Abstract
We show that nucleic acid structures may be conveniently and inexpensively characterized by their UV thermal difference spectra. A thermal difference spectrum (TDS) is obtained for a nucleic acid by simply recording the ultraviolet absorbance spectra of the unfolded and folded states at temperatures above and below its melting temperature (Tm). The difference between these two spectra is the TDS. The TDS has a specific shape that is unique for each type of nucleic acid structure, a conclusion that is based on a comparison of >900 spectra from 200 different sequences. The shape of the TDS reflects the subtleties of base stacking interactions that occur uniquely within each type of nucleic acid structure. TDS provides a simple, inexpensive and rapid method to obtain structural insight into nucleic acid structures, which is applicable to both DNA and RNA from short oligomers to polynucleotides. TDS complements circular dichroism as a tool for the structural characterization of nucleic acids in solution.
INTRODUCTION
DNA and RNA may adopt a number of unusual conformations. Several laboratories have shown the existence and functional importance of nucleic acid structures that are fundamentally different from the classical, canonical B-form duplex. A few years ago, Alexander Rich wrote: ‘DNA comes in many forms’ (1). This is true at all levels of structure. Different base pairs can form (G.G, A.A, etc.) and the helicity and/or orientation of the strands can be altered such as in Z-DNA (2), parallel-stranded (3) and Hoogsteen duplexes. One can also observe nucleic acid structures composed of more than two strands such as triple- (4,5) and quadruple- (6–12) helices. All these structures rely upon the formation of specific base-association schemes (base pairs, triplets or quartets) (1). The stability of these various conformational forms can be measured by a thermal denaturation experiment. Heating a structured nucleic acid leads to changes in its ultraviolet absorbance, which reflect the conformational change of the molecule in solution, specifically a disruption of base-stacking interactions. Such melting experiments can be analyzed to determine the thermodynamic basis of DNA or RNA structure stability. In most cases, one may perform thermal denaturation experiments by recording absorbance at 260 nm as a function of temperature. However, it may also be preferable to record absorbance at other wavelengths. For example, quadruplex denaturation does not lead to large variation in absorbance at 260 nm. It is, however, possible to follow this denaturation at other wavelengths such as 295 nm (13). This observation prompted us to determine the optimal wavelengths for thermal denaturation experiments for each type structure. In the course of our investigations, we realized that thermal difference spectra (TDS) provided a distinctive spectroscopic signature for each nucleic acid structure. We describe the method and its utility here.
MATERIALS AND METHODS
Nucleic acids samples
Most oligonucleotides presented here were synthesized by Eurogentec (Belgium) or Genosys on the 40–1000 nmol scale and resuspended in 200–500 µl of bi-distilled water. We checked the purity of most samples by mass spectrometry, HPLC or gel electrophoresis. Polynucleotide samples were purchased from Amersham-Pharmacia (Piscataway, NJ) and were used without further purification.
Determination of extinction coefficients
Concentration of all oligodeoxynucleotides were estimated by UV absorption using published sequence-dependent extinction coefficients (14). In selected cases, molar extinction coefficients were determined by a colorimetric phosphate analysis (15). One should not underestimate the difficulty in obtaining reliable extinction coefficients for all oligonucleotides. For example, we measured for the human telomeric repeat sequence 5′-AGGG(TTAG3)3, a molar extinction coefficient of 196 479 ± 5675 M−1 cm−1 by a colorimetric phosphate assay. Using two commonly used algorithms, values for the molar extinction coefficient of 251 800 and 228 500 M−1 cm−1 were calculated. The calculated values differ by 16–28% from the more accurate analytically determined value.
Absorbance spectra
All experiments presented here were performed on either Kontron Uvikons 940/943 or a Cary 3E UV/Vis spectrophotometer using quartz cuvettes with an optical pathlength of 0.2–1 cm (16). Absorbance spectra were recorded in the 220–335 nm range, with a scan speed of 200–1000 nm/min, with a data interval of 1 nm. In most cases, using cuvettes of 0.05–1 cm pathlength, maximum absorbance of the sample was 0.5–2.0.
