Abstract
The control of gene transcription is dependent on DNA-binding and coregulatory proteins that assemble in distinct regions of the cell nucleus. We use multispectral wide-field microscopy of cells expressing transcriptional coregulators labeled with fluorescent proteins (FP) to study the subnuclear localization and function of these factors in living cells. In coexpression studies, the glucocorticoid receptor interacting protein (GRIP) coactivator protein and the silencing mediator of retinoid and thyroid (SMRT) corepressor protein form spherical subnuclear focal bodies that are spatially distinct, suggesting that specific protein interactions concentrate these divergent proteins in separate subnuclear regions. However, the variability of these subnuclear bodies between cells within the population makes analysis based on “representative images” difficult, if not impossible. To address this issue, we develop a protocol for unbiased selection of cells from the population, followed by the automated quantification of the subnuclear organization of the labeled proteins. Statistical methods identify a significant linear correlation between the FP-coregulator expression level and subnuclear focal body formation for both FP-GRIP and FP-SMRT. Importantly, we confirm that these changes in subnuclear organization could be statistically normalized for differences in coregulator expression level. This integrated quantitative image analysis method will allow the rigorous comparison of different experimental cell populations that express variable levels of FP fusion proteins.
Keywords: bioimaging, fluorescence microscopy, green fluorescent protein, nuclear structure, nuclear coactivator, glucocorticoid receptor interacting protein
1 Introduction
The application of fluorescence microscopy and advanced digital imaging to the investigation of dynamic processes inside living cells is a rapidly evolving field. The recent development of new fluorescent probes, coupled with advances in digital image acquisition and analysis, has transformed studies in cell biology by allowing the behavior of proteins to be tracked in their natural environment within the living cell. The challenge now confronting cell biologists is how to extract the biologically significant information from very large and complex digital imaging datasets. For example, a single high-resolution digital image commonly contains more than one million data points, and multidimensional imaging experiments may produce hundreds or thousands of these images.1Because of this complexity, it is often difficult to compare and contrast the morphometric information in multiple images without advanced informatics tools.2
Fortunately, automated computer algorithms have been developed to support the quantitative analysis of subcellular morphologies captured in large digital image datasets, reviewed by Ref. 3. These automated approaches typically involve the segmentation of the images, followed by quantitative measurement of specific features. The application of this method effectively reduces millions of data points into a few thousand morphometric measurements. However, even these simplified morphometric datasets contain many interrelated parameters, and the relationships between parameters are often difficult to interpret. To address this issue, we have developed a quantitative image analysis and statistical modeling approach, allowing us to begin to establish links between the subcellular distribution of proteins and their function in populations of living cells.
We are applying this image analysis approach to the investigation of gene regulatory proteins in the interphase nucleus. It is well established that the cell nucleus is organized in numerous distinct subcompartments that consist of specific ensembles of interacting proteins.4–6 The partitioning of these different subcompartments without intervening membranes indicates that the proteins that form these structures must self-organize.7 For example, transcriptional coregulatory proteins, which function to modify chromatin structure and recruit the general transcription apparatus to target genes, assemble in subnuclear focal bodies.8–14 The direct visualization of these subnuclear foci has been achieved by labeling the coregulatory proteins with the visible fluorescent proteins (VFPs).8–11,14,15 This approach has also been used to demonstrate that these highly ordered subnuclear foci are dynamic, rapidly exchanging with proteins in the surrounding nucleoplasm.16 Defining the mechanisms that control the formation of these higher-order protein assemblies within the context of the intact cell nucleus will be necessary to understand fully the regulation of gene expression.
