1 | This study |
| BY228 | MATa leu2 trp1 ura3 kar1 | This study |
| BY241 | MATa leu2 trp1 ura3 PDAL5:ADE2 PDAL5:CAN1 kar1 | This study |
| BY245 | MATα his3 leu2 trp1 PDAL5:ADE2 PDAL5:CAN1 kar1 | This study |
| BY248 | MATα his3 leu2 trp1 PDAL5:ADE2 PDAL5:CAN1 kar1 | This study |
| BY250 | MATα his3 leu2 trp1 PDAL5:ADE2 PDAL5:CAN1 kar1 | This study |
| BY251 | MATα his3 leu2 trp1 PDAL5:ADE2 PDAL5:CAN1 kar1 | This study |
| BY252 | MATα his3 leu2 trp1 PDAL5:ADE2 PDAL5:CAN1 kar1 | This study |
| BY254 | MATα leu2 trp1 ura3 ure2Δ::kanMX PDAL5:ADE2 PDAL5:CAN1 kar1 | This study |
| BY256 | MATa his3 leu2 trp1 ure2Δ::kanMX PDAL5:ADE2 PDAL5:CAN1 kar1 | This study |
| BY259 | MATα leu2 trp1 ura3 ure2fs PDAL5:ADE2 PDAL5:CAN1 kar1 | This study |
| BY277 | MATα leu2 trp1 ura3 URE290−354 PDAL5:ADE2 PDAL5:CAN1 kar1 | This study |
| BY315 | MATα leu2 PDAL5:ADE2 PDAL5:CAN1 kar1 | This study |
| BY317 | MATa PDAL5:ADE2 PDAL5:CAN1 kar1 | This study |
| BY327 | MATa trp1 ura2 PDAL5:ADE2 PDAL5:CAN1 kar1 | This study |
| BY334 | MATα leu2 ura2 PDAL5:ADE2 PDAL5:CAN1 kar1 | This study |
| BY337 |
MATα trp1 ura2 PDAL5:ADE2 PDAL5:CAN1 kar1 |
This study |
| aOriginal [URE3] isolate from Lacroute (1971). | ||
Spheroplast preparation and transformation procedure
Yeast strains were grown in 50 ml YPAD at 30°C to OD600=0.5, pelleted at 1500 g for 5 min at room temperature, and washed twice with 25 ml ST buffer (1 M sorbitol, 10 mM Tris–HCl, pH 7.5). Cells were resuspended in 5 ml ST buffer and spheroplasted with 10 μl lyticase (10 U/μl in 20% glycerol; Sigma, L-5263) for 40 min at 30°C. Spheroplasts were pelleted at 250 g for 5 min at room temperature, washed twice with 10 ml ST buffer, and then resuspended in 1 ml STC buffer (1 M sorbitol, 10 mM Tris–HCl, 10 mM CaCl2, pH 7.5). A 100 μl portion of this spheroplast suspension was mixed with 5 μl of salmon sperm DNA (2 mg/ml; Sigma, D-1626), 1 μl plasmid pH125 (0.5 mg/ml), and 5 μl solution containing prion particles, either in vitro-formed filaments or whole cell extracts. This mixture was incubated for 10 min at room temperature. Fusion was induced by addition of 900 μl PTC buffer (20% (w/v) PEG 8000, 10 mM Tris–HCl, 10 mM CaCl2, pH 7.5) and incubation for 20 min at room temperature. Spheroplasts were collected by centrifugation at 500 g for 5 min at room temperature, resuspended in 200 μl SOS+HLUW buffer (1 M sorbitol, 7 mM CaCl2, 1/3 YPAD, 20 mg/l histidine, 100 mg/l leucine, 20 mg/l uracil, 20 mg/l tryptophan), and incubated for 30 min at 30°C.
