Abstract
We recently described a novel megaplasmid-encoded adhesin produced by certain Shiga toxigenic Escherichia coli (STEC) strains that lack the locus for enterocyte effacement (LEE) pathogenicity island. This adhesin, designated Saa (STEC autoagglutinating adhesin), may be a marker for a subset of LEE-negative STEC strains capable of causing severe gastrointestinal and systemic diseases in humans. In this study, we developed a pentavalent PCR assay for the detection of saa as well as other proven and putative STEC virulence genes (stx1, stx2, eae, and ehxA). The five primer pairs used in the assay do not interfere with each other and generate amplification products of 119, 180, 255, 384, and 534 bp.
Shiga toxigenic Escherichia coli (STEC) strains are an important cause of gastrointestinal diseases in humans, particularly since these infections may result in life-threatening sequelae, such as hemolytic uremic-syndrome (HUS) (8, 12, 19). The STEC family is very diverse, and strains belonging to a broad range of O:H serotypes have been associated with human diseases (8). However, epidemiological evidence indicates that certain STEC subsets (for example, strains belonging to serotype O157:H7) account for a disproportionately large number of serious infections. STEC strains produce one or both of two major types of Shiga toxin, designated Stx1 and Stx2, and the production of the latter is associated with an increased risk of developing HUS (2, 9, 13). In addition, a subset of STEC strains considered to be highly virulent for humans has the capacity to produce attaching and effacing lesions on intestinal mucosa, a property encoded on a pathogenicity island termed the locus for enterocyte effacement (LEE). LEE encodes a type III secretion system and E. coli secreted proteins, which deliver effector molecules to the host cell and disrupt the host cytoskeleton (4, 5, 20). LEE also carries eae, which encodes an outer membrane protein (intimin) required for intimate attachment to epithelial cells (22); eae has been used as a convenient diagnostic marker for LEE-positive STEC strains (7, 11, 14). However, the presence of eae is not absolutely linked to human virulence, as some sporadic cases of severe STEC disease, including HUS, as well as occasional outbreaks have been caused by LEE-negative strains (18, 19). Most STEC strains isolated from humans (both LEE positive and LEE negative) also carry large (>90-kb) plasmids encoding proteins such as the enterohemorrhagic E. coli enterohemolysin (EhxA) (21) and an extracellular serine protease (EspP) (3), both of which may be accessory virulence factors.
Direct PCR analysis is increasingly being used for the detection of STEC in primary cultures of feces or foods. A positive reaction with primers specific for stx1 or stx2 is sufficient to confirm the presence of STEC in a sample, but the use of primers capable of detecting accessory virulence genes provides additional clinically relevant information that may also have great epidemiological value. Indeed, we previously described a multiplex PCR assay specific for stx1, stx2, eae, and ehxA genes. This assay permits the direct detection and characterization of STEC in crude fecal culture extracts without the need for the isolation of STEC itself (14). Recently, Paton et al. (17) described a gene, designated saa, which is carried on the large plasmid of certain LEE-negative but not LEE-positive STEC strains. This gene encodes a novel outer membrane protein which appears to function as an autoagglutinating adhesin, and the introduction of cloned saa confers a semilocalized adherence phenotype on E. coli K-12 strains (17). The saa gene was originally isolated from 98NK2, a LEE-negative O113:H21 STEC strain responsible for an outbreak of HUS in Adelaide, South Australia, in 1998 (18). It is also present in other LEE-negative STEC strains from HUS cases in our collection, including B2F1, an O91:H21 strain with very high virulence in a mouse model of STEC disease (10). Thus, it is possible that saa is a marker for the hitherto ill-defined subset of LEE-negative STEC strains capable of causing life-threatening disease in humans. Accordingly, in the present study, we modified our multiplex PCR assay for stx1, stx2, eae, and ehxA to include additional primers for the detection of saa.
saa-specific PCR.
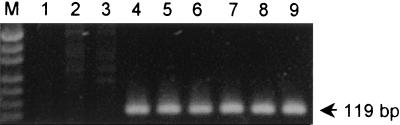
The saa-specific PCR primers used were 5′-CGTGATGAACAGGCTATTGC-3′ and 5′-ATGGACATGCCTGTGGCAAC-3′ (designated SAADF and SAADR, respectively). These primers direct the amplification of a 119-bp portion of the saa gene (nucleotides 2652 to 2770 in the sequence deposited in GenBank [accession number AF325220]) which is absolutely conserved among diverse STEC strains and avoid a region containing two to four copies of a direct 111-bp repeat sequence (17). The saa-specific PCR was initially characterized by using crude DNA extracts from 32 STEC strains in our collection, prepared as described previously (16). These strains had previously been tested for the presence of saa-related genes by Southern hybridization analysis, and 19 were known to be saa positive (17). The STEC strains tested included representatives of serogroups O113 (5 strains), O111 (3 strains), O157 (2 strains), and O91 (2 strains); 1 strain each from serogroups O6, O23, O26, O48, O82, O98, O128, O141, O159, and OX3; and 10 STEC strains which were O nontypeable or of unknown type. Samples (3 μl) of each extract were amplified by PCR under previously described conditions (14). PCR products were then electrophoresed on 2% agarose gels and stained with ethidium bromide. All known saa-positive STEC extracts yielded strong 119-bp PCR products (results for six of these are shown in Fig. 1, lanes 4 to 9), whereas this result was not seen for extracts from any of the saa-negative STEC strains (results for two such strains are shown in Fig. 1, lanes 2 and 3).
