In Albania in 1996, a large outbreak of poliomyelitis, caused by a wild type 1 poliovirus, lasted from May to November, starting soon after the National Immunization Days (April to May 1996) and 15 years after the apparent interruption of circulation of indigenous wild-type strains of poliovirus (3). Genomic sequencing of the VP1-2A junction performed for all 74 isolates of wild poliovirus type 1 from poliomyelitic patients demonstrated 95% homology with a wild strain of poliovirus type 1 isolated in Pakistan in 1995 (3). Sequence of the VP1 region (nucleotides 2531 to 2830) had also shown 99 to 100% homology among all strains isolated during the epidemic, thus demonstrating the involvement of a unique genotype throughout the outbreak (3).
To better investigate the possible circulation of recombinant polioviruses among the hit population during the outbreak, as found by Divizia et al. (2) for environmental samples collected before the outbreak, a larger sequence analysis was carried out, which included sequencing of the 5′ untranslated region (UTR) and the 3D coding regions of all 74 isolates of wild poliovirus type 1 and of the full VP1 sequences of 10 isolates representative of the outbreak. These new sequences were then compared with those of the poliovirus strains isolated in Pakistan in 1995, which were kindly provided by Olen Kew (Centers for Disease Control and Prevention, Atlanta, Ga.). The study was also precipitated by the need to gather as much information as possible from this important sample collection before elimination of wild polioviruses, as recommended by the World Health Organization for containment procedures (7).
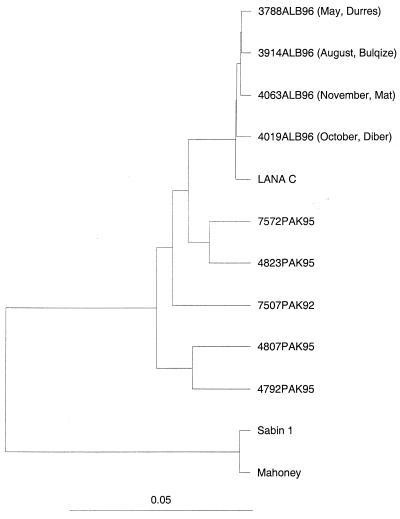
As with the VP1-2A region and complete VP1 sequences, comparison of the 5′ UTR sequences of all Albanian isolates with those of isolates found in Pakistan in 1995 and with those of environmental isolates of wild type 1 polioviruses revealed a 97 to 100% homology among these strains, confirming that the poliovirus involved in the outbreak was imported directly from Pakistan (Fig. 1). The 90% sequence homology in the 5′ UTR reported for the environmental isolates (2) with respect to the poliovirus type 3 isolate gb/L76413/POL5-UTRN-50/URSS/87 and the poliovirus type 1 isolate gb/L76402/POL5UTRF-15/Hong Kong/81 was also shown by the strains circulating in Pakistan in 1995 and all strains isolated from the Albanian poliomyelitic patients. This evidence seems to exclude the possibility of two wild poliovirus serotypes circulating before the outbreak, suggested by Divizia and coworkers (2). Alignment studies performed with the BLAST software package should be carefully interpreted, because similar levels of homology and other BLAST parameters (such as the Expect value) were also found after alignment of the sequence of the Albanian isolates with those of poliovirus type 3 isolate USOL-D-BAC and other enterovirus strains (Coxsackie A21, Coxsackie A24, and several strains of Enterovirus 70).
FIG. 1.
Dendrogram of sequence relatedness in the 5′ UTR of four representative poliovirus type 1 strains isolated in Albania, environmental strain Lana C (3), strains isolated in Pakistan in 1995, wild poliovirus type 1 strain Mahoney, and a vaccine reference strain (accession no. gi/61252/emb7V01149/POLIO1B and gi/61257/emb7V01150/POLIOS1, respectively). The Albanian virus strains were selected by place and time of isolation.
Recombination was further analyzed by comparing sequences of the 3D polymerase region (frequently involved in recombination events) of all Albanian strains with those of strains found in Pakistan in 1995. No 3D sequences were available for environmental isolates. The homologies between Albanian and Pakistani strains for the 3D region were much lower (85 to 87%) than those for the other regions described. Even when the sequence were submitted to an accessible databank (GenBank), for comparison with sequences of other enterovirus strains reported no better alignment was found. We cannot exclude the possibility that recombination may have occurred in the past, but with the data presently available, it was not possible to identify the donor, if such exists.