A quantitative imaging approach to analyze the assembly of VFP-fusion proteins into complexes in the nucleus requires an unbiased method for selection of the cells to be imaged within the population. In this regard, we reported earlier the use of monomeric red fluorescent protein (mRFP) as a noninvasive cell selection marker that allows identification of cells expressing other VFP-fusion proteins.15 Once the cells have been selected based on diffuse mRFP, then the subcellular features of the coexpressed VFP-fusion protein can be automatically quantified using a computerized image analysis algorithm. In the current study, we used this integrated analytical method to characterize in detail the subnuclear organization of the nuclear receptor coactivator glucocorticoid receptor interacting protein (GRIP).17 This quantitative image analysis approach is important for understanding the actions of the coregulatory proteins, since earlier studies demonstrated that there was substantial heterogeneity from cell-to-cell in the subnuclear organization of GRIP.8,10,11
The results presented here provide a rigorous analysis of the subnuclear organization of green fluorescent protein (GFP)-labeled GRIP within the transfected cell population. Significantly, we then compare the results obtained for GRIP with the quantitative image analysis of cells expressing a different class of coregulatory protein, the silencing mediator of retinoid and thyroid hormone receptors (SMRT) transcriptional corepressor.18 We show that although the subnuclear focal bodies formed by GRIP and SMRT were separate and distinct, similar mechanisms likely regulate their formation. In both cases, there was a significant correlation between increasing protein expression levels and the formation of larger, more distinct subnuclear focal bodies. Our novel numerical method takes advantage of this linear relationship to normalize measurements of subnuclear morphology for differences in VFP-fusion protein expression level in individual cells. By correcting for the effects of variable fusion-protein expression level within cell populations, it will be possible to evaluate the effect of changing experimental conditions on the formation of these nuclear structures. Together, these results demonstrate the utility of quantitative image analysis and statistical modeling techniques to rigorously define the mechanisms that control subnuclear protein organization and function.
2 Materials and Methods
2.1 Expression Vectors and Cell Transfection
Nucleotide sequences encoding the monomeric variant of Discosoma sp: red, mRFP,19 kindly provided by Tsien, University of California, San Diego, was substituted for the yellow fluorescent protein (YFP) encoding sequence in the EYFP-C2 vector (BD Biosciences Clontech, Palo Alto, California) to generate the mRFP expression vector. The expression vector encoding EGFP fused to the amino terminus of GRIP (GFP-GRIP) has been previously described.8 The cDNA encoding the human SMRT repression and nuclear receptor interaction domains (AA 237-1495)18 was inserted to the 3′ end of the cDNA encoding EYFP (BD Biosciences Clontech) in the pNAss expression vector.20 The expression vectors were verified by automated nucleotide sequencing. The mouse embryonic pituitary GHFT1-5 cells were transfected by electroporation, and cultured for 24 h on glass coverslips as described earlier.21
2.2 Digital Imaging of Protein Organization in Living Cells
The cover glass with the monolayer of cells was transferred to a medium-filled chamber that was fitted to the stage on the microscope.21 The wide-field fluorescence microscopy (WFM) images were acquired using an inverted Olympus IX-70 microscope equipped with a 1.2 numerical aperture, 60× aqueous-immersion objective lens. A 75-W xenon/mercury combination lamp was used to illuminate the samples. An Opti-Quip (Highland Hills, New York) model 1962 long-term stabilizer was used to keep light intensity constant for accurate quantitative data collection. The GFP filter combination used 470/20-nm excitation with 510/20-nm emission, the YFP filter combination used 510/20 excitation and 560/40 emission, and the RFP filter combination used 560/40-nm excitation with 630/60-nm emission. Grayscale images with no saturated pixels were obtained using a cooled digital interline camera (Orca-200, Hamamatsu, Bridgewater, New Jersey). All images were collected at a similar gray-level intensity by controlling the excitation intensity with constant neutral density filtration, and by varying the on-camera integration time (0.1 to 8 s). Reference images of standard fluorescent beads were acquired to monitor consistency of microscope performance for all quantitative imaging experiments. All image files were processed for presentation using ISee software (ISee Imaging Systems, Raleigh, North Carolina) and Canvas 8.0 software (Deneba, Incorporated, Miami, Florida).
2.3 Automated Image Analysis
The automated image analysis algorithm was described earlier.12 Briefly, the ISee graphical programming software (ISee Imaging Systems) was used to integrate a series of computerized image analysis functions into a single algorithm. The first subroutine uses a histogram-based statistical method to optimally threshold the image acquired in the RFP channel to identify the whole cell region of interest (ROI). The mean intensity of the area outside the whole nucleus ROI was measured in both the red and green channel images to define the background fluorescence. Optimal thresholding of the green fluorescence channel image was then used to select the whole nucleus ROI. Measurements of mean fluorescence intensity in the whole cell ROI and whole nucleus ROI were used to estimate the relative expression of mRFP and GFP-GRIP in each cell. The normalized relative fluorescence intensity for all images was expressed as gray-level per second exposure time.
