Abstract
Telomeres protect chromosomes from end-to-end fusions. In yeast Saccharomyces cerevisiae, the protein Rap1 directly binds telomeric DNA. Here, we use a new conditional allele of RAP1 and show that Rap1 loss results in frequent fusions between telomeres. Analysis of the fusion point with restriction enzymes indicates that fusions occur between telomeres of near wild-type length. Telomere fusions are not observed in cells lacking factors required for nonhomologous end joining (NHEJ), including Lig4 (ligase IV), KU and the Mre11 complex. SAE2 and TEL1 do not affect the frequency of fusions. Together, these results show that Rap1 is essential to block NHEJ between telomeres. Since the presence of Rap1 at telomeres has been conserved through evolution, the establishment of NHEJ suppression by Rap1 could be universal.
Keywords: ATM, chromosome fusion, genomic instability, NHEJ, telomere
Introduction
Telomeres are the DNA–protein complexes found at the ends of linear chromosomes. As double-strand ends, telomeres could get involved in DNA double-strand break repair. In most cells, two pathways efficiently repair double-strand breaks: nonhomologous end joining (NHEJ) and homologous recombination. NHEJ is essentially a direct religation between the two ends (Wilson et al, 2003). Homologous recombination is a more complex process that uses a template sequence for repair (Symington, 2002). When these pathways act on telomeres, the consequences are utterly different. Homologous recombination events between telomeres can elongate or shorten telomeres but cannot fuse them since telomeric DNA are tandem arrays of short duplex repeats always in the same orientation relative to the chromosome ends. Homologous recombination at telomeres is only partially repressed in normal cells and may play a significant role in telomere length homeostasis, in particular when telomere length has shifted far away from equilibrium (Walmsley et al, 1983; Dunn et al, 1984; Lundblad and Blackburn, 1993; Li and Lustig, 1996; Le et al, 1999; Dunham et al, 2000; Teng et al, 2000; Cerone et al, 2001; Grandin et al, 2001; Lustig, 2003; Bailey et al, 2004; Londono-Vallejo et al, 2004; Tarsounas et al, 2004; Teixeira et al, 2004; Wang et al, 2004). By contrast, a single NHEJ event between two telomeres fuses the involved chromosomes, a gross rearrangement that can initiate a cycle of genomic instability. In normal cells, such event is rare (Ferreira et al, 2004). For instance, in wild-type yeast cells, fusions involving a telomere seem to occur in less than 1 every 107 cells (DuBois et al, 2002; Chan and Blackburn, 2003; Mieczkowski et al, 2003). In humans, cytogenetic analyses of normal lymphocytes suggest that chromosome end fusions occur at a frequency lower than 10−3 per cell (Prieur et al, 1988). Interestingly, end-to-end fusions leading to chain multicentric chromosomes were seen once in lymphocytes from a single patient (Dutrillaux et al, 1977). This observation remained exceptional but suggests that dysfunction of NHEJ suppression at telomeres can happen spontaneously in humans.
In fission yeast Schizosaccharomyces pombe, the Taz1 protein binds telomeric DNA (Cooper et al, 1997). Cells lacking Taz1 accumulate telomere fusions, which required KU and ligase IV, two essential components of the NHEJ machinery (Ferreira and Cooper, 2001, 2004). In mammals, TRF1 and TRF2 are the two telomere-binding proteins orthologous to Taz1 (Chong et al, 1995; Bilaud et al, 1996, 1997; Broccoli et al, 1997). A dominant negative allele of TRF2 displacing the protein from telomeres causes telomere fusions (van Steensel et al, 1998). In cells lacking ligase IV, fusions induced by TRF2 loss of function are not observed (Smogorzewska et al, 2002). Thus, Taz1 and TRF2 establish NHEJ suppression at telomeres. Taz1 and TRF2 interact at telomeres with a conserved protein, Rap1, which is required for proper telomere length regulation (Li et al, 2000; Chikashige and Hiraoka, 2001; Kanoh and Ishikawa, 2001; Li and de Lange, 2003). A role for Rap1 in telomere protection against NHEJ remained to be addressed.
In the budding yeast Saccharomyces cerevisiae, there is no ortholog of Taz1/TRF1/TRF2 and Rap1 directly binds the TG1–3 telomere sequences (Conrad et al, 1990; Konig et al, 1996). Rap1 establishes a negative feedback loop on telomere elongation by telomerase (Kyrion et al, 1992; Krauskopf and Blackburn, 1996; Marcand et al, 1997; Ray and Runge, 1999; Grossi et al, 2001; Teixeira et al, 2004). For this pathway, Rap1 acts through its carboxy-terminal domain by recruiting two factors, Rif1 and Rif2, whose mode of action is still unknown (Hardy et al, 1992; Buck and Shore, 1995; Wotton and Shore, 1997; Levy and Blackburn, 2004; Teixeira et al, 2004). Rap1 also establishes transcriptional silencing on the adjacent chromatin by recruiting a different set of factors through the same domain (Kyrion et al, 1993; Moretti and Shore, 2001; Luo et al, 2002). In addition, Rap1 binds the promoters of a large fraction of genes expressed during exponential growth, where it seems to play an essential role in transcriptional activation (Lieb et al, 2001). Possibly because of its role in transcription, Rap1 is essential for viability in budding yeast, precluding the use of a simple gene knockout to study its functions (Shore and Nasmyth, 1987).
An indirect approach is to study mutations in the telomerase RNA template that are translated by the recurrent action of telomerase into mutations within the distal repeats of telomeres (Yu et al, 1990; Singer and Gottschling, 1994; McEachern and Blackburn, 1995). In budding yeast Kluyveromyces lactis, changes in the telomerase RNA template disrupting Rap1 binding site can result in telomere fusions (McEachern et al, 2000). This suggests that Rap1 plays a role in telomere protection, although the mutations that abolish Rap1 binding could also impact other pathways. Similar approaches were carried out in S. cerevisiae but did not show a requirement for Rap1 in telomere protection against fusions (Prescott and Blackburn, 2000; Alexander and Zakian, 2003; Brevet et al, 2003; Lin et al, 2004). We addressed this issue differently by looking at the direct consequences of Rap1 protein loss in S. cerevisiae using a conditional allele.
Results
Rap1 loss causes telomere fusions
First, we constructed a degron allele of RAP1: rap1-(Δ). This allele is constructed by the ‘double shut off' method and is integrated at the endogenous RAP1 locus (supplementary data). rap1-(Δ) is controlled by the ANB1 promoter, which is repressible by Rox1. rap1-(Δ) encodes a Rap1 protein with an amino-terminal fusion making it a target for Ubr1 and degradation by the N-end rule. Induction of Rox1 and Ubr1 by elevated copper concentration allows the simultaneous transcriptional repression of the gene and proteolysis of the existing molecules. Copper addition to a growing rap1-(Δ) cell culture causes Rap1 loss, telomere elongation and cell growth arrest (supplementary data).
Without copper addition to the medium, rap1-(Δ) cells grow exponentially with a slightly higher doubling time and with a lower Rap1 steady-state level than isogenic wild-type cells (Figure 1A and B). This lower amount of Rap1 drops even further when cells exit exponential phase. Telomere length is slightly more heterogeneous in rap1-(Δ) cells than in wild type with a mean at 300 and 320 nt, respectively, and remains unchanged during progression toward stationary phase (data not shown). In this study, we used the rap1-(Δ) allele in basal conditions (i.e. without copper addition) and took advantage of the protein loss in the mutant to address the contribution of Rap1 to telomere protection.