The P1 and P2 coding regions and almost all of the 5′ UTR and P3 coding regions of three representative virus strains were completely sequenced. These strains were selected because they had been isolated in different districts and at different times during the epidemic (strain 3788 was isolated in May in Durres, strain 3914 was isolated in August in Bulqize, and strain 4019 was isolated in Diber in October). The virus strains showed 97 to 100% homology between each other for all regions analyzed. Comparison of the 2C, 3A, 3B, and 3C region sequences with those of other circulating wild poliovirus strains was not possible because data were not available in accessible data banks.
It is worthy of note that although almost 700,000 doses of trivalent oral poliovirus vaccine were distributed between May and November, before and during the outbreak, no wild-vaccine recombinant strains were found among the 74 poliovirus strains isolated from poliomyelitic patients. This was despite the isolation of a recombinant strain (poliovirus type 2 vaccine-poliovirus type 1 wild strain) from an environmental sample collected before the outbreak (3). Even when poliovirus mixtures were contemporaneously isolated from stool samples, isolates showed genomes almost identical to those of either vaccine reference strains or wild type 1 poliovirus strains.
Although inter- and intratypic recombinations among Sabin strains in vaccinees (4) and among vaccine and wild-type virus strains (5) have been demonstrated to occur in vivo, this was not evidenced in our study. It is possible that the infectivity of recombinant strains found in the environment was much lower than that of the parental strains.
This study once again indicates that effective vaccination programs are essential to eliminate gaps in immunity and to achieve the goal of poliomyelitis global eradication (6). It also points to the need to coordinate environmental surveillance with acute flaccid paralysis surveillance (1) to have a better overview of the progress in polio eradication.
Nucleotide sequence accession numbers.
Sequence data have been deposited in GenBank; the accession numbers are AY056620 to AY056693, AY056697, AY056699 to AY056703, and AY056707 to AY056777.
Acknowledgments
This study was partially supported by a grant from the WHO (“Characterization of polio and other enteroviruses associated with paralytic disease in Italy, Albania, Bosnia and Malta.” 2000–2001 I8/181/838(1), HQ/00/684769).
We thank Romina Tomasetto for editorial assistance.
REFERENCES
- 1.Diamanti, E., B. Ibrahimi, F. Tafaj, E. Mezini, A. Dodbiba, V. Dobi, S. Catone, D. Genovese, P. Simeoni, and L. Fiore. 1998. Surveillance of suspected poliomyelitis in Albania, 1980–1995: suggestion of increased risk of vaccine associated poliomyelitis. Vaccine 16:940–948. [DOI] [PubMed] [Google Scholar]
- 2.Divizia, M., L. Palombi, E. Buonomo, D. Donia, V. Ruscio, M. Equestre, L. Leno, A. Pana, and A. M. Degener. 1999. Genomic characterization of human and environmental polioviruses isolated in Albania. Appl. Environ. Microbiol. 65:3534–3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fiore, L., D. Genovese, E. Diamanti, S. Catone, B. Ridolfi, B. Ibrahimi, R. Konomi, H. G. van der Avoort, T. Hovi, R. Crainic, P. Simeoni, and C. Amato. 1998. Antigenic and molecular characterization of wild type 1 poliovirus causing outbreaks of poliomyelitis in Albania and neighboring countries in 1996. J. Clin. Microbiol. 36:1912–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furione, M., S. Guillot, D. Otelea, J. Balanant, A. Candrea, and R. Crainic. 1993. Polioviruses with natural recombinant genomes isolated from vaccine-associated paralytic poliomyelitis. Virology 196:199–208. [DOI] [PubMed] [Google Scholar]
- 5.Guillot, S., V. Caro, N. Cuervo, E. Korotkova, M. Combiescu, A. Persu, A. Aubert-Combiescu, F. Delpeyroux, and R. Crainic. 2000. Natural genetic exchanges between vaccine and wild poliovirus strains in humans. J. Virol. 74:8434–8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization. 1990. Resolution of the 41st World Health Assembly (WHO 41.28) 1988. Global eradication of poliomyelitis by the year 2000. World Health Organization, Geneva, Switzerland.
- 7.World Health Organization. 2000. Guidelines for the implementation of laboratory containment of wild poliovirus. World Health Organization, Copenhagen, Denmark.