Abstract
The presence of active copper- and zinc-containing superoxide dismutase in isolates of the cryptic genospecies of Haemophilus, responsible for urogenital, neonatal, and mother-infant infections, can be used as a biochemical marker to discriminate them from H. influenzae sensu stricto strains.
Noncapsulate Haemophilus influenzae strains of biotype IV (and, to a lesser extent, biotype II [7]) are a significant cause of urogenital, neonatal, and mother-infant infections (15, 16, 20). Quentin and colleagues (13, 15) have shown that a subset of these strains belongs to a genetically distinct group (as adjudged by multilocus enzyme electrophoresis) which has been described as a cryptic genospecies related to H. influenzae and H. haemolyticus. These strains have increased sensitivity to quinolones (14) and a characteristic outer membrane protein profile (16), express a particular variant of outer membrane protein P6 (9), and possess peritrichous pili and exhibit tissue tropism for host cells of genital origin (18). While the lack of hemolytic activity and a positive ornithine decarboxylase test for the cryptic genospecies are used to separate them from H. haemolyticus (12), the cryptic genospecies can be discriminated from H. influenzae sensu stricto only by a PCR-based test that exploits the differences in their 16S ribosome sequences; no discriminating biochemical marker has yet been identified (17). In the work described here we have established that the presence of the enzyme copper- and zinc-containing superoxide dismutase (CuZnSOD) can be used to differentiate H. influenzae sensu stricto strains and those of the cryptic genospecies.
Bacterial CuZnSODs are involved in the protection of cells against free radical-mediated damage, a threat to pathogens derived from host defenses (2, 21). In capsulate strains of H. influenzae, which can be divided on the basis of multilocus electrophoretic typing into two widely separated phylogenetic divisions, divisions I and II, sodC (which encodes CuZnSOD) is present only in division II and type e strains (type e strains are distantly related to strains of both phylogenetic divisions). The gene is not present in division I strains (3). However, CuZnSOD activity cannot be detected in strains from either division. In one division II strain in which the lack of CuZnSOD activity was examined in depth (strain NCTC 8648, biotype IV), this was considered to be the result of the replacement of a critical histidine residue at the active site with tyrosine as the result of a mutation (3). In a follow-up study we found the presence of sodC and active enzyme both in human and animal oropharyngeal commensal species (including H. haemolyticus) and in pathogenic Haemophilus species (6). At that time four strains of noncapsulate H. influenzae were screened. Two of these (strains 11PS and 26E) contained sodC and produced active enzyme, and both are representatives of genital isolates of cryptic genospecies biotype IV. We have now extended these studies to screen further examples of the cryptic genospecies, capsulate division I and II strains (including biotype IV strains), and a defined collection of noncapsulate H. influenzae sensu stricto strains (10) for the prevalence of sodC and the ability to produce active CuZnSOD activity.
A collection of noncapsulate H. influenzae strains that were characterized by multilocus enzyme electrophoresis and that comprised electrotypes (ETs) 11 to 13, 26 to 27, 29 to 32, 35 to 45, 49 to 51, 53 to 55, 57 to 61, and 63 to 76 (10) was kindly provided by Terence Stull (Department of Pediatrics, University of Oklahoma College of Medicine). The Haemophilus cryptic genospecies biotype IV strains (strains 10N, 12N, 15N, 16N, 11PS, 26E, 189, 422, 427, 799, 847, and 911) have been described previously (13, 16). H. influenzae sensu stricto strains of serotypes a (n = 0), b (n = 5), e (n = 2), and f (n = 0) (the number of biotype IV strains is given in parentheses) were from our collection. All strains were grown in brain heart infusion broth supplemented with 1 μg of NAD ml−1 and 10 μg of hemin ml−1 at 37°C on an orbital shaker (200 rpm) for 18 h. Whole-cell sonicates were prepared and separated, and superoxide dismutase (SOD) activity was visualized in nondenaturing polyacrylamide gels as described previously (3, 4, 6). In some experiments SOD activity was visualized in samples separated in isoelectric focusing (IEF) pH 3-10 Ready gels (Bio-Rad) according to the manufacturer’s instructions. The copper chelator diethyl dithiocarbamic acid (DEDC; 10 mM) or potassium cyanide (2 mM) was used as the inhibitor of CuZnSOD activity (1, 3). Chromosomal DNA was prepared from 3-ml broth cultures (8) or with the Qiagen Genomic-tip kit according to the manufacturer’s instructions, and standard methods were used for restriction endonuclease digestion and Southern blotting, with washing to 80% stringency (19). Southern blots were probed with either a 32P- or a digoxigenin (Roche)-labeled Haemophilus DNA insert of pJSK114 (3, 6) consisting of a 360-bp HindIII-NcoI fragment, the 5′ part of the H. influenzae NCTC 8648 sodC gene, or a 537-bp probe of the H. parainfluenzae sodC gene. A standard PCR with oligonucleotides 5′-CTTAGCATTAGCAATCAGCGG-3′ and 5′-CACACCACATGCCATACGTG-3′ and with plasmid pJSK130 (3) as the DNA template was used to obtain an H. parainfluenzae sodC PCR product for labeling. The sodC genes from H. influenzae and H. parainfluenzae are virtually identical at the nucleotide level (3).