A second subroutine then takes the whole nucleus ROI as input and optimally thresholds this region using an iterative method to separate areas of bright fluorescence from surrounding regions. Next, the subroutine measures the shape of each identified bright ROI using several parameters. For the studies of GRIP focal bodies described here, ROIs were empirically defined as statistically significant regions of elevated fluorescent intensities that have a contiguous size between 10 and 3000 pixels. Additionally, the spherical GRIP foci were defined by a roundness value between 0.9 and 1.5 [roundness=(4*pi*total area)/(perimeter2), roundness of perfect circle=1], and an axial ratio value between 1 and 1.3. The algorithm automatically selects the ROIs that meet empirically determined shape parameters of GRIP protein focal bodies for further analysis. If the ROI does not meet the requirements of spherical GRIP foci, then the ROI is reanalyzed by automatic thresholding and shape measurements to determine if GRIP foci are located within the original ROI. The process is repeated until all ROIs are evaluated.
The area and fluorescence intensity of each selected focal body ROI is automatically measured and recorded. The center position of the selected GRIP focal body is then used to place a second rectangular ROI that measures the fluorescence intensity of the nucleoplasm surrounding the focal body. The size of the surrounding square ROI is four times that of the selected focal body. All the selected ROIs are marked in the image and each is annotated with the acquired data. All the measurements were automatically exported to text files, and further analysis was performed using spreadsheet software (Microsoft, Excel) to determine the relationship between the labeled protein expression levels and subnuclear organization.
3 Results
3.1 Unbiased Selection of Transfected Cells and Automated Image Analysis
The organization of transcriptional coregulatory proteins into highly ordered focal bodies within the cell nucleus is well documented, but little is known of biological mechanisms that regulate this organization.8–13,22 The nuclear receptor coactivator GRIP forms well-defined focal bodies in the nucleus, but there is also substantial variability in its distribution within the cell population, ranging from a diffuse nucleoplasmic distribution to an arrangement of highly concentrated focal bodies.8,11 This variability is exemplified by images of two cells taken from the same population expressing GFP-GRIP (Fig. 1). This cell-to-cell heterogeneity in GRIP distribution makes any analysis of the biological mechanisms that regulate GRIP subnuclear positioning based on “representative images” difficult, if not impossible. Therefore, it is necessary to use quantitative imaging techniques and statistical methods to describe the biology that underlies differences between the cells expressing GRIP. This rigorous statistical analysis of protein distribution also requires the random sampling of the cell population during image acquisition.
Fig. 1.
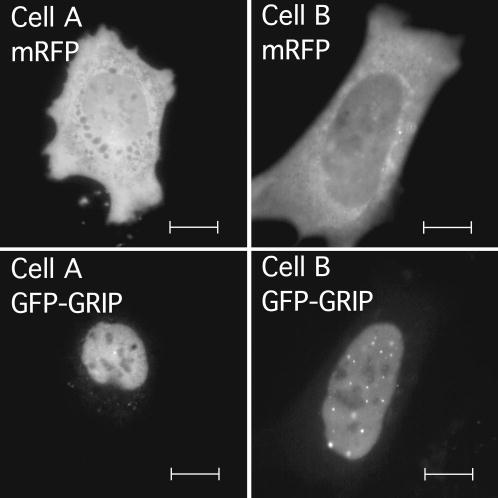
Unbiased cell selection. Pituitary GHFT1-5 cells were cotransfected with vectors encoding mRFP and GFP-GRIP. The living cells expressing the fusion proteins were selected for imaging using the RFP signal. Images of two example cells are shown with mRFP and GFP fluorescence channels displayed separately as labeled. Scale bars are 10 μm in length.
To accomplish this, we have taken advantage of the observation that when cells are cotransfected with plasmids encoding two or more protein fusions to VFP-color variants, most all the transfected cells express each different color protein.15 Using this approach, cells are cotransfected with an expression plasmid encoding mRFP19 and a second plasmid encoding the protein of interest, GFP-GRIP. This allowed the selection of cells for imaging based solely on the expression of the diffuse cellular mRFP, with no prior knowledge of the subnuclear organization of the coexpressed GFP-GRIP. The images shown in Fig. 1 demonstrate the selection of cells based on the expression of mRFP, and the subsequent detection of GFP-GRIP among the cells in the transfected population. Importantly, the examination of 45 randomly selected mRFP-expressing cells revealed that more than 95% also contained a detectable nuclear GFP-GRIP fluorescence signal. This confirmed that images of transfected cells expressing a protein of interest could be obtained using the mRFP channel without user bias to particular patterns of GFP-labeled protein distribution or expression level.