Transformation reactions were mixed with 10 ml liquid top agar at 50°C and immediately poured onto plates with 20 ml of solidified bottom agar. Both top and bottom agar consisted of Hartwell Complete medium (Amberg et al, 2005) with 1 M sorbitol (HCS), lacking leucine, and with either 5 mg/l adenine (HCS+A5−L) for selection of Leu+ colonies or 0.1 mg/l adenine (HCS+A.1−L) for selection of Ade+ Leu+ transformants. Typically, three selection plates were prepared for each individual transformation: 1/10 of the transformation mixture would be plated on HCS+A5−L, and 1/2 and 1/10 plated on HCS+A.1−L. Colony numbers were determined after 6 days incubation at 30°C. Using this method, transformation efficiency of pH125 into BY241 was typically on the order of 5 × 104 Leu+ colonies per μg plasmid. [URE3] transformants on HCS+A.1−L plates were verified by purifying 48 randomly chosen clones on SC−Ade plates (KD Medical Inc.). These plates were replica plated onto YPG plates to determine whether the clones were still ρ+ (this step was necessary since strain BY241 becomes ρ− at an unusually high spontaneous frequency, and this interfered with color determination on 1/2 YPD plates), and onto 1/2 YPD plates with and without 3 mM guanidine on which loss of [URE3] could be scored by reversion to red colony color. With few exceptions, all clones tested were Ade+ and curable, and usually more than 80% of all colonies were ρ+. For every transformation, a negative control without filaments or extracts was performed, which only rarely gave rise to spontaneous [URE3] clones.
Protein preparation and filament formation
Expression and purification of His6-tagged proteins was as described earlier (Baxa et al, 2002, 2003). Filament formation was performed by incubating the filaments at 4°C for about 16 h to 3 days directly in the buffer used for elution from the Ni-NTA column (50 mM sodium phosphate, 300 mM NaCl, 240 mM imidazole, pH 8.0) with agitation for at least 16 h. Proteinase K treatment for digested Ure2p filaments was performed as described earlier (Baxa et al, 2003; Ross et al, 2004). Preparation of yeast extracts is described in Supplementary data.
Filament and extract fractionation
A 1 ml aliquot of filaments or cell extracts was first sedimented at 13 000 g for 1 min at 4°C. Aliquots of the supernatant fractions were then filtered through either 200 or 20 nm Anatop 10 filters (Whatman). The pellet fraction was washed twice with 1 ml H2O and resuspended in 1 ml H2O. Pellets were sonicated two times for 15 s each at 60 W on a Kontes model 9110001 sonicator. Protein in different fractions was quantified using a BCA assay (Pierce). Absolute amounts of Ure2p protein in cell extract fractions were estimated by spotting aliquots on a PVDF membrane (Millipore) together with a concentration series of purified recombinant Ure2p and reacting with anti-Ure2p antibody as described (Baxa et al, 2003). Ure2p was about 0.007% of total yeast cell protein, independent of whether the cells were [ure-o] or [URE3]. This corresponds to approximately 5000 Ure2p monomers per cell, which is similar to previously published data (Ghaemmaghami et al, 2003).
Analysis of prion variants
The spectrum of [URE3] variants obtained in a transformation was typically assessed by streaking 16 randomly chosen clones on 1/2 YPD plates (Quality Biologics) and incubating at 30°C for 5 days. To further distinguish between [URE3] variants 1, 2, and 3, successive 10-fold cell dilutions were spotted onto Synthetic Complete plates lacking adenine (SC−Ade; KD Medical Inc.) and Synthetic Complete plates lacking arginine (SC−R; KD Medical Inc.) and containing 200 mg/l canavanine. For Figure 1B, cells were also spotted onto both Synthetic Dextrose plates (Amberg et al, 2005) containing 20 mg/l adenine, histidine, leucine, and tryptophan (SD+Ade, HLW), and 33 mg/l ureidosuccinate, and onto Hartwell Complete plates lacking arginine (HC−R; Amberg et al, 2005) and containing 200 mg/l canavanine. Plates were incubated for 3 days at 30°C unless otherwise noted. All images of yeast plates were captured using an HP Scanjet 5550c flatbed scanner and processed using Photoshop 7.0 (Adobe) and Canvas 6.0 (Deneba) software.
Electron and fluorescence light microscopy
EM of negatively stained filament preparations has been described (Baxa et al, 2003). To visualize Ure2p distribution in yeast cells, BY241 strains with either [ure-o] or one of the three [URE3] variants were transformed with plasmid pH458 expressing a Ure2p1−80-GFP fusion protein under control of the URE2 promoter (H Edskes, unpublished), and grown in 2 ml SD medium containing leucine, tryptophan, and uracil at concentrations of 20 mg/ml each at 30°C to OD600=0.5. Light microscopy was performed using an Axiovert 200M microscope (Zeiss) with a Coolsnap fx camera (Photometrix) in conjunction with the program IPLab (Scanalytics). GFP fluorescence was detected with a specific filter set.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Table I
Supplementary Methods
Acknowledgments
We are grateful to Dr Alasdair Steven for support of UB, Motomasa Tanaka and Jonathan S Weissman for initial help with the transformation procedure, Shawn Burgess for assistance with microscopy, Eric Ross and Herman Edskes for plasmids, Martin Schlumpberger for strain YMS23, Jack F Kirsch for the AAT plasmid, W-M Yau and Robert Tycko for Ure2p10−39 peptide, and Michael Feldbrügge, Andrea Patten, and members of LBG for helpful comments on the manuscript.