Choice of the upper and lower temperatures
A melting curve is usually measured first to learn the width of the transition. The difference in spectra weakly depends on the difference in temperature. Using the narrowest possible temperature range is, in principle, recommended. However, one can still have an excellent superposition for most structures if one compares various temperature windows [see Supplementary Figure S1 which presents the TDS (raw data or normalized] for an i-DNA sample, using a number of different temperatures. Clearly, once the curves are normalized, the shapes are almost superimposable (Supplementary Figure S1B), demonstrating that the choice of the exact temperatures does not play a great role.
Experimental constraints
Reproducibility of the TDS is excellent even if spectra are recorded in a relatively crude way. There is no need for very slow scans and 1 pt/nm is enough for most applications. Any high-quality single- or double-beam spectrophotometer may be used, provided that its performance and stability over time in the far-UV region (and at high temperature) is checked. Either a Peltier thermoelectric temperature controller or a circulating water bath may be used, as long as a wide temperature range can be used. Appropriate precautions should be taken below room temperature to avoid condensation, and at high temperature to avoid evaporation and air bubbles. Thorough degassing of solutions helps to minimize bubble formation at high temperatures. Layering of solutions with mineral oil and tightly stoppering cuvettes help to minimize evaporation. Condensation at low temperatures can be minimized by blowing a stream of dry air on the outer cuvette surfaces. It is, of course, essential to choose a buffer that does not absorb light in the far-UV region. Cacodylate (pKa: 6.14 at 25°C) and acetate (pKa: 4.62) were preferred over phosphate or Tris buffer for experiments performed at neutral or slightly acidic pH. Their pKa is almost temperature independent.
RESULTS AND DISCUSSION
An example of the determination of a TDS is shown in Figure 1. The structure in this determination is the antiparallel ‘basket’ G-quadruplex form observed in Na+ solution for the human telomere repeat sequence 5′-AGGG(TTAGGG)3 (17). Figure 1A shows the circular dichroic spectra of the quadruplex. Figure 1B shows UV absorbance spectra at temperatures well above (90°C) and below (20°C) the melting temperature (56°C). Figure 1C shows the TDS obtained by subtracting the low temperature spectrum from the high temperature spectrum. In this case, the TDS is displayed as the difference between molar extinction coefficients between the unfolded and folded forms. Other methods for displaying the TDS that are convenient and informative are described below.
Figure 1.
Example of a thermal difference spectrum. The structure in this determination is the antiparallel ‘basket’ G-quadruplex form observed in Na+ solution for the human telomere repeat sequence 5′-AGGG(TTAGGG)3. (A) Circular dichroic spectrum, expressed as molar ellipticity. (B) UV absorbance spectra (expressed as molar extinction, M−1 cm−1) at 20°C (solid line) and 90°C (dashed line). (C). Thermal difference spectrum resulting from the subtraction of the 20°C spectrum in (B) from the 90°C spectrum.
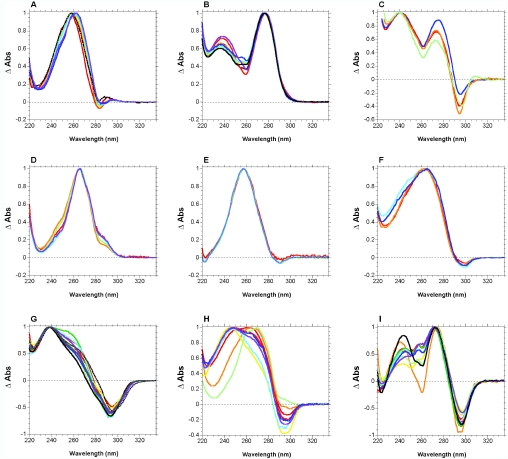
Examples of TDS for nine different structure types are shown in Figure 2. The curves are simply the result of the arithmetic difference between high and low temperature absorbance spectra (example shown in Figure 1B). For most experiments, high temperature was chosen >80°C, low temperature 20°C or below. These difference spectra were then normalized in order to facilitate the comparison of the spectral shapes, simply by dividing the raw data by its maximum value, so that the highest positive peak gets a Y-value of +1. This normalization, which unfortunately leads to a loss of information, gives in our hands the most reproducible results. In principle, molar extinction differences should also be interesting. However, our opinion is that, unless analytically determined extinction coefficients are available for all oligonucleotides (see Materials and Methods section), it is preferable and more reliable to use the normalization procedure. Analysis of the normalized TDS offers another advantage, as it allows to deal with partial transitions when (i) the Tm is too low or too high (no spectra of the pure folded/unfolded species) (ii) two transitions are overlapping. One may also analyze the shape of the ‘beginning’ of a transition (i.e. the part corresponding to partial dissociation at low temperature, below Tm) and compare it with the global shape or the ‘end’ (see Supplementary Figure S1 for an example).