FIG. 1.
saa-specific PCR analysis of reference STEC strains. Lanes: M, DNA size markers (pUC19 DNA digested with HpaII; fragment sizes visible are 501 or 489, 404, 331, 242, 190, 147, and 111 bp); 1, negative control; 2 and 3, saa-negative STEC strains PH (O111:H−) and 95SF2 (O157:H−), respectively; 4 to 9, saa-positive STEC strains B2F1 (O91:H21), 94CR (O48:H21), 98NK2 (O113:H21), 97MW1 (O113:H21), 95HE4 (O91), and 99AM1 (O23), respectively. The expected mobility for the saa-specific PCR product is also indicated.
Sensitivity of the saa-specific PCR.
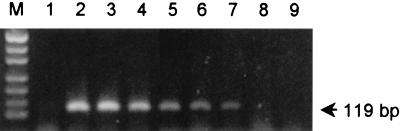
To assess the sensitivity of the saa-specific PCR, a fresh overnight broth culture of E. coli K-12 was spiked with serial 10-fold dilutions of a broth culture of the O113:H21 STEC strain 98NK2. Extracts of these samples were then subjected to the saa-specific PCR assay (Fig. 2). A 119-bp PCR product could still be seen in the sample that contained a 106-fold-diluted STEC culture (equivalent to <102 STEC CFU per assay) but not in the sample containing the 107- or 108-fold dilution.
FIG. 2.
Sensitivity of saa-specific PCR. A culture of E. coli K-12 was spiked with serial 10-fold dilutions of a culture of O113:H21 STEC strain 98NK2, and extracts of these samples were subjected to the saa-specific PCR assay. Lanes: M, DNA size markers (as for Fig. 1); 1, negative control (unspiked E. coli K-12 extract); 2 to 9, extracts of an E. coli K-12 culture spiked with 101-, 102-, 103-, 104-, 105-, 106-, 107-, and 108-fold dilutions of a 98NK2 culture, respectively. The expected mobility for the saa-specific PCR product is also indicated.
Multiplex PCR analysis of STEC strains and primary fecal cultures.
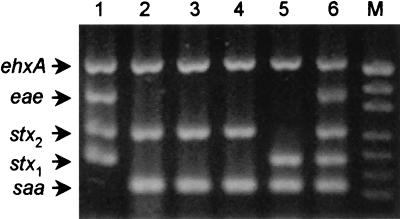
Having confirmed the specificity of the saa-specific primers, we combined SAADF and SAADR with four primer pairs described previously for the detection of stx1, stx2, eae, and ehxA; the latter primer pairs generate amplification products of 180, 255, 384, and 534 bp, respectively (14). The pentavalent PCR assay was initially tested by using chromosomal DNA extracted from previously characterized STEC strains as a template. Reaction mixtures contained a 250 nM concentration of each of the 10 primers, and amplification conditions were as described previously (14). The PCR product profiles for five of the previously characterized STEC strains are shown in Fig. 3, lanes 1 to 5; in each case, the pattern obtained was consistent with previous analyses. The O111 STEC strain shown in Fig. 3, lane 1, was known to be positive for stx1, stx2, eae, and ehxA and yielded four PCR products of the expected sizes (180, 255, 384, and 534 bp, respectively). The strains shown in Fig. 3, lanes 2 to 5, were known to be saa positive, and each contained a PCR product of the expected size for this gene (119 bp) as well as products of the expected sizes for ehxA (534 bp) and either stx2 (255 bp) or stx1 (180 bp). Figure 3, lane 6, shows the pattern obtained when pooled DNA from the STEC strains tested in lanes 1 and 2 was analyzed; clear PCR products were obtained for all five genes, indicating a lack of interference between any of the primer pairs or amplicons. The remaining STEC strains also yielded multiplex PCR profiles which were consistent with previous characterizations (results not shown).
FIG. 3.
Multiplex PCR analysis of chromosomal DNA from reference STEC strains. Lanes: 1, saa-negative STEC strain 96RO1 (O111:H−; 2 to 5, saa-positive STEC strains 98NK2 (O113:H21), B2F1 (O91:H21), 99AM1 (O23), and 95HE4 (O91), respectively; 6, pooled DNA from strains 96RO1 and 98NK2; M, DNA size markers (as for Fig. 1). The expected mobilities for the various PCR products are also indicated.