References
- Aigle M, Lacroute F (1975) Genetical aspects of [URE3], a non-Mendelian, cytoplasmically inherited mutation in yeast. Mol Gen Genet 136: 327–335 [DOI] [PubMed] [Google Scholar]
- Amberg D, Burke DJ, Strathern JN (2005) Methods in Yeast Genetics. Woodbury, NY, USA: Cold Spring Harbor Laboratory Press [Google Scholar]
- Baxa U, Cheng N, Winkler DC, Chiu TK, Davies DR, Sharma D, Inouye H, Kirschner DA, Wickner RB, Steven AC (2005) Filaments of the Ure2p prion protein have a cross-beta core structure. J Struct Biol 150: 170–179 [DOI] [PubMed] [Google Scholar]
- Baxa U, Speransky V, Steven AC, Wickner RB (2002) Mechanism of inactivation on prion conversion of the Saccharomyces cerevisiae Ure2 protein. Proc Natl Acad Sci USA 99: 5253–5260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxa U, Taylor KL, Wall JS, Simon MN, Cheng N, Wickner RB, Steven A (2003) Architecture of Ure2p prion filaments: the N-terminal domain forms a central core fiber. J Biol Chem 278: 43717–43727 [DOI] [PubMed] [Google Scholar]
- Bessen RA, Marsh RF (1992) Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J Virol 66: 2096–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousset L, Redeker V, Decottignies P, Dubois S, Le Marechal P, Melki R (2004) Structural characterization of the fibrillar form of the yeast Saccharomyces cerevisiae prion Ure2p. Biochemistry 43: 5022–5032 [DOI] [PubMed] [Google Scholar]
- Bousset L, Thomson NH, Radford SE, Melki R (2002) The yeast prion Ure2p retains its native α-helical conformation upon assembly into protein fibrils in vitro. EMBO J 21: 2903–2911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce ME (2003) TSE strain variation: an investigation into prion disease diversity. Br Med Bull 66: 99–108 [DOI] [PubMed] [Google Scholar]
- Caughey B, Lansbury PT (2003) Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible prion aggregates from innocent bystanders. Annu Rev Neurosci 26: 267–298 [DOI] [PubMed] [Google Scholar]
- Caughey B, Raymond GJ, Bessen RA (1998) Strain-dependent differences in β-sheet conformations of abnormal prion protein. J Biol Chem 273: 32230–32235 [DOI] [PubMed] [Google Scholar]
- Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ (2002) Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295: 851–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TG (2002) Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol Rev 26: 223–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustou V, Deleu C, Saupe S, Begueret J (1997) The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc Natl Acad Sci USA 94: 9773–9778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox BS (1965) PSI, a cytoplasmic suppressor of super-suppressor in yeast. Heredity 20: 505–521 [Google Scholar]
- Cox BS (1993) Psi phenomena in yeast. In The Early Days of Yeast Genetics, Hall MN, Linder P (eds) pp 219–239. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press [Google Scholar]
- Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW (1996) Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 144: 1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes HK, Gray VT, Wickner RB (1999) The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc Natl Acad Sci USA 96: 1498–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Gibbink MFBG, Claessen D, Bouma B, Dijkhuizen L, Wosten HAB (2005) Amyloids—a functional coat for microorganisms. Nat Rev Microbiol 3: 333–341 [DOI] [PubMed] [Google Scholar]
- Grenson M, Mousset M, Wiame JM, Bechet J (1966) Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. I. Evidence for a specific arginine-transporting system. Biochim Biophys Acta 127: 325–338 [DOI] [PubMed] [Google Scholar]
- Jayasinghe SA, Langen R (2004) Identifying structural features of fibrillar islet amyloid polypeptide using site-directed spin labeling. J Biol Chem 279: 48420–48425 [DOI] [PubMed] [Google Scholar]
- King CY (2001) Supporting the structural basis of prion strains: induction and identification of [PSI] variants. J Mol Biol 307: 1247–1260 [DOI] [PubMed] [Google Scholar]
- King CY, Diaz-Avalos R (2004) Protein-only transmission of three yeast prion strains. Nature 428: 319–323 [DOI] [PubMed] [Google Scholar]