The Haemophilus sodC DNA-specific probes failed to hybridize to any of the defined noncapsulate collection of strains or to H. influenzae sensu stricto division I strains. In contrast, they hybridized to the division II, the type e, and all of the cryptic genospecies strains (Table 1). However, CuZnSOD activity was found only in the cryptic genospecies isolates (Table 1 and Fig. 1). In our hands the test is 100% sensitive and 100% specific. Thus, CuZnSOD activity is a useful biochemical marker for discrimination of the cryptic genospecies from H. influenzae sensu stricto. It should be noted that it is important to clarify initially that isolates are H. influenzae since it is known that other Haemophilus species isolated from humans can also produce active CuZnSOD under the growth conditions used in the present study (6).
TABLE 1.
CuZnSOD activity (potassium cyanide or DEDC inhibitable) and hybridization of Haemophilus sodC probes to chromosomal DNA
| Strain (no. of isolates) | Phylogenetic divisiona | Hybridization to sodC probeb | CuZnSOD activityb |
|---|---|---|---|
| Noncapsulate ET (45) | NA | − | − |
| Serotype a (2) | I | − | − |
| Serotype a (3) | II | + | − |
| Serotype b (5) | II | + | − |
| Serotype e (6) | * | + | − |
| Serotype f (6) | II | + | − |
| Cryptic genospecies (12) | NA | + | + |
I and II correspond to phylogenetic divisions I and II, respectively, of capsulate H. influenzae (11). NA, not applicable; *, distantly related to division I and II strains.
+ and −, positive and negative, respectively, for probe hybridization or CuZnSOD activity.
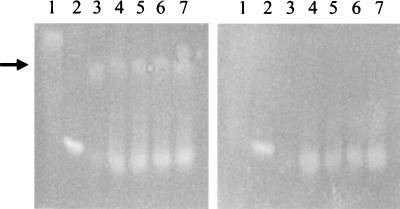
FIG. 1.
Paired IEF gels stained to show SOD activity as an achromatic zone without selective CuZnSOD inactivation (left) and in the presence of the CuZnSOD-inhibitor DEDC (right). Lanes contain whole-cell protein extracts from bacterial species, as follows: lane 1, Actinobacillus pleuropneumoniae (positive control) (5); lane 2, H. influenzae Eagan (negative control) (3); lanes 3 to 7, strains of the cryptic genospecies (strains 10N [lane 3], 12N [lane 4], 15N [lane 5], 11PS [lane 6], and 26E [lane 7]). The achromatic bands of activity corresponding to proteins with greater isoelectric points (lanes 1 and the approximate position indicated by an arrow for lanes 3 to 7), which are absent in the presence of DEDC, represents CuZnSOD. The zones unaffected by DEDC are not inhibited by 5 mM hydrogen peroxide (data not shown) and represent manganese-containing SODs.
The procedures used in the present study to detect CuZnSOD activity are too time-consuming to become part of a routine laboratory protocol. However, the finding that CuZnSOD activity can be used as a marker for strains of the cryptic genospecies of Haemophilus opens the way for the development of a rapid test.
The presence of active CuZnSOD in the cryptic genospecies raises the intriguing question as to whether the enzyme is involved in the virulence of these strains. Its periplasmic location can protect bacteria against exogenously derived superoxide such as that produced during the respiratory burst of phagocytic cells (3), and comparison of wild-type and defined sodC mutants of Salmonella enterica and Neisseria meningitidis in mouse models of infections confirms that the enzyme can contribute to the virulence of human pathogens (2, 21). As yet no information on whether bacterial sodC can confer a survival advantage in a genital setting is available, and in the case of the cryptic genospecies of Haemophilus, such information awaits the development of a suitable in vivo model.
Acknowledgments
This work was partly supported by awards from the Wellcome Trust and BBSRC (to P.R.L. and J.S.K.).