Figure 1.
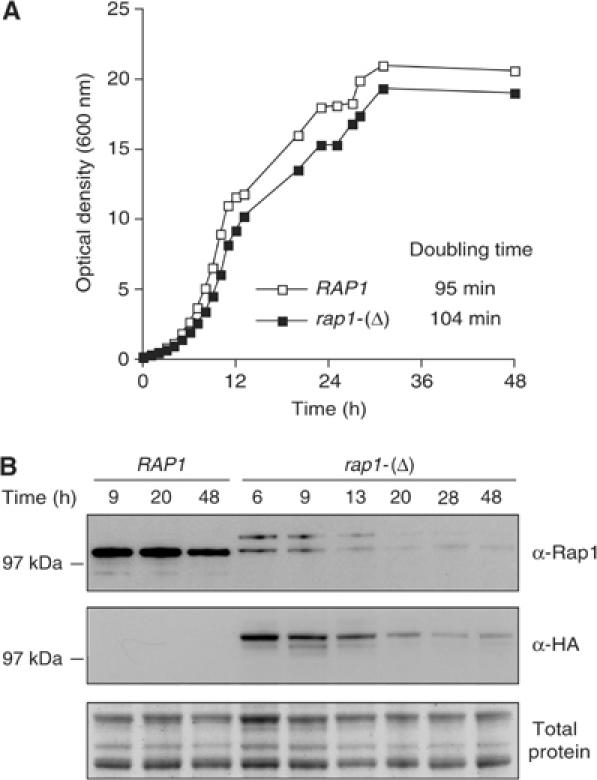
Rap1 loss in rap1-(Δ) cells progressing toward stationary phase. (A) Growth curve of wild-type and rap1-(Δ) cells in rich medium at 30°C. Yeast strains ZMY60 (wild type) and Lev391 (rap1-(Δ)) were maintained in exponential phase by successive dilutions in rich medium for 2 days and at time 0 allowed to exhaust the medium. The doubling time is estimated from the initial exponential growth. (B) Immunoblot showing Rap1 level in wild-type and rap1-(Δ) cells. Rap1 in rap1-(Δ) cells is tagged with four HA epitopes. To detect Rap1 and Rap1 tagged with HA epitopes, we used, respectively, a rabbit polyclonal antibody directed against the carboxy-terminal region of Rap1 (α-Rap1) and a monoclonal antibody directed against the HA epitope (α-HA). Loading was controlled by gel staining.
Fusion events between telomeres were looked at using a PCR strategy similar to the one previously described (Mieczkowski et al, 2003). In S. cerevisiae, a conserved element, X, is located adjacent to every telomere. In the sequenced yeast genome, 17 out of the 32 chromosome ends display a second element, Y′, inserted between X and the telomere. We chose two primers annealing with X and Y′, respectively, located at about 500 bp from the terminal telomeric repeats (Figure 2A). As shown in Figure 2B, a smearing PCR signal centered at about 1540 bp appears and accumulates in rap1-(Δ) cells a few hours after exit from exponential phase, concomitant to Rap1 loss. This signal is not observed in wild-type cells and in rap1-(Δ) cells with an integrated copy of the wild-type gene (Figure 2B and C). PCR with only one of the two primers X or Y′ fails to amplify a signal (Figure 2C). This does not rule out fusions between telomeres of the same class (i.e. between two X telomeres and between two Y′ telomeres), which, as palindromes, are likely to resist PCR amplification.
Figure 2.
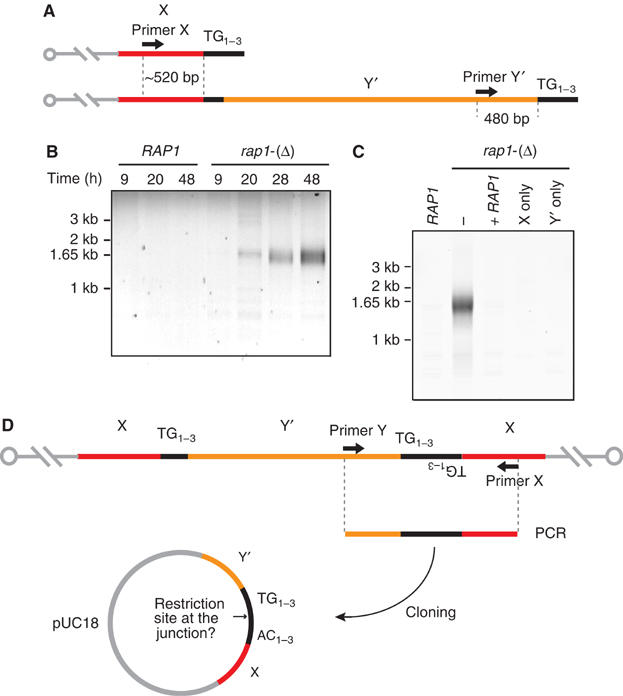
Telomere fusions in rap1-(Δ) cells progressing toward stationary phase. (A) Schematic representation of X and Y′ telomeres in S. cerevisiae and relative positions of the primers used for PCR amplification. In the sequenced yeast genome, there are 15 X telomeres and 17 Y′ telomeres. The fusion of two telomeres should give a PCR product of about 1000 bp plus the TG1–3 telomeric repeats at the junction. Fusions between two X or two Y′ telomeres form quasiperfect palindromes and are unlikely to be amplified. (B) Detection by PCR of telomere fusions in rap1-(Δ) cells reaching stationary phase. Time points are the same as in Figure 1. (C) Complementation of rap1-(Δ) in Lev391 with a wild-type copy of RAP1 integrated at URA3. Cells were grown to saturation in rich medium for 5 days. Telomere fusions are not amplified with only one of the two primers X or Y′. (D) Schematic representation of PCR amplification, cloning and restriction analysis of telomere fusions.
The PCR products were cloned, amplified in Escherichia coli and sequenced (Figure 2D). The mean length of the cloned fragments is 1540±120 bp (n=83). All the clones contain an X element end coming from one of 13 different chromosome ends, a Y′ element end on the other side and TG1–3 repeats pointing at each other. The quasi-palindromic TG1–3 fusions could not be sequenced through the fusion point. To determine the sequence at the junctions, we looked for restriction sites that would be formed by joining inverted TG1–3 repeats. Tested sites were found with occurrences not statistically different from those expected from a random fusion between two telomeres (Table I). Among the clones with a restriction site at the junction, the mean length of the telomeres trapped in the fusion is 240±80 bp (n=58). Together, these results show that telomeres of near wild-type length fuse with each other when Rap1 disappears in rap1-(Δ) cells.
Table 1.