Within the transiently transfected cell population, individual cells express different relative levels of both mRFP and GFP-GRIP. In addition, as mentioned before, there is also substantial cell-to-cell variability in the subnuclear organization of the GFP-GRIP, and we want to quantify this organization of GRIP in a consistent and unbiased way. This was achieved by acquiring images using the GFP channel from each of the cells that were selected based on mRFP expression. The images of GFP-GRIP from many mRFP-selected cells were then batch analyzed using a computer algorithm designed to identify and measure the subnuclear distribution of proteins without any user intervention (see Sec. 2, Materials and Methods). The images shown in Fig. 1 illustrate the highly variable subnuclear organization of GFP-GRIP (Fig. 1 and Table 1). Despite this, the algorithm identified GFP-GRIP focal body ROIs in the nuclei of both cells [Fig. 2(a)], and the morphometric data describing each ROI were automatically generated (summarized in Table 1).
Table 1.
Summary of morphometric data from cells coexpressing mRFP and GFP-GRIP. All intensity data are relative fluorescence intensity with gray level per second camera time. EF is the mean foci intensity/surrounding intensity, and OF is the mean foci size×mean EF.
| Morphometric data | Cell A | Cell B |
|---|---|---|
| Mean whole nucleus GFP intensity | 83 | 177 |
| Mean whole cell mRFP intensity | 268 | 257 |
| Mean foci area (pixels) | 29 | 70 |
| Mean foci intensity | 101 | 336 |
| Mean surrounding intensity | 91 | 213 |
| Mean enrichment factor (EF) | 1.11 | 1.57 |
| Mean organization factor (OF) | 32 | 110 |
Fig. 2.
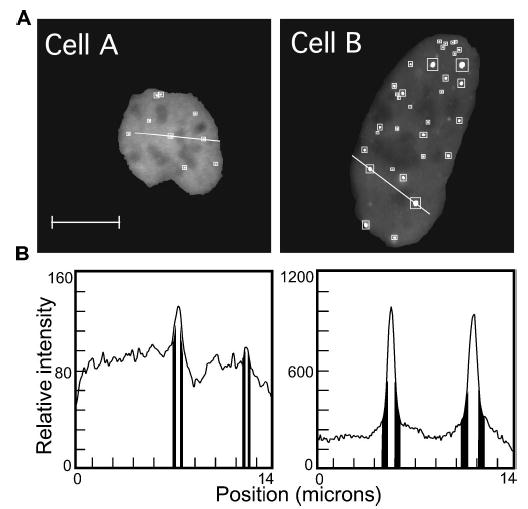
Computer-assisted image analysis of nuclear GFP-GRIP focal body organization. The computer algorithm was used to analyze the images of the cell nuclei that were shown in Fig. 1. (a) The whole nucleus is shown in gray and the autoselected GFP-GRIP foci are highlighted with white. The white boxes indicate the surrounding ROI that was automatically assigned to each focal body. The relative fluorescence intensity along the white line in each image is displayed in the profile plot. Scale bar indicates 10 μm. (b) In the profile plot, regions in the foci ROIs are highlighted by the surrounding ROIs that are indicated with black vertical bars. The morphometric data that were automatically extracted from these cells are summarized in Table 1.
Each bright focal body is a region of highly concentrated GRIP protein, which is surrounded by regions that contain a lower concentration of GRIP, as illustrated by the intensity profile plots [Fig. 2(b)]. The ratio of fluorescence signal originating from the foci to that from the surrounding region [white squares, Fig. 2(a)] defines the enrichment factor (EF), the steady-state concentration of protein maintained in the focal body. Quantifying the relative intensities of the focal bodies in the two cells in Fig. 2 revealed that the average enrichment factor for the foci in cell B was 1.4-fold higher than that for cell A (Fig. 2 and Table 1). Finally, the algorithm determined an organization factor (OF) for each cell, which is the product of foci size and the enrichment factor. The greater the mean OF value, the larger and more distinct the focal bodies are in the cell nucleus (Fig. 2 and Table 1).