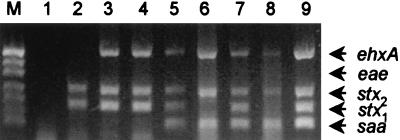
To demonstrate the diagnostic utility of the assay, crude DNA extracts of primary fecal cultures from seven patients (one with uncomplicated diarrhea, five with bloody diarrhea, and one with HUS) were examined (Fig. 4). Each of these samples had previously been found positive by PCR for the presence of the stx1 and/or stx2 genes (16), confirming the diagnosis of STEC infection. All of the samples were found negative for eae, indicating that the various causative STEC strains were LEE negative. Three of the samples (from two of the patients with bloody diarrhea and the one with uncomplicated diarrhea) were found negative for saa (Fig. 4, lanes 2 to 4). The crude extracts from the patient with HUS (Fig. 4, lane 5) and the three remaining patients with bloody diarrhea (lanes 6 to 8) were found saa positive. In all of the above cases, the multiplex PCR profile of the crude extract matched that of the STEC strains isolated from that sample (results not presented, except for the O48:H21 isolate from the HUS patient, shown in Fig. 4, lane 9). Analysis of crude fecal culture extracts from 12 healthy controls yielded negative results for all target genes (results not shown).
FIG. 4.
Multiplex PCR analysis of crude DNA extracts from primary fecal cultures. Lanes: M, DNA size markers (as for Fig. 1); 1, negative control; 2 to 8, extracts from patients with culture-proven STEC infection; 9, positive control (chromosomal DNA from saa-positive O48:H21 STEC strain 94CR). The expected mobilities for the various PCR products are also indicated.
Conclusions.
PCR is one of the most sensitive yet relatively rapid means of determining whether a fecal specimen or a food sample contains STEC (19). Although direct extracts of feces or foods can be used as templates for PCR, the best results are usually obtained by testing extracts of primary broth cultures (1, 6, 16, 19). The broth enrichment step can involve as little as 4 h of incubation and serves two purposes: inhibitors in the sample are diluted, and bacterial growth increases the number of copies of the target sequence, enhancing sensitivity. Detection of either the stx1 or the stx2 gene confirms the presence of STEC, but testing for the presence of additional gene sequences can provide clinically and epidemiologically important additional information about the infecting strain. For example, testing for eae and ehxA confirms the presence of the LEE pathogenicity island and the large virulence plasmid, respectively, both of which are more commonly found in STEC strains associated with severe human disease (2, 7, 21).
In the present study, we designed PCR primers based on the saa gene, which is encoded on the large virulence plasmid of a subset of LEE-negative STEC strains. This subset includes six of the seven strains in our collection from cases of HUS caused by LEE-negative STEC strains (17). Thus, saa may be an important marker for human-virulent LEE-negative STEC strains. However, the absence of an appropriate animal model prevents direct assessment of the contribution of saa to virulence. Demonstration of a link between carriage of this gene and the capacity to cause severe disease will require analysis of much larger STEC strain collections, a task for which PCR is well suited. In the present study, the saa-specific primers were combined with primers specific for other proven or putative STEC virulence genes (stx1, stx2, eae, and ehxA) in a multiplex format. The various primers were designed such that the PCR products differed in size (119, 180, 255, 384, and 534 bp, respectively) and so could be readily distinguished by agarose gel electrophoresis. The specificity of the saa-specific primers was confirmed by testing DNA extracted from a wide range of STEC strains previously characterized for the presence of saa by Southern hybridization analysis. The saa-specific PCR assay was also very sensitive, and a saa-negative E. coli culture spiked with a 106-fold-diluted saa-positive STEC culture (i.e., the STEC comprised 0.0001% of the total flora) generated a PCR product which was visible on an ethidium bromide-stained agarose gel.
The pentavalent multiplex PCR assay described in this article is clearly a useful tool for the molecular analysis of fecal samples from patients with suspected STEC disease. We also previously described another multiplex PCR assay directed at serogroup-specific genes within the rfb loci of E. coli O111, O113, and O157 (15). Collectively, these two assays can provide comprehensive information on the genotype of an infecting STEC strain within 24 h of receipt of a specimen. Detection of a similar PCR profile in crude fecal extracts from more than one patient within a given period of time may provide the earliest (albeit circumstantial) evidence for a link between cases consistent with a common-source outbreak. Confirmation of an outbreak would ultimately depend on isolation and genotyping of the causative STEC strain or strains, but this process could take weeks. Moreover, given the sensitivity of PCR screens, there is a likelihood that a proportion of genuine STEC PCR-positive specimens will not yield a strain even after heroic efforts. In such circumstances, multiplex PCR analysis is a valuable diagnostic and epidemiological tool. This multiplex assay will also facilitate the analysis of large STEC culture collections to further examine the linkage between the carriage of saa and the capacity of a given strain to cause severe disease in humans.