Restriction site occurrences at the junction among the cloned fusions
| Enzyme name | Enzyme site | Occurrence | Expecteda |
|---|---|---|---|
| Hpy8I | -GTNNAC- | 28 | 27.9 |
| RsaI | -GTAC- | 6 | 11.6 |
| ApaLI | -GTGCAC- | 10 | 11.6 |
| KpnI | -GGTACC- | 1 | 1.8 |
| Total number of clones | 83 | ||
| The expected occurrences are calculated with a native 350 bp telomeric sequence displaying a GC content of 62%. | |||
The rap1-(Δ) allele expresses a protein with an amino-terminal tag. Telomere fusions observed in rap1-(Δ) cells could be a consequence of Rap1 loss, Rap1 tagging or both. To address this, we used a strain in which RAP1 is simply deleted and viability rescued by an ectopic copy of the wild-type gene. The RAP1 sequence is either on a plasmid or integrated in a chromosome (Figure 3A). Among these cells, telomere fusions occur when RAP1 is on a plasmid but not when RAP1 is integrated (Figure 3B). We propose that the fusions are caused by continuous loss of the plasmid among the growing cell population, producing doomed cells that lose wild-type Rap1, indicating that Rap1 loss is sufficient to cause telomere fusions. The PCR used to detect telomere fusions is semiquantitative. We estimated that fusions are about 80 times more frequent in rap1-(Δ) cells than in rap1-Δ cells with RAP1 on a plasmid (Figure 3C). In the latter situation, RAP1 is lost only in a small fraction of cells and the wild-type protein is itself more stable, probably explaining why the fusions are less frequent.
Figure 3.
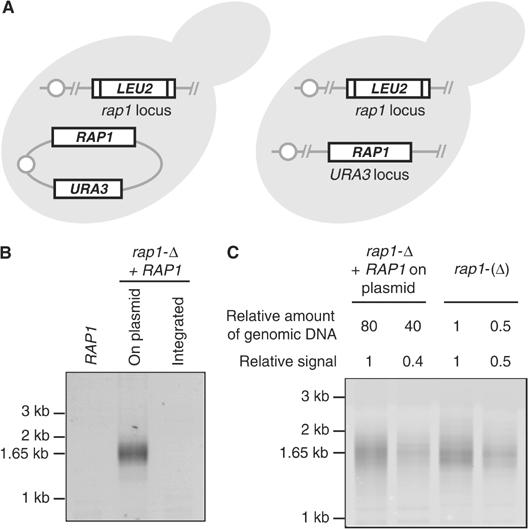
Telomere fusions among cells with RAP1 on a plasmid. (A) Schematic representation of the yeast strains deleted for RAP1 and carrying a wild-type copy of RAP1 either on a plasmid or integrated in a chromosome. (B) Continuous loss of wild-type RAP1 gene on a plasmid causes telomere fusions. In the yeast strain Lev9, plasmid pCEN-Sup4-RAP1 was replaced by centromeric plasmid pRS316-RAP1 or by pRS306-RAP1 integrated at URA3. The control RAP1 strain is W303-1a. Cells were grown to saturation in rich medium for 5 days. Telomere fusions were amplified by PCR. (C) Relative quantification of telomere fusions. Strains Lev9 shuffled with pRS316-RAP1 and Lev391 (rap1-(Δ)) were grown to saturation in rich medium for 5 days. Fusions were amplified by PCR and quantified as described in Materials and methods.
Formally, Rap1 could establish two different types of mechanisms: it could prevent the fusions from occurring and it could revert telomere fusions once they have occurred. We looked at the outcome of a telomere fusion once it is formed. A fusion was cloned from rap1-(Δ) cells into a centromeric yeast plasmid and reintroduced into wild-type cells. Southern analysis shows that this fusion on a circular plasmid can be maintained for many generations prior to rearrangements within the fusion or linearization of the plasmid (data not shown). This relative stability suggests that Rap1 must act before the fusion to prevent it.
Telomere fusions are produced by NHEJ
NHEJ requires the DNA end-binding proteins Yku70 and Yku80 that form the KU heterodimer, as well as the ligase Lig4 and its associated factor Lif1 (ligase IV and XRCC4 respectively in mammals) (Milne et al, 1996; Schar et al, 1997; Teo and Jackson, 1997; Wilson et al, 1997; Herrmann et al, 1998; Lee et al, 1998; Teo and Jackson, 2000; Chen et al, 2001; Walker et al, 2001). In addition, at least in S. cerevisiae, NHEJ required the Lif1-interacting factor Lif2 (also called Nej1) and the Mre11 complex made of Mre11, Rad50 and Xrs2 (Moore and Haber, 1996; Boulton and Jackson, 1998; Frank-Vaillant and Marcand, 2001; Kegel et al, 2001; Ooi et al, 2001; Valencia et al, 2001). To address the pathway responsible for fusing Rap1-defective telomeres, we tested the deletion of genes required for NHEJ. As shown in Figure 4, in rap1-(Δ) cells lacking Yku70, Yku80, Lig4, Lif1, Lif2, Mre11, Rad50 and Xrs2, telomere fusions could not be detected by PCR. Complementation of the lif2 disruption by a centromeric plasmid encoding Lif2 restores the appearance of telomere fusions in rap1-(Δ) cells (Figure 4A). Thus, telomere fusions caused by Rap1 loss are produced by NHEJ.
Figure 4.
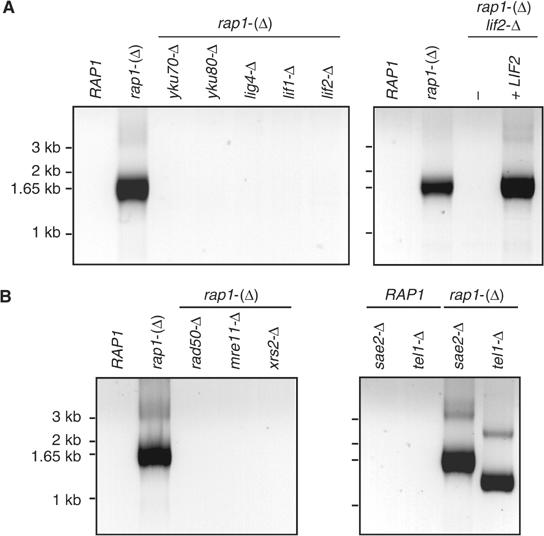
NHEJ factors are required for telomere fusions. (A) Yeast strains ZMY60 (wild type), Lev391 (rap1-(Δ)), Lev397 (rap1-(Δ) yku70-Δ), Ybp43 (rap1-(Δ) yku80-Δ), Ybp11 (rap1-(Δ) lig4-Δ), Lev396 (rap1-(Δ) lif1-Δ) and Ybp14 (rap1-(Δ) lif2-Δ) were grown to saturation in rich medium for 7 days. Strains ZMY60, Lev391 and Ybp14 transformed with plasmid pRS314 and strain Ybp14 transformed with plasmid pRS314-LIF2 were grown to saturation in synthetic medium lacking tryptophan for 5 days. Telomere fusions were amplified by PCR. (B) Yeast strains ZMY60, Lev391, Ybp29 (rap1-(Δ) mre11-Δ), Ybp27 (rap1-(Δ) rad50-Δ), Ybp9 (rap1-(Δ) xrs2-Δ), Ybp40 (sae2-Δ), Ybp41 (tel1-Δ), Ybp31 (rap1-(Δ) sae2-Δ) and Ybp23 (rap1-(Δ) tel1-Δ) were grown to saturation in rich medium for 7 days. Telomere fusions were amplified by PCR.