REFERENCES
- 1.Benov, L. T., and I. Fridovich. 1994. Escherichia coli expresses a copper- and zinc-containing superoxide dismutase. J. Biol. Chem. 269:25310–25314. [PubMed] [Google Scholar]
- 2.Farrant, J. L., A. Sansone, J. R. Canvin, M. J. Pallen, P. R. Langford, T. S. Wallis, G. Dougan, and J. S. Kroll. 1997. Bacterial copper- and zinc-cofactored superoxide dismutase contributes to the pathogenesis of systemic salmonellosis. Mol. Microbiol. 25:785–796. [DOI] [PubMed] [Google Scholar]
- 3.Kroll, J. S., P. R. Langford, and B. M. Loynds. 1991. Copper-zinc superoxide dismutase of Haemophilus influenzae and H. parainfluenzae. J. Bacteriol. 173:7449–7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kroll, J. S., P. R. Langford, K. E. Wilks, and A. D. Keil. 1995. Bacterial [Cu,Zn]-superoxide dismutase: phylogenetically distinct from the eukaryotic enzyme, and not so rare after all! Microbiology 141:2271–2279. [DOI] [PubMed] [Google Scholar]
- 5.Langford, P. R., B. M. Loynds, and J. S. Kroll. 1996. Cloning and molecular characterization of Cu,Zn superoxide dismutase from Actinobacillus pleuropneumoniae. Infect. Immun. 64:5035–5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langford, P. R., B. M. Loynds, and J. S. Kroll. 1992. Copper-zinc superoxide dismutase in Haemophilus species. J. Gen. Microbiol. 138:517–522. [DOI] [PubMed] [Google Scholar]
- 7.Martinez, M. A., A. Ovalle, M. T. Ulloa, and R. M. Vidal. 1999. Role of Haemophilus influenzae in intra-amniotic infection in patients with preterm rupture of membranes. Eur. J. Clin. Microbiol. Infect. Dis. 18:890–892. [DOI] [PubMed] [Google Scholar]
- 8.Moxon, E. R., R. A. Deich, and C. Connelly. 1984. Cloning of chromosomal DNA from Haemophilus influenzae. Its use for studying the expression of type b capsule and virulence. J. Clin. Investig. 73:298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy, T. F., C. Kirkham, and D. J. Sikkema. 1992. Neonatal, urogenital isolates of biotype 4 nontypeable Haemophilus influenzae express a variant P6 outer membrane protein molecule. Infect. Immun. 60:2016–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Musser, J. M., S. J. Barenkamp, D. M. Granoff, and R. K. Selander. 1986. Genetic relationships of serologically nontypable and serotype b strains of Haemophilus influenzae. Infect. Immun. 52:183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Musser, J. M., J. S. Kroll, E. R. Moxon, and R. K. Selander. 1988. Evolutionary genetics of the encapsulated strains of Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 85:7758–7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quentin, R., I. Dubarry, C. Martin, B. Cattier, and A. Goudeau. 1992. Evaluation of four commercial methods for identification and biotyping of genital and neonatal strains of Haemophilus species. Eur. J. Clin. Microbiol. Infect. Dis. 11:546–549. [DOI] [PubMed] [Google Scholar]
- 13.Quentin, R., A. Goudeau, R. J. Wallace, Jr., A. L. Smith, R. K. Selander, and J. M. Musser. 1990. Urogenital, maternal and neonatal isolates of Haemophilus influenzae: identification of unusually virulent serologically non-typable clone families and evidence for a new Haemophilus species. J. Gen. Microbiol. 136:1203–1209. [DOI] [PubMed] [Google Scholar]
- 14.Quentin, R., N. Koubaa, B. Cattier, M. Gavignet, and A. Goudeau. 1988. In vitro activities of five new quinolones against 88 genital and neonatal Haemophilus isolates. Antimicrob. Agents Chemother. 32:147–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quentin, R., C. Martin, J. M. Musser, N. Pasquier-Picard, and A. Goudeau. 1993. Genetic characterization of a cryptic genospecies of Haemophilus causing urogenital and neonatal infections. J. Clin. Microbiol. 31:1111–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quentin, R., J. M. Musser, M. Mellouet, P. Y. Sizaret, R. K. Selander, and A. Goudeau. 1989. Typing of urogenital, maternal, and neonatal isolates of Haemophilus influenzae and Haemophilus parainfluenzae in correlation with clinical source of isolation and evidence for a genital specificity of H. influenzae biotype IV. J. Clin. Microbiol. 27:2286–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quentin, R., R. Ruimy, A. Rosenau, J. M. Musser, and R. Christen. 1996. Genetic identification of cryptic genospecies of Haemophilus causing urogenital and neonatal infections by PCR using specific primers targeting genes coding for 16S rRNA. J. Clin. Microbiol. 34:1380–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenau, A., P. Y. Sizaret, J. M. Musser, A. Goudeau, and R. Quentin. 1993. Adherence to human cells of a cryptic Haemophilus genospecies responsible for genital and neonatal infections. Infect. Immun. 61:4112–4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Wallace, R. J., Jr., C. J. Baker, F. J. Quinones, D. G. Hollis, R. E. Weaver, and K. Wiss. 1983. Nontypable Haemophilus influenzae (biotype 4) as a neonatal, maternal, and genital pathogen. Rev. Infect. Dis. 5:123–136. [DOI] [PubMed] [Google Scholar]
- 21.Wilks, K. E., K. L. Dunn, J. L. Farrant, K. M. Reddin, A. R. Gorringe, P. R. Langford, and J. S. Kroll. 1998. Periplasmic superoxide dismutase in meningococcal pathogenicity. Infect. Immun. 66:213–217. [DOI] [PMC free article] [PubMed] [Google Scholar]