Figure 2.
Normalized differential absorbance signatures: (A) DNA self-complementary duplexes, 100% AT; (B) DNA self-complementary duplexes 100% GC; (C) Z-DNA; (D) Parallel-stranded DNA; (E) GA DNA duplexes; (F) Hoogsteen DNA duplexes; (G) i-DNA; (H) Pyrimidine triplexes; (I) DNA G-quadruplexes in Na+. The complete legend as well as expanded spectra and relevant references may be found as Supplementary Figure S2.
The resulting normalized TDS are concentration independent (data not shown). Except for pyrimidine triplexes (Figure 2H) and to a lesser extent G-quadruplexes (Figure 2I), one cannot but notice that all spectra within a structural family are strikingly similar (e.g. Figure 2A, B and G). Sometimes spectra are essentially identical (Figure D and E). Comparison of these different structures reveals reproducible differences in TDS shapes, allowing an investigator to determine the structural type by the spectrum alone.
The robustness of this method was analyzed by several control experiments (see Figure 3). A simple DNA self-complementary duplex (5′-d-AAATTTAATTAAATTT) was chosen; its differential absorbance spectra was determined at various pH (5.5–7.5), salt concentration, in the presence or absence of magnesium and with different monocations (sodium, lithium or potassium). All spectra were nearly superimposable, despite the fact that the Tm of the duplex may depend on these conditions: as long as the melting transition occurs in the temperature interval defined by the low and high-temperature spectra, very reproducible differences were found (Figure 3A). Similar observations were found for most structures, with a few notable exceptions. For G-quadruplexes, this profile was independent on pH, magnesium, ionic strength, but not on the nature of the monocation [different quadruplex conformations may be observed in Na+ and K+ (12,17–19)]. A systematic difference was found between the spectra recorded in sodium and in potassium (Figure 3B). For i-DNA, the spectra appear to be different when working below pH 6: at this pH, a significant proportion of cytosines are protonated in the unfolded form, and C+ has a different absorbance from unprotonated C [data not shown and (20)].
Figure 3.
Experimental condition dependency of the differential absorbance shapes. (A) For a 16-base long self-complementary DNA duplex 5′-AAATTTAATTAAATTT; pH 5.5–7.2, with or without 10 mM MgCl2, in a sodium cacodylate buffer with 0.1–1 M NaCl, KCl or LiCl. All curves are superimposable. (B) For the 5′-AGGGTTAGGGTTAGGGTTAGGG intramolecular DNA quadruplex pH 6.0–7.2, in a sodium cacodylate buffer with 0.1–1 M NaCl (in blue), or 0.03–0.6 M KCl (in red). Note that CD spectra of the same sequence are shown as Supplementary Figure S3, and error bars at each wavelength (average of 11 independent measurements) as Supplementary Figure S4.