We examined the involvement in telomere fusion of two known regulators of the Mre11 complex that could affect the processing of telomere ends, thereby influencing fusions by NHEJ. Sae2 is an activator of the Mre11 nuclease activity (Rattray et al, 2001; Lobachev et al, 2002). Tel1 is the yeast ortholog of the human DNA damage checkpoint kinase ATM and is required for the function of the Mre11 complex in telomere elongation by telomerase (Greenwell et al, 1995; Ritchie and Petes, 2000; Tsukamoto et al, 2001). tel1-Δ cells display short telomeres (Greenwell et al, 1995). As shown in Figure 4B, Sae2 and Tel1 are not required for fusion in rap1-(Δ) cells. As expected for fusions between shorter telomeres, rap1-(Δ) tel1-Δ double mutant cells display shorter fusion products.
In mammals, the ERCC1-XPF nuclease is required for the telomere fusions induced by TRF2 inhibition (Zhu et al, 2003). The yeast homolog of ERCC1-XPF is Rad1-Rad10. In a rap1-(Δ) rad1-Δ double mutant, telomere fusions are still observed but the double mutant displays a significant growth defect compared to rap1-(Δ) and rad1-Δ single mutants (data not shown). This negative genetic interaction is surprising and will require further investigation to be explained.
To estimate the absolute frequencies of fusions, a centromeric plasmid with a fusion cloned from rap1-(Δ) cells was reintroduced into wild-type cells (Figure 5A). Its average copy number was estimated by Southern analysis at about 1.9 copies per genome (data not shown). The increase in copy number is probably a consequence of the antagonism between the telomere sequences and the centromere (Enomoto et al, 1994). The signal from this plasmid provides an external control for quantification by PCR of the fusions in rap1-(Δ) and rap1-(Δ) sae2-Δ cells. To quantify fusions in rap1-(Δ) tel1-Δ cells, a second plasmid with a short fusion cloned from rap1-(Δ) tel1-Δ cells was used. This plasmid is maintained at about 1.3 copies per genome (data not shown). As shown in Figure 5, in cells in stationary phase, we observe about one fusion between an X telomere and a Y′ telomere every two genomes. The number of fusions is not significantly different in the absence of Sae2 or Tel1 (Figure 4B and C). Since fusions can presumably also occur between telomeres of the same class, we probably underestimate the actual frequencies of fusions by about two-fold.
Figure 5.
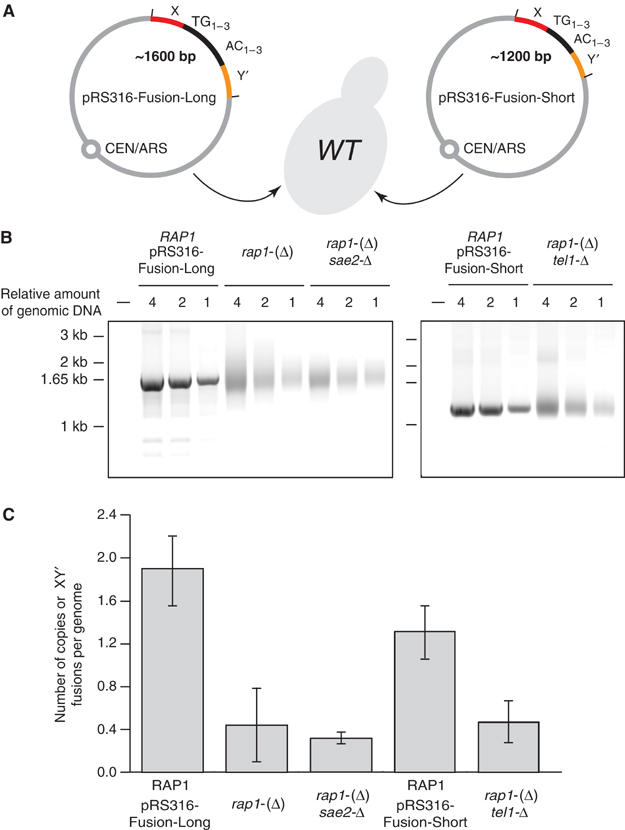
Quantification of telomere fusions. (A) Schematic map of the pRS316-Fusion-Long and pRS316-Fusion-Short plasmids. In pRS316-Fusion-Long, the X element comes from the right end of chromosome II. An ApaLI site is present at the junction and the two TG1–3 repeats pointing at each other are about 350 and 270 bp. In pRS316-Fusion-Short, the X element comes from the right end of chromosome IX. The sum of the TG1–3 inverted repeats is about 200 bp. pRS316-Fusion-Long and pRS316-Fusion-Short were separately reintroduced into wild-type cells. (B) Quantification of fusions between X and Y′ telomeres. Yeast strains ZMY60 (wild type) transformed with pRS316-Fusion-Long and pRS316-Fusion-Short were grown to saturation for 5 days in medium lacking uracil. Lev391 (rap1-(Δ)), Ybp31 (rap1-(Δ) sae2-Δ) and Ybp23 (rap1-(Δ) tel1-Δ) were grown to saturation for 5 days in rich medium. Fusions were amplified by PCR and quantified as described in Materials and methods. (C) Number of fusions per genome. Plasmid copy number was estimated by Southern analysis (data not shown). The signals from strains Lev391 (rap1-(Δ)) and Ybp31 (rap1-(Δ) sae2-Δ) were normalized with the signals from strain ZMY60 (wild type) transformed with pRS316-Fusion-Long. The signals from strain Ybp23 (rap1-(Δ) tel1-Δ) were normalized with the signals from strain ZMY60 (wild type) transformed with pRS316-Fusion-Short. Mean and standard deviation were calculated by averaging 12 amplifications (three dilutions of four independent samples).
Telomere fusions reduce cell viability
These relatively high frequencies of fusions per cell prompted us to look for an impact on cell viability. Cells were grown to saturation in liquid media for 3–4 weeks and individually placed on plates. When each cell resumed growth, the two products of its first division were separated and one was placed at an adjacent position (Figure 6A). Then, each cell was allowed to form a colony. Four different outcomes are observed and represented in Figure 6B: (a) the products of the first division give rise to two viable colonies, (b) the initial cell fails to re-enter the cell cycle or to finish the first division, (c) only one of the two products of the first division gives rise to a viable colony and, finally, (d) the two products of the first division fail to produce a viable colony. In this one-generation pedigree, we observed that, in a wild-type strain, a majority of cells form two viable colonies, about a third failed to resume growth and only a small fraction produced unviable microcolonies (Figure 6D). Compared to the wild-type strain, the rap1-(Δ) strain displays a higher frequency of cells that re-entered the cell cycle and failed to form viable colonies and increased asymmetric lethality, when only one of the two products of the first division gives rise to a viable colony. The rap1-(Δ) lif1-Δ double mutant defective for NHEJ behaves as the wild-type strain (Figure 6D), suggesting that increased failure to form a viable colony in rap1-(Δ) cells is a consequence of telomere fusions.
Figure 6.