Acknowledgments
This work was supported by a grant from the National Health and Medical Research Council of Australia.
REFERENCES
- 1.Begum, D., and M. P. Jackson. 1995. Direct detection of Shiga-like toxin-producing Escherichia coli in ground beef using the polymerase chain reaction. Mol. Cell Probes 9:259–264. [DOI] [PubMed] [Google Scholar]
- 2.Boerlin, P., S. A. McEwen, F. Boerlin-Petzold, J. B. Wilson, R. P. Johnson, and C. L. Gyles. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37:497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunder, W., H. Schmidt, and H. Karch. 1997. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol. Microbiol. 24:767–778. [DOI] [PubMed] [Google Scholar]
- 4.Donnenberg, M. S., J. B. Kaper, and B. B. Finlay. 1997. Interactions between enteropathogenic Escherichia coli and host epithelial cells. Trends Microbiol. 5:109–114. [DOI] [PubMed] [Google Scholar]
- 5.Elliott, S. J., L. A. Wainwright, T. K. McDaniel, K. G. Jarvis, Y. K. Deng, L.-C. Lai, B. P. McNamara, M. S. Donnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1–4. [DOI] [PubMed] [Google Scholar]
- 6.Gannon, V. P. J., R. K. King, J. Y. Kim, and E. J. Thomas. 1992. Rapid and sensitive method for detection of Shiga-like toxin-producing Escherichia coli in ground beef using the polymerase chain reaction. Appl. Environ. Microbiol. 58:3809–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gannon, V. P. J., M. Rashed, R. K. King, and E. J. G. Thomas. 1993. Detection and characterization of the eae gene of Shiga-like toxin-producing Escherichia coli using polymerase chain reaction. J. Clin. Microbiol. 31:1268–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karmali, M. A. 1989. Infection by verotoxin-producing Escherichia coli. Clin. Microbiol. Rev. 2:15–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleanthous, H., H. R. Smith, S. M. Scotland, R. J. Gross, B. Rowe, C. M. Taylor, and D. V. Milford. 1990. Haemolytic uraemic syndromes in the British Isles, 1985-8: association with Verocytotoxin producing Escherichia coli. Part 2: microbiological aspects. Arch. Dis. Child. 65:722–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindgren, S. W., A. R. Melton, and A. D. O’Brien. 1993. Virulence of enterohemorrhagic Escherichia coli O91:H21 clinical isolates in an orally infected mouse model. Infect. Immun. 61:3832–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Louie, M., J. De-Azavedo, R. Clarke, A. Borczyk, H. Lior, M. Richter, and J. Brunton. 1994. Sequence heterogeneity of the eae gene and detection of verotoxin-producing Escherichia coli using serotype-specific primers. Epidemiol. Infect. 112:449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ostroff, S. M., P. I. Tarr, M. A. Neill, J. H. Lewis, N. Hargrett-Bean, and J. M. Kobayashi. 1989. Toxin genotypes and plasmid profiles as determinants of systemic sequelae in Escherichia coli O157:H7 infections. J. Infect. Dis. 160:994–999. [DOI] [PubMed] [Google Scholar]
- 14.Paton, A. W., and J. C. Paton. 1998. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 36:598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paton, A. W., and J. C. Paton. 1999. Direct detection of Shiga toxigenic Escherichia coli strains belonging to serogroups O111, O157, and O113 by multiplex PCR. J. Clin. Microbiol. 37:3362–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paton, A. W., J. C. Paton, P. N. Goldwater, and P. A. Manning. 1993. Direct detection of Escherichia coli Shiga-like toxin genes in primary fecal cultures using the polymerase chain reaction. J. Clin. Microbiol. 31:3063–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paton, A. W., P. Srimanote, M. C. Woodrow, and J. C. Paton. 2001. Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 69:6999–7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paton, A. W., M. C. Woodrow, R. M. Doyle, J. A. Lanser, and J. C. Paton. 1999. Molecular characterization of a Shiga toxigenic Escherichia coli O113:H21 strain lacking eae responsible for a cluster of cases of hemolytic-uremic syndrome. J. Clin. Microbiol. 37:3357–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perna, N. T., G. F. Mayhew, G. Posfai, S. J. Elliott, M. S. Donnenberg, J. B. Kaper, and F. R. Blattner. 1998. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 66:3810–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt, H., L. Beutin, and H. Karch. 1995. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL933. Infect. Immun. 63:1055–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu, J., and J. B. Kaper. 1992. Cloning and characterization of the eae gene of enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 6:411–417. [DOI] [PubMed] [Google Scholar]