The first obvious application of this analysis is to help in the choice of appropriate wavelength(s) for a thermal denaturation experiment for more detailed thermodynamic studies. These results have been collected in Table 1. It should be noted that the relation between a differential peak and a ‘good’ wavelength is not always direct. In some cases, the observed hypochromicity does not result from the melting of a structure, but is the consequence of temperature-dependent extinction coefficient of the folded and/or the unfolded forms. For example, the positive peak at ∼273 nm observed in most quadruplexes (Figure 2I) results from a significant temperature-dependent increase of absorbance of the single strands. This is related to the classical problem of the sloping baseline analysis of melting profiles: all species exhibit temperature-dependent absorbance properties. One would then argue that the most convenient differential spectra should be isothermal; however, this requires the ability to record the absorbance of the folded and the unfolded states at the same temperature. This is possible in only a limited number of cases (21), when association and dissociation of the structure are very slow. Otherwise, T-jump or fast mixing experiments are required. Despite this limitation, this simple dual-temperature measurement is sufficient for most determinations.
Table 1.
Useful wavelengths for each nucleic acid structure
| Structure | Major positive peak | Other peaks/features |
|---|---|---|
| Duplexes | ||
| DNA duplex 0% GC | 259.5 ± 2 nma | 284 nm (−0.03) |
| RNA duplex 0% GC | 259 ± 2 nm | 284 nm (−0.28) |
| DNA duplex 50% GC | 267 ± 2 nm | |
| DNA duplex 100% GC | 276.5 ± 0.5 nm | 237.5 ± 1 nm (±0.66) |
| Parallel AT Duplex | 265.5 ± 0.5 nmb | shoulder at 288 nmc |
| Hoogsteen Duplex | 264 ± 1 nm | 299 ± 1 nm (−0.08)d |
| GA Duplex | 257.5 ± 0.5 nm | 293 ± 1 nm (−0.06) |
| 223 ± 1 nm (0)e | ||
| Z-DNA | 241 ± 2 nm | 295 ± 1 nm (−0.37) |
| Triplexes | ||
| Triplex TCf | 247 ± 1 nm | 295.5 ± 1 nm (−0.30)d,g |
| Quadruplexes | ||
| i-motif | 239 ± 1 nm | 294.5 nm (−0.60)d |
| G-quartet | 243 ± 2 nmh | 295 ± 1 nm (−0.73) |
| 273 ± 1 nmh | ||
For each structure, the wavelength of the highest positive TDS peak used for normalization is indicated in the first column, in bold. For the other peaks, their relative value is provided. (+) and (−) indicate positive and negative peaks, respectively. A negative peak indicates that upon melting a cooperative decrease in absorbance should be obtained.
aValid for B-DNA; A-DNA exhibits a different absorbance spectra, as shown by Jose and Pörschke (36). 0, 50 or 100% GC refers to the relative amounts of GC and AT base pairs. The exact position of the positive peak depends on sequence: alternating AT motifs have a maximum at ∼258 nm, whereas An.Tn blocks lead a maximum at ∼262 nm (see Figure 2A).
bIn agreement with Ramsing and Jovin (27).
cAlso see Pohl and Jovin (37).
dThis negative peak at ∼295 nm is the result of partial cytosine deprotonation upon unfolding (20).
eThis is the only structure for which an annulation of the differential spectra is observed in this region (see Figure 2E).
fValid for triplexes involving mixed TAT and CGC+ triplets. This spectra corresponds to a triplex-to-duplex conversion. For RNA polynucleotide triplexes solely involving U.A*U triplets, see (25). Poly dA. 2 poly dT gives a TDS close to dA18 2dT18 (Figure 2H).
gLavelle and Fresco (29).
hThe exact position and relative contribution of the peaks depend on the nature of the cation. The positive peak at ∼273 nm is attributed in large part to the variation in absorbance of the single-strand; the depth of the valley between the 243 and 273 nm peaks is strongly sequence dependent (see the thrombin aptamer, GGTTGGTGTGGTTGG for an atypical spectrum). The negative value at 295 nm may vary significantly. Also see (18,21) for the signature of other quadruplexes.