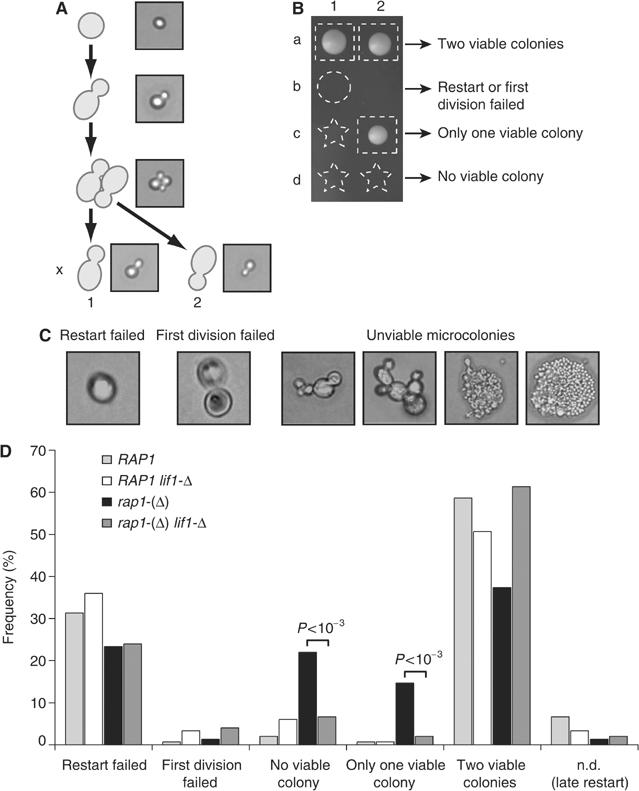
Telomere fusions reduce cell viability. (A) Schematic representation of single cell micromanipulation during its first division. Yeast strains were grown to saturation in rich medium for 3–4 weeks. Cells were spread onto plates, micromanipulated onto a grid and left at 30°C. The two products of the first division were separated as budded cells and placed adjacent to each other on the same line. An example has been recorded at each step and is shown. (B) After micromanipulation, cells were grown for 4 days at 30°C. Possible outcomes are shown. Dashed rectangles denote the cell that gave rise to a viable colony, the dashed circle denotes the initial cell that failed to restart growth or achieve its first division and dashed stars denote the cell that failed to achieve its first division or formed an unviable microcolony of two to a few hundred cells. (C) Morphology of cells that failed to form a viable colony. (D) Cell outcomes after stationary phase. Yeast strains ZMY60 (wild type), Lev391 (rap1-(Δ)), Lev396 (rap1-(Δ) lif1-Δ) and Lev398 (lif1-Δ) were grown and micromanipulated as described in panel A. The data come from two independent experiments (ntotal=150 cells). P-values were calculated using a Student's test.
To explain asymmetric lethality, we propose the following scenario. A dicentric chromosome, formed by a telomere fusion in stationary phase and duplicated in S phase, will break during anaphase if the centromeres present on the same chromatid are pulled apart to opposite poles. Two breaks, one on each sister chromatid, will occur. Irrespective of the break positions, one cell will have lost at least one chromosome fragment and will be unlikely to form a viable colony. The other cell will receive a complete set of genes and maintain the possibility of forming a viable colony. This could occur through repair by break-induced replication (BIR) until the fusion is resolved, for instance through degradation from a break up to the fused telomeres. If during the first division the two centromeres present on the same chromatid go toward the same pole, the two cells will inherit the dicentric chromosome. Breakage will be postponed to later divisions, increasing the probability of giving rise to viable colonies. If more than one fusion is formed in stationary phase, the possibility of a viable outcome will be significantly reduced, explaining that the two products of the first division can fail to form viable colonies. In conclusion, telomere fusions occurring in stationary phase reduce cell viability when cells resume cell division, but a single telomere fusion in a haploid cell does not seem to be necessarily a lethal event.
Discussion
We describe here that Rap1 loss in cells exiting the cell cycle results in telomere fusions by NHEJ, indicating that Rap1 prevents this pathway at telomeres. The timing of Rap1 loss in rap1-(Δ) mutant cells separates the functions of Rap1. Indeed, another function of Rap1 at telomeres is to negatively regulate telomere elongation by telomerase (Kyrion et al, 1992; Marcand et al, 1997). Telomere elongation is restricted to late S phase when telomeres are replicated (Marcand et al, 2000; Taggart et al, 2002). By causing Rap1 loss in cells that do not progress through S phase, the rap1-(Δ) mutant allows the progressive accumulation of fusions between telomeres that remain at normal lengths. In addition, Rap1 involvement in transcriptional activation during exponential growth is probably relieved when cells are out of the cell cycle as suggested by the ability of rap1-(Δ) cells to normally enter and exit stationary phase. Thus, Rap1 role on NHEJ suppression can be studied in cells that are not grossly challenged in housekeeping genes expression.
In yeast, in the absence of telomerase, telomeres get progressively very short and fuse by NHEJ at increasing frequencies (Chan and Blackburn, 2003; Mieczkowski et al, 2003). We propose that in this situation fusions occur when the remaining Rap1 binding sites become insufficient to insure full NHEJ inhibition. Interestingly, Tel1 loss in cells lacking telomerase increases the frequency of fusions (Chan and Blackburn, 2003). Here we show that a tel1-Δ mutant does not affect fusions caused by Rap1 loss (Figure 5). It suggests that, among the mechanisms established by Rap1 to suppress NHEJ, one involves Tel1. Another explanation would be that Tel1 loss alleviates a checkpoint and in the absence of telomerase increases the frequency of critically short telomeres that can be subjected to NHEJ, a situation that would not happen when telomerase is present as in rap1-(Δ) cells.
It would be interesting to know what is the minimum number of Rap1 molecules sufficient to establish NHEJ suppression at telomeres. Rap1 binds telomeric DNA at a density of 1 per 18 bp (Gilson et al, 1993). In tel1-Δ cells, telomere length is between 100 and 150 bp, which corresponds to five to nine Rap1 binding sites, and telomeres do not seem to fuse (DuBois et al, 2002; Mieczkowski et al, 2003). A study by Chan and Blackburn (2003) shows that, in telomerase-defective cells with a mean telomere length of 230 bp, fusions involve very short telomeres of about 33 bp, that is two Rap1 binding sites. This strong bias suggests that three Rap1 molecules are enough to repress NHEJ significantly.
Our results do not directly address the mechanism(s) by which Rap1 establishes NHEJ suppression at telomeres. KU is constitutively bound to telomeres (Gravel et al, 1998), suggesting that Rap1 or factors recruited by Rap1 act downstream of KU binding to inhibit the synapsis, processing and ligation steps of NHEJ. This could be mediated through the formation of a DNA secondary structure that escapes NHEJ, as previously suggested in higher eukaryotes (Griffith et al, 1999; Zhu et al, 2003), and through protein–protein interactions. The experimental approach presented here should allow us to address these models in budding yeast.
In fission yeast S. pombe and in mammals, Rap1 is present at telomeres through interactions with the telomeric DNA binding factors Taz1 and TRF2, respectively (Li et al, 2000; Chikashige and Hiraoka, 2001; Kanoh and Ishikawa, 2001; Li and de Lange, 2003). In S. pombe, the absence of Rap1 results in telomere fusions by NHEJ (Miller et al, 2005). Thus, despite differences in the assembly of telomeric protein–DNA complexes between budding yeasts and other eukaryotes, Rap1 role in establishing NHEJ inhibition at telomeres appears to be conserved. Understanding how Rap1 acts will be relevant to the biology of human telomeres.