3.2 Quantitative Analysis of Fluorescent Fusion Protein Organization and Expression Level in Cell Populations
To validate this approach, images of 45 cells were collected based solely on the mRFP signal, and were then analyzed for GFP-GRIP organization and protein expression using the automated algorithm. Statistical linear regression and ANOVA analysis confirmed that foci size, EF, and OF parameters were all significantly related to the GFP-GRIP expression level in each cell (F test p-values<0.05). The strong positive correlation between GFP-GRIP expression level and GFP-GRIP subnuclear organization in the cell population is illustrated by the calculated best-fit line in Fig. 3(a) (F test p-value=0.0025). For the proteins studied in this report, we found that the changing OF values reflect significant changes in both focal body size and protein enrichment. However, this dependence of OF on both foci size and enrichment should be confirmed when applying this method to other experimental systems. Once this behavior is established, the OF value provides a convenient way to summarize the morphology of focal bodies. By revealing the statistically significant relationship between GFP-GRIP fluorescence intensity and OF values, this method firmly establishes that the formation of the foci is related to the amount of coactivator protein that is expressed.
Fig. 3.
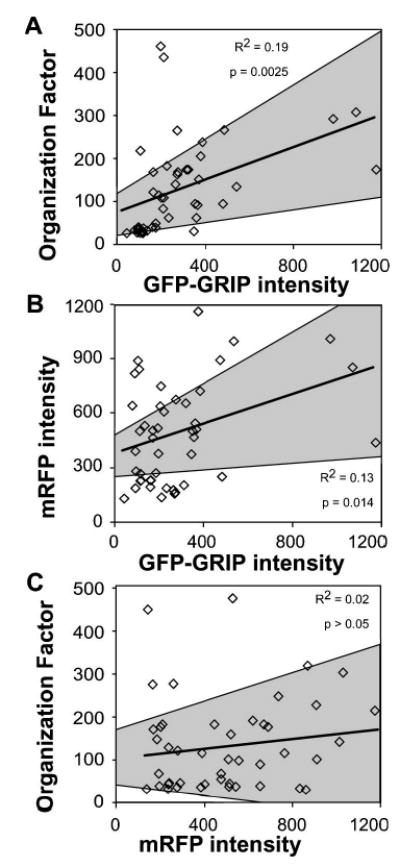
Cell population studies using the computer-assisted image analysis protocol. Images of 45 cells expressing mRFP and GFP-GRIP were acquired and analyzed. In the plots, each square represents data from a single cell. The best-fit line in each graph shows the relationship for the cell population between (a) GFP-GRIP subnuclear organization and relative GFP-GRIP expression level, (b) GFP-GRIP and mRFP expression levels, and (c) GFP-GRIP subnuclear organization and mRFP expression level. The gray areas define the 95% confidence intervals for the best-fit lines. The R2 value and the ANOVA F test p-value estimate the correlation between the parameters as calculated by linear regression analysis.
The ratio of GFP to mRFP intensity for individual cells within the selected population was highly variable, and was distributed over a 250-fold range. This was reflected in the modest correlation of the expression levels of the cotransfected GFP-GRIP and mRFP [F test p-value=0.014, Fig.3(b)]. Moreover, since the mRFP did not colocalize with GFP-GRIP (Fig. 1), we would not expect mRFP to influence GRIP subnuclear organization. To verify this, we quantitatively compared mRFP fluorescence intensity to the GFP-GRIP organization in each cell of the population. Statistical analysis revealed that there was no significant relationship between these two parameters [F test p-value=0.36, Fig. 3(c)], eliminating the possibility that mRFP expression influenced the organization of GFP-GRIP. This supplies further evidence that mRFP expression is well tolerated by the cells, similar to other FP spectral variants.23–27
Together, these results indicated that while most all transfected cells expressed both mRFP and GFP-GRIP, mRFP expression was not a good predictor of GFP-GRIP expression level. This allowed the selection of cells expressing very low levels of GFP-GRIP by using the brighter mRFP signal, which is more easily detected by the user’s eye. Further, the weak correlation of the expression levels of the cotransfected GFP-GRIP and mRFP allowed a more random selection of cells expressing different levels of GFP-GRIP in the population chosen using the mRFP signal. Accordingly, the sample of 45 cells selected for analysis included a 40-fold range in GFP-GRIP fluorescence intensity per nucleus (Fig. 3). Since the level