These spectra provide information complementary to circular dichroism. The UV absorbance of nucleic acids from 200 to 300 nm is exclusively due to transitions of the planar purine and pyrimidine bases. The phosphate backbone begins to contribute at ∼190 nm. The CD spectrum is probing the asymmetry of bases. At wavelengths >200 nm, the asymmetry arises from CD induced into the bases as a result of stacking interactions and coupling with backbone transitions. Hypochromicity is mainly the result of base stacking (22). Therefore, one can expect that the relative positioning of the bases will play a crucial role in this differential spectra. The theory of hypochromicity is complex, the changes in light absorption occurring upon denaturation is attributed to the interaction between the dipoles induced in the chromophore by the light (22). It is striking that a number of very different structures involve a negative differential absorbance at ∼295 nm (Table 1). This might be the result of the significant contribution of n → π* transition moments at this wavelength: as these transition moments are parallel to the helix axis (23), increased stacking resulting from the formation of a folded structure may result in hyperchromism. However, the absorbance spectra of nucleic acid bases are deceptively simple: they actually correspond to a number of different π → π* and n → π* transitions. Hypochromism at a given wavelength will not only depend on the intrinsic transition moments of each base, but also on the relative moment of the interacting bases (base-paired or stacked). Such complexity will make an accurate calculation of predicted differential absorbances a difficult task.
Although the dissociation of all G-quadruplexes analyzed so far show an inverted transition at 295 nm (13), this property is not unambiguously indicative of G-quadruplexes. Z-DNA (Figure 2C) i-motif (Figure 2G) Hoogsteen duplexes (Figure 2F) and pyrimidine triplex (Figure 2H) formation also leads to an inverted transition at 295 nm. In Figure 2F–H, this is the result of the differences in absorbance between C and C+ (20). A simple comparison of these spectra indicates that the global shape rather than the wavelength of the peak is specific for each structure (Figure 2).
These observations suggest that the simple recording of two absorbance spectra might be sufficient to obtain qualitative structural information on the identity of the folded form. This was a common accepted practice in the sixties, when the melting of polynucleotides [or guanylic acid (6)] was analyzed [see for example (24–26)]. Unfortunately, with a few notable exceptions (27–32), complete spectra are rarely presented in contemporary reports. We demonstrate here that in many cases this difference is very informative. Of course, this requires that the high and low temperatures correspond to the unfolded and folded forms, respectively. In other words, the Tm of the structure must lie between these two values. Although the spectra of different nucleic acids that fold into the same type of structure are not identical, significant homology was found between the spectra. The uniqueness of these spectra is sufficient, in most cases, to suggest a type of folding. However, we could not confidently distinguish the differential spectra of a purine-rich triplex (the so-called GA or GT motif) from the melting of a duplex (data not shown). This is in agreement with the conclusions that the formation of these triplexes occurs without significant hypochromicity (33). Similarly, no significant difference was found between a mismatched and a fully complementary DNA duplex (data not shown).
We have now collected >900 spectra, corresponding to >200 different nucleic acid sequences (mostly DNA), each tested 2–26 times under different experimental conditions. This is, to our knowledge, the first attempt to systematically collect thermal difference spectra as initially proposed 40 years ago by Fresco et al (34). Our goal is to first build an open access database of the spectra of nucleic acids and to propose a predictive algorithm that provides a probability function of a given structure. This should be helpful in the discovery of new nucleic acid folding schemes, and in the crude determination of the global folding of an oligonucleotide (which, for example, might have been found by SELEX). We are now applying this method to the analysis of trinucleotide repeats (35) and to samples which may adopt complex conformations, involving more than one simple structure. Efforts are also being made on RNA structures (and corresponding DNA strands carrying deoxyuracyl bases): RNA duplex and quadruplex structures exhibit related but distinct signatures from the corresponding DNA duplexes (data not shown) and quadruplexes (18,21), respectively. We also wish to analyze the signature of modified bases (such as 5-methyl cytosines) and the impact of modifying the sugar/phosphate backbone (such as 2′-O-methyl phosphorothioates, LNA or PNA).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank M. Rougée, T. Garestier, L. Guittat, P. Arimondo, P. Alberti, B. Saccà (MNHN, Paris, France) M. Mills (UCT, Cape Town, Republic of South Africa) for helpful discussions. This work was supported by an ARC grant (#3365 to J.L.M), a EU FP6 MOLCANCERMED grant (to J.L.M.), a fondation Jérome Lejeune research grant (to S.A.) and National Cancer Institute grant CA35635 (to J.B.C). Funding to pay the Open Access publication charges for this article was provided by INSERM.