Materials and methods
Amplification and analysis of telomere–telomere fusions
Telomere fusions were amplified by PCR. Genomic DNA was prepared by phenol–chloroform extraction and resuspended in TE pH 8.0 buffer. One primer (X; AGGGTATAGACCGCTGAGGCAAGTG) had a sequence from X elements (e.g. coordinates 649–673 of chromosome IV; Stanford Genome Database) and the second (Y′; AGCGATGATGTTCCAGACGGTAGAT) had a sequence from Y′ elements (e.g. coordinates 314–338 of chromosome V). PCR reactions (30 μl) contained genomic DNA ∼10 ng, ProofStart buffer 1 × supplemented with MgSO4 0.83 mM, dNTP 0.3 mM each, primers 1 μM each, HotStarTag 1.2 U (Qiagen™) and ProofStart 0.12 U (Qiagen™). The conditions were as follows: 95°C 15 min; then 25–30 cycles of 94°C 30 s, 68°C 30 s, 72°C 1 min 30 s; followed by 72°C 3 min. The products were run through a 1% agarose gel and visualized by ethidium bromide staining. Amplified fusions were cloned by HindIII–EcoRI digestion into pUC18 using primers with added restriction sites. The clones were amplified in XL1-blue cells grown at 25°C, analyzed by restriction and sequenced. The restriction analysis for Hpy8I, RsaI, ApaLI and KpnI was facilitated by the X and Y′ sequence alignments provided by Dr E Louis at http://www.le.ac.uk/genetics/ejl12/index.html.
Semiquantitative PCR
Quantifications were carried out with the same PCR conditions. The relevant genomic DNA was diluted with genomic DNA from a wild-type strain to maintain the total DNA concentration constant at ∼10 ng. The products were run through a 1% agarose gel containing 0.1 μg/ml ethidium bromide and fluorescence was quantified using a Typhoon™ imager and ImageQuant™ 5.0 software (Amersham Biosciences™). The PCR used to detect fusions remains sensitive at at least three orders of magnitude below the level of fusions observed in rap1-(Δ) cells (data not shown).
Strains and plasmids
The strains used in this study are listed in Table II. The rap1-(Δ) allele was generated in strain ZMY60 by PCR-mediated transformation using the method described by Dubacq et al (2002). Gene deletions were made by PCR-mediated transformation (Longtine et al, 1998).
Table 2.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| ZMY60 (Moqtaderi et al, 1996) | MATa ura3-52 trp1-Δ1 ade2-101 pACE1-UBR1 pACE1-ROX1 |
| Lev391 | ZMY60 rap1-(Δ)∷KANr (KANr-ANB-UB-R-lacI-4HA-RAP1) |
| Lev397 | Lev391 yku70-Δ∷klURA3 |
| Ybp43 | Lev391 yku80-Δ∷klURA3 |
| Ybp11 | Lev391 lig4-Δ∷klURA3 |
| Lev396 | Lev391 lif1-Δ∷klURA3 |
| Ybp14 | Lev391 lif2-Δ∷klURA3 |
| Ybp29 | Lev391 mre11-Δ∷klURA3 |
| Ybp27 | Lev391 rad50-Δ∷klURA3 |
| Ybp9 | Lev391 xrs2-Δ∷klURA3 |
| Ybp40 | ZMY60 sae2-Δ∷klURA3 |
| Ybp41 | ZMY60 tel1-Δ∷klURA3 |
| Ybp31 | Lev391 sae2-Δ∷klURA3 |
| Ybp23 | Lev391 tel1-Δ∷klURA3 |
| Lev398 | ZMY60 lif1-Δ∷klURA3 |
| W303-1a | MATa ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 can1-100 rad5-535 |
| YLS85 (Sussel and Shore, 1991) | W303-1a rap1∷LEU2 pCEN-Sup4-RAP1 |
The RAP1 gene (a SacI–XbaI genomic fragment) was inserted into pRS306 (integrative, URA3) and pRS316 (CEN, URA3) creating plasmids pRS306-RAP1 and pRS316-RAP1, respectively. pRS306-RAP1 was digested by NdeI prior to transformation into yeast. A telomere fusion that was cloned into pUC18 and analyzed was subcloned using EcoRI and HindIII into pRS316 to create plasmid pRS316-Fusion. The LIF2 gene (including 360 bp upstream of the start codon and 330 bp downstream of the stop codon) was inserted into pRS314 (CEN, TRP1) creating plasmid pRS314-LIF2.
Telomere length
Telomere length in wild-type and rap1-(Δ) cells was measured by PCR after end labeling with terminal transferase as described (Teixeira et al, 2004). The mean length of the X telomeres was 320 and 330 nt in wild-type and rap1-(Δ) cells, respectively. The mean length of the Y′ telomeres was 320 and 270 nt in wild-type and rap1-(Δ) cells, respectively (data not shown). The mean length of the telomeres in the cloned fusions with a restriction site at the junction was 270±80 bp (n=29) on the X side and 220±70 bp on the Y′ side (n=29).
Western blot
Proteins were extracted at 95°C in urea 8 M, SDS 2% and Tris–HCl pH 7.5 100 mM followed by vortexing at 2500 r.p.m. with glass beads. A 10 μg portion of proteins was run through a 4–12% NuPage gel (Invitrogen™) and blotted onto a nitrocellulose membrane. To detect Rap1 and Rap1 tagged with HA epitopes, we used, respectively, a rabbit polyclonal antibody directed against the carboxy-terminal region of Rap1 (sc-20167; SantaCruz™; 1:500) and a monoclonal antibody directed against the HA epitope (12CA5; 1:10 000). Loading was controlled by gel staining with Simple Blue™ (Invitrogen™).
Supplementary Material
Supplementary Data
Acknowledgments
We thank Julia P Cooper, Miguel G Ferreira and Kyle Miller for discussions and personal communication prior to publication, Emilie Ma for assistance, Carl Mann and Caroline Dubacq for plasmids and strains and Emmanuelle Martini, Ariane Gratias, Bernard Dutrillaux, Madalena Tarsounas, Francis Fabre, Serge Gangloff, Xavier Veaute, Laure Sabatier and Bernard Lopez for suggestions and comments. This work was supported by grants from Association pour la recherche sur le cancer (ARC), Fondation de France (programme Tumeurs) and Ministère délégué à la recherche (ACI jeunes chercheurs).