- Kirschner DA, Damas A, Teplow D (2000) Special issue on amyloid: twist and sheet. J Struct Biol 130: 101–383 [DOI] [PubMed] [Google Scholar]
- Lacroute F (1971) Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J Bacteriol 106: 519–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legname G, Baskakov IV, Nguyen H-OB, Reisner D, Cohen FE, DeArmond SJ, Prusiner SB (2004) Synthetic mammalian prions. Science 305: 673–676 [DOI] [PubMed] [Google Scholar]
- Maddelein ML, Dos Reis S, Duvezin-Caubet S, Coulary-Salin B, Saupe SJ (2002) Amyloid aggregates of the HET-s prion protein are infectious. Proc Natl Acad Sci USA 99: 7402–7407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magasanik B, Kaiser CA (2002) Nitrogen regulation in Saccharomyces cerevisiae. Gene 290: 1–18 [DOI] [PubMed] [Google Scholar]
- Masison DC, Maddelein M-L, Wickner RB (1997) The prion model for [URE3] of yeast: spontaneous generation and requirements for propagation. Proc Natl Acad Sci USA 94: 12503–12508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masison DC, Wickner RB (1995) Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science 270: 93–95 [DOI] [PubMed] [Google Scholar]
- Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R (2002) A structural model for Alzheimer's beta-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci USA 99: 16742–16747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova AT, Leapman RD, Guo Z, Yau WM, Mattson MP, Tycko R (2005) Self-propagating, molecular-level polymorphism in Alzheimer's beta-amyloid fibrils. Science 307: 262–265 [DOI] [PubMed] [Google Scholar]
- Podrabsky JE, Carpenter JF, Hand SC (2001) Survival of water stress in annual fish embryos: dehydration avoidance and egg amyloid fibers. Am J Physiol Regul Integr Comp Physiol 280: R123–R131 [DOI] [PubMed] [Google Scholar]
- Prusiner SB (1982) Novel proteinaceous infectious particles cause scrapie. Science 216: 136–144 [DOI] [PubMed] [Google Scholar]
- Prusiner SB, Safar J, DeArmond SJ (2004) Bioassays of prions. In Prion Biology and Diseases, Prusiner SB (ed) pp 14–186. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press [Google Scholar]
- Ripaud L, Maillet L, Immel-Torterotot F, Durand F, Cullin C (2004) The [URE3] yeast prion results from protein aggregates that differ from amyloid filaments formed in vitro. J Biol Chem 279: 50962–50968 [DOI] [PubMed] [Google Scholar]
- Rizet G (1952) Les phenomenes de barrage chez Podospora anserina: analyse genetique des barrages entre les souches s et S. Rev Cytol Biol Veg 13: 51–92 [Google Scholar]
- Ross ED, Baxa U, Wickner RB (2004) Scrambled prion domains form prions and amyloid. Mol Cell Biol 24: 7206–7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlumpberger M, Prusiner SB, Herskowitz I (2001) Induction of distinct [URE3] yeast prion strains. Mol Cell Biol 21: 7035–7046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speransky V, Taylor KL, Edskes HK, Wickner RB, Steven A (2001) Prion filament networks in [URE3] cells of Saccharomyces cerevisiae. J Cell Biol 153: 1327–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Chien P, Naber N, Cooke R, Weissman JS (2004) Conformational variations in an infectious protein determine prion strain differences. Nature 428: 323–328 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chien P, Yonekura K, Weissman JS (2005) Mechanism of cross-species prion transmission: an infectious conformation compatible with two highly divergent yeast prion proteins. Cell 121: 49–62 [DOI] [PubMed] [Google Scholar]
- Taylor KL, Cheng N, Williams RW, Steven AC, Wickner RB (1999) Prion domain initiation of amyloid formation in vitro from native Ure2p. Science 283: 1339–1343 [DOI] [PubMed] [Google Scholar]
- Turoscy V, Cooper TG (1987) Ureidosuccinate is transported by the allantoate transport system in Saccharomyces cerevisiae. J Bacteriol 169: 2598–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB (1994) [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 264: 566–569 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Table I
Supplementary Methods