Conflict of interest statement: None declared.
REFERENCES
- 1.Rich A. DNA comes in many forms. Gene. 1993;135:99–109. doi: 10.1016/0378-1119(93)90054-7. [DOI] [PubMed] [Google Scholar]
- 2.Rich A., Zhang S.G. Z-DNA: the long road to biological function. Nature Rev. Genet. 2003;4:566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- 3.Jovin T.M., Rippe K., Ramsing N.B., Klement R., Elhorst W., Vojtisova M. Parallel stranded DNA. In: Sarma R.H., Sarma M.H., editors. Structure & Methods, DNA & RNA. Vol 3. Schenectady, New York: Adenine Press; 1990. pp. 155–174. [Google Scholar]
- 4.Moser H.E., Dervan P.B. Sequence specific cleavage of double helical DNA by triple helix formation. Science. 1987;238:645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- 5.Le Doan T., Perrouault L., Praseuth D., Habhoub N., Decout J.-L., Thuong N.T., Lhomme J., Hélène C. Sequence specific recognition, photocrosslinking and cleavage of the DNA double helix by an oligo thymidylate covalently linked to an azidoproflavine derivative. Nucleic Acids Res. 1987;15:7749–7760. doi: 10.1093/nar/15.19.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gellert M., Lipsett M.N., Davies D.R. Helix formation by guanylic acid. Proc. Natl Acad. Sci. USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gehring K., Leroy J.L., Guéron M. A tetrameric structure with protonated cytosine.cytosine base pairs. Nature. 1993;363:561–565. doi: 10.1038/363561a0. [DOI] [PubMed] [Google Scholar]
- 8.Henderson E., Hardin C.C., Walk S.K., Tinoco I., Jr, Blackburn E.H. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine.guanine base pairs. Cell. 1987;51:899–908. doi: 10.1016/0092-8674(87)90577-0. [DOI] [PubMed] [Google Scholar]
- 9.Sundquist W.I., Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- 10.Williamson J.R., Raghuraman M.K., Cech T.R. Monovalent cation induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 11.Kang C.H., Berger I., Lockshin C., Radliff R., Moyzis R., Rich A. Crystal structure of intercalated four stranded d(C3T) at 1.4 Å resolution. Proc. Natl Acad. Sci. USA. 1994;91:11636–11640. doi: 10.1073/pnas.91.24.11636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parkinson G.N., Lee M.P.H., Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 13.Mergny J.L., Phan A.T., Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 14.Cantor C.R., Warshaw M.M., Shapiro H. Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 15.Plum G.E. Current Protocols in Nucleic Acid Chemistry. New York: Wiley; 2000. Determination of oligonucleotide molar extinction coefficients; pp. 7.3.1–7.3.17. [Google Scholar]
- 16.Mergny J.L., Lacroix L. Analysis of thermal melting curves. Oligonucleotides. 2003;13:515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y., Patel D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 18.Saccà B., Lacroix L., Mergny J.L. The effect of chemical modifications on the thermal stability of different G-quadruplexes-forming oligonucleotides. Nucleic Acids Res. 2005;33:1182–1192. doi: 10.1093/nar/gki257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J., Correia J.J., Wang L., Trent J.O., Chaires J.B. Not so crystal clear: the structure of the human telomeric G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mergny J.L., Lacroix L., Han X., Leroy J.L., Hélène C. Intramolecular folding of pyrimidine oligodeoxynucleotides into an i-DNA motif. J. Am. Chem. Soc. 1995;117:8887–8898. [Google Scholar]