References
- Alexander MK, Zakian VA (2003) Rap1p telomere association is not required for mitotic stability of a C(3)TA(2) telomere in yeast. EMBO J 22: 1688–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey SM, Brenneman MA, Goodwin EH (2004) Frequent recombination in telomeric DNA may extend the proliferative life of telomerase-negative cells. Nucleic Acids Res 32: 3743–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilaud T, Brun C, Ancelin K, Koering CE, Laroche T, Gilson E (1997) Telomeric localization of TRF2, a novel human telobox protein. Nat Genet 17: 236–239 [DOI] [PubMed] [Google Scholar]
- Bilaud T, Koering CE, Binet-Brasselet E, Ancelin K, Pollice A, Gasser SM, Gilson E (1996) The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res 24: 1294–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP (1998) Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J 17: 1819–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brevet V, Berthiau AS, Civitelli L, Donini P, Schramke V, Geli V, Ascenzioni F, Gilson E (2003) The number of vertebrate repeats can be regulated at yeast telomeres by Rap1-independent mechanisms. EMBO J 22: 1697–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broccoli D, Smogorzewska A, Chong L, de Lange T (1997) Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet 17: 231–235 [DOI] [PubMed] [Google Scholar]
- Buck SW, Shore D (1995) Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev 9: 370–384 [DOI] [PubMed] [Google Scholar]
- Cerone MA, Londono-Vallejo JA, Bacchetti S (2001) Telomere maintenance by telomerase and by recombination can coexist in human cells. Hum Mol Genet 10: 1945–1952 [DOI] [PubMed] [Google Scholar]
- Chan SW, Blackburn EH (2003) Telomerase and ATM/Tel1p protect telomeres from nonhomologous end joining. Mol Cell 11: 1379–1387 [DOI] [PubMed] [Google Scholar]
- Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE (2001) Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell 8: 1105–1115 [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Hiraoka Y (2001) Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr Biol 11: 1618–1623 [DOI] [PubMed] [Google Scholar]
- Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T (1995) A human telomeric protein. Science 270: 1663–1667 [DOI] [PubMed] [Google Scholar]
- Conrad MN, Wright JH, Wolf AJ, Zakian VA (1990) RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell 63: 739–750 [DOI] [PubMed] [Google Scholar]
- Cooper JP, Nimmo ER, Allshire RC, Cech TR (1997) Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385: 744–747 [DOI] [PubMed] [Google Scholar]
- Dubacq C, Guerois R, Courbeyrette R, Kitagawa K, Mann C (2002) Sgt1p contributes to cyclic AMP pathway activity and physically interacts with the adenylyl cyclase Cyr1p/Cdc35p in budding yeast. Eukaryot Cell 1: 568–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBois ML, Haimberger ZW, McIntosh MW, Gottschling DE (2002) A quantitative assay for telomere protection in Saccharomyces cerevisiae. Genetics 161: 995–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham MA, Neumann AA, Fasching CL, Reddel RR (2000) Telomere maintenance by recombination in human cells. Nat Genet 26: 447–450 [DOI] [PubMed] [Google Scholar]
- Dunn B, Szauter P, Pardue ML, Szostak JW (1984) Transfer of yeast telomeres to linear plasmids by recombination. Cell 39: 191–201 [DOI] [PubMed] [Google Scholar]
- Dutrillaux B, Aurias A, Couturier J, Croquette MF, Viegas-Pequignot E (1977) Multiple telomeric fusions and chain configurations in human somatic chromosomes. Chromosomes Today 6: 37–44 [Google Scholar]
- Enomoto S, Longtine MS, Berman J (1994) TEL+CEN antagonism on plasmids involves telomere repeat sequences tracts and gene products that interact with chromosomal telomeres. Chromosoma 103: 237–250 [DOI] [PubMed] [Google Scholar]
- Ferreira MG, Cooper JP (2001) The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol Cell 7: 55–63 [DOI] [PubMed] [Google Scholar]
- Ferreira MG, Cooper JP (2004) Two modes of DNA double-strand break repair are reciprocally regulated through the fission yeast cell cycle. Genes Dev 18: 2249–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MG, Miller KM, Cooper JP (2004) Indecent exposure: when telomeres become uncapped. Mol Cell 13: 7–18 [DOI] [PubMed] [Google Scholar]
- Frank-Vaillant M, Marcand S (2001) NHEJ regulation by mating type is exercised through a novel protein, Lif2p, essential to the ligase IV pathway. Genes Dev 15: 3005–3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E, Roberge M, Giraldo R, Rhodes D, Gasser SM (1993) Distortion of the DNA double helix by RAP1 at silencers and multiple telomeric binding sites. J Mol Biol 231: 293–310 [DOI] [PubMed] [Google Scholar]
- Grandin N, Damon C, Charbonneau M (2001) Cdc13 prevents telomere uncapping and Rad50-dependent homologous recombination. EMBO J 20: 6127–6139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Larrivee M, Labrecque P, Wellinger RJ (1998) Yeast Ku as a regulator of chromosomal DNA end structure. Science 280: 741–744 [DOI] [PubMed] [Google Scholar]
- Greenwell PW, Kronmal SL, Porter SE, Gassenhuber J, Obermaier B, Petes TD (1995) TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell 82: 823–829 [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T (1999) Mammalian telomeres end in a large duplex loop. Cell 97: 503–514 [DOI] [PubMed] [Google Scholar]
- Grossi S, Bianchi A, Damay P, Shore D (2001) Telomere formation by rap1p binding site arrays reveals end-specific length regulation requirements and active telomeric recombination. Mol Cell Biol 21: 8117–8128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy CF, Sussel L, Shore D (1992) A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev 6: 801–814 [DOI] [PubMed] [Google Scholar]
- Herrmann G, Lindahl T, Schar P (1998) Saccharomyces cerevisiae LIF1: a function involved in DNA double-strand break repair related to mammalian XRCC4. EMBO J 17: 4188–4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J, Ishikawa F (2001) spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol 11: 1624–1630 [DOI] [PubMed] [Google Scholar]
- Kegel A, Sjostrand JO, Astrom SU (2001) Nej1p, a cell type-specific regulator of nonhomologous end joining in yeast. Curr Biol 11: 1611–1617 [DOI] [PubMed] [Google Scholar]
- Konig P, Giraldo R, Chapman L, Rhodes D (1996) The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell 85: 125–136 [DOI] [PubMed] [Google Scholar]
- Krauskopf A, Blackburn EH (1996) Control of telomere growth by interactions of RAP1 with the most distal telomeric repeats. Nature 383: 354–357 [DOI] [PubMed] [Google Scholar]
- Kyrion G, Boakye KA, Lustig AJ (1992) C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Mol Cell Biol 12: 5159–5173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrion G, Liu K, Liu C, Lustig AJ (1993) RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev 7: 1146–1159 [DOI] [PubMed] [Google Scholar]
- Le S, Moore JK, Haber JE, Greider CW (1999) RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152: 143–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE (1998) Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94: 399–409 [DOI] [PubMed] [Google Scholar]
- Levy DL, Blackburn EH (2004) Counting of Rif1p and Rif2p on Saccharomyces cerevisiae telomeres regulates telomere length. Mol Cell Biol 24: 10857–10867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, de Lange T (2003) Rap1 affects the length and heterogeneity of human telomeres. Mol Biol Cell 14: 5060–5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Lustig AJ (1996) A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev 10: 1310–1326 [DOI] [PubMed] [Google Scholar]
- Li B, Oestreich S, de Lange T (2000) Identification of human Rap1: implications for telomere evolution. Cell 101: 471–483 [DOI] [PubMed] [Google Scholar]
- Lieb JD, Liu X, Botstein D, Brown PO (2001) Promoter-specific binding of Rap1 revealed by genome-wide maps of protein–DNA association. Nat Genet 28: 327–334 [DOI] [PubMed] [Google Scholar]
- Lin J, Smith DL, Blackburn EH (2004) Mutant telomere sequences lead to impaired chromosome separation and a unique checkpoint response. Mol Biol Cell 15: 1623–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev KS, Gordenin DA, Resnick MA (2002) The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108: 183–193 [DOI] [PubMed] [Google Scholar]
- Londono-Vallejo JA, Der-Sarkissian H, Cazes L, Bacchetti S, Reddel RR (2004) Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res 64: 2324–2327 [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Lundblad V, Blackburn EH (1993) An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 73: 347–360 [DOI] [PubMed] [Google Scholar]
- Luo K, Vega-Palas MA, Grunstein M (2002) Rap1–Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev 16: 1528–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig AJ (2003) Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nat Rev Genet 4: 916–923 [DOI] [PubMed] [Google Scholar]
- Marcand S, Brevet V, Mann C, Gilson E (2000) Cell cycle restriction of telomere elongation. Curr Biol 10: 487–490 [DOI] [PubMed] [Google Scholar]
- Marcand S, Gilson E, Shore D (1997) A protein-counting mechanism for telomere length regulation in yeast. Science 275: 986–990 [DOI] [PubMed] [Google Scholar]
- McEachern MJ, Blackburn EH (1995) Runaway telomere elongation caused by telomerase RNA gene mutations. Nature 376: 403–409 [DOI] [PubMed] [Google Scholar]
- McEachern MJ, Iyer S, Fulton TB, Blackburn EH (2000) Telomere fusions caused by mutating the terminal region of telomeric DNA. Proc Natl Acad Sci USA 97: 11409–11414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski PA, Mieczkowska JO, Dominska M, Petes TD (2003) Genetic regulation of telomere–telomere fusions in the yeast Saccharomyces cerevisae. Proc Natl Acad Sci USA 100: 10854–10859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Ferreira MG, Cooper JP (2005) Taz1, Rap1 and Rif1 act both inter-dependently and independently to maintain telomeres. EMBO J [E-pub ahead of print: 11 August 2005; doi:10.1038/sj.emboj.7600779] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne GT, Jin S, Shannon KB, Weaver DT (1996) Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol Cell Biol 16: 4189–4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, Haber JE (1996) Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol 16: 2164–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z, Bai Y, Poon D, Weil PA, Struhl K (1996) TBP-associated factors are not generally required for transcriptional activation in yeast. Nature 383: 188–191 [DOI] [PubMed] [Google Scholar]
- Moretti P, Shore D (2001) Multiple interactions in Sir protein recruitment by Rap1p at silencers and telomeres in yeast. Mol Cell Biol 21: 8082–8094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SL, Shoemaker DD, Boeke JD (2001) A DNA microarray-based genetic screen for nonhomologous end-joining mutants in Saccharomyces cerevisiae. Science 294: 2552–2556 [DOI] [PubMed] [Google Scholar]
- Prescott JC, Blackburn EH (2000) Telomerase RNA template mutations reveal sequence-specific requirements for the activation and repression of telomerase action at telomeres. Mol Cell Biol 20: 2941–2948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieur M, Al Achkar W, Aurias A, Couturier J, Dutrillaux AM, Dutrillaux B, Flüry-Herard A, Gerbault-Seureau M, Hoffschir F, Lamoliatte E, Lefrançois D, Lombard M, Murelis M, Ricoul M, Sabatier L, Viegas-Péquignot E (1988) Acquired chromosome rearrangements in human lymphocytes: effect of aging. Hum Genet 79: 147–150 [DOI] [PubMed] [Google Scholar]
- Rattray AJ, McGill CB, Shafer BK, Strathern JN (2001) Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae: a role for SAE2/COM1. Genetics 158: 109–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Runge KW (1999) The yeast telomere length counting machinery is sensitive to sequences at the telomere–nontelomere junction. Mol Cell Biol 19: 31–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie KB, Petes TD (2000) The Mre11p/Rad50p/Xrs2p complex and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics 155: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schar P, Herrmann G, Daly G, Lindahl T (1997) A newly identified DNA ligase of Saccharomyces cerevisiae involved in RAD52-independent repair of DNA double-strand breaks. Genes Dev 11: 1912–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D, Nasmyth K (1987) Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell 51: 721–732 [DOI] [PubMed] [Google Scholar]
- Singer MS, Gottschling DE (1994) TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266: 404–409 [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, Karlseder J, Holtgreve-Grez H, Jauch A, de Lange T (2002) DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr Biol 12: 1635–1644 [DOI] [PubMed] [Google Scholar]
- Sussel L, Shore D (1991) Separation of transcriptional activation and silencing functions of the RAP1-encoded repressor/activator protein 1: isolation of viable mutants affecting both silencing and telomere length. Proc Natl Acad Sci USA 88: 7749–7753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS (2002) Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev 66: 630–670, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart AK, Teng SC, Zakian VA (2002) Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297: 1023–1026 [DOI] [PubMed] [Google Scholar]
- Tarsounas M, Munoz P, Claas A, Smiraldo PG, Pittman DL, Blasco MA, West SC (2004) Telomere maintenance requires the RAD51D recombination/repair protein. Cell 117: 337–347 [DOI] [PubMed] [Google Scholar]
- Teixeira MT, Arneric M, Sperisen P, Lingner J (2004) Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117: 323–335 [DOI] [PubMed] [Google Scholar]
- Teng SC, Chang J, McCowan B, Zakian VA (2000) Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol Cell 6: 947–952 [DOI] [PubMed] [Google Scholar]
- Teo SH, Jackson SP (1997) Identification of Saccharomyces cerevisiae DNA ligase IV: involvement in DNA double-strand break repair. EMBO J 16: 4788–4795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo SH, Jackson SP (2000) Lif1p targets the DNA ligase Lig4p to sites of DNA double-strand breaks. Curr Biol 10: 165–168 [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y, Taggart AK, Zakian VA (2001) The role of the Mre11–Rad50–Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr Biol 11: 1328–1335 [DOI] [PubMed] [Google Scholar]
- Valencia M, Bentele M, Vaze MB, Herrmann G, Kraus E, Lee SE, Schar P, Haber JE (2001) NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature 414: 666–669 [DOI] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T (1998) TRF2 protects human telomeres from end-to-end fusions. Cell 92: 401–413 [DOI] [PubMed] [Google Scholar]
- Walker JR, Corpina RA, Goldberg J (2001) Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412: 607–614 [DOI] [PubMed] [Google Scholar]
- Walmsley RM, Szostak JW, Petes TD (1983) Is there left-handed DNA at the ends of yeast chromosomes? Nature 302: 84–86 [DOI] [PubMed] [Google Scholar]
- Wang RC, Smogorzewska A, de Lange T (2004) Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119: 355–368 [DOI] [PubMed] [Google Scholar]
- Wilson TE, Grawunder U, Lieber MR (1997) Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature 388: 495–498 [DOI] [PubMed] [Google Scholar]
- Wilson TE, Topper LM, Palmbos PL (2003) Non-homologous end-joining: bacteria join the chromosome breakdance. Trends Biochem Sci 28: 62–66 [DOI] [PubMed] [Google Scholar]
- Wotton D, Shore D (1997) A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev 11: 748–760 [DOI] [PubMed] [Google Scholar]
- Yu GL, Bradley JD, Attardi LD, Blackburn EH (1990) In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature 344: 126–132 [DOI] [PubMed] [Google Scholar]
- Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T (2003) ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol Cell 12: 1489–1498 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data