- 21.Mergny J.L., de Cian A., Ghelab A., Saccà B., Lacroix L. Kinetics of tetramolecular quadruplexes. Nucleic Acids Res. 2005;33:81–94. doi: 10.1093/nar/gki148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tinoco I., Jr Hypochromism in polynucleotides. J. Am. Chem. Soc. 1960;82:4785–4790. [Google Scholar]
- 23.Rich A., Kasha M. The n->pi* transition in nucleic acids and polynucleotides. J. Am. Chem. Soc. 1960;82:6197–6199. [Google Scholar]
- 24.Riley M., Maling B., Chamberlin M.J. Physical and chemical characterization of two and three stranded adenine thymine and adenine uracil homopolymer complexes. J. Mol. Biol. 1966;20:359–389. doi: 10.1016/0022-2836(66)90069-6. [DOI] [PubMed] [Google Scholar]
- 25.Blake R.D., Massoulié J., Fresco J.R. A spectral approach to the equilibria between polyriboadenylate and polyribouridylate and their complexes. J. Mol. Biol. 1967;30:291–308. [PubMed] [Google Scholar]
- 26.Martin F.H., Uhlenbeck O.C., Doty P. Self complementary oligoribonucleotides: adenylic acid–uridylic acid block copolylers. J. Mol. Biol. 1971;57:201–215. doi: 10.1016/0022-2836(71)90341-x. [DOI] [PubMed] [Google Scholar]
- 27.Ramsing N.B., Jovin T.M. Parallel stranded duplex DNA. Nucleic Acids Res. 1988;16:6659–6676. doi: 10.1093/nar/16.14.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van de Sande J.H., Jovin T.M. Z* DNA, the left handed helical form of poly d(G–C) in MgCl2-ethanol is biologically active. EMBO J. 1982;1:115–120. doi: 10.1002/j.1460-2075.1982.tb01133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lavelle L., Fresco J.R. Enhanced stabilization of the triplexes d(C+-T)(6):d(A-G)(6).d(C-T)(6), d(T)(21):d(A)21.d(T)(21) and poly r(U:A.U) by water structure-making solutes. Biophys. Chem. 2003;105:701–720. doi: 10.1016/s0301-4622(03)00098-x. [DOI] [PubMed] [Google Scholar]
- 30.Davis T.M., McFail-Isom L., Keane E., Williams L.D. Melting of a DNA hairpin without hyperchromism. Biochemistry. 1998;37:6975–6978. doi: 10.1021/bi980047l. [DOI] [PubMed] [Google Scholar]
- 31.Kankia B.I. Mg2+-induced triplex formation of an equimolar mixture of poly(rA) and poly(rU) Nucleic Acids Res. 2003;31:5101–5107. doi: 10.1093/nar/gkg698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haq I., Chowdhry B.Z., Chaires J.B. Singular value decomposition of 3-D DNA melting curves reveals complexity in the melting process. Eur. Biophys. J. 1997;26:419–426. doi: 10.1007/s002490050096. [DOI] [PubMed] [Google Scholar]
- 33.Mills M., Arimondo P., Lacroix L., Garestier T., Hélène C., Klump H.H., Mergny J.L. Energetics of strand displacement reactions in triple helices: a spectroscopic study. J. Mol. Biol. 1999;291:1035–1054. doi: 10.1006/jmbi.1999.3014. [DOI] [PubMed] [Google Scholar]
- 34.Fresco J.R., Klotz L.C., Richards E.G. A new spectroscopic approach to the determination of helical secondary structure in ribonucleic acids. Cold Spring Harbor Symp. Quant. Biol. 1963;28:83–90. [Google Scholar]
- 35.Amrane S., Saccà B., Mills M., Chauhan M., Klump H.H., Mergny J.L. Length-dependent energetics of (CTG)n and (CAG)n trinucleotide repeats. Nucleic Acids Res. 2005;33:4065–4077. doi: 10.1093/nar/gki716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jose D., Pörschke D. Dynamics of the B-A transition of DNA double-helices. Nucleic Acids Res. 2004;32:2251–2258. doi: 10.1093/nar/gkh551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pohl F.M., Jovin T.M. Salt-induced co-operative conformational change of a synthetic DNA: equilibrium and kinetic studies with poly(dG-dC) J. Mol. Biol. 1972;67:375–396. doi: 10.1016/0022-2836(72)90457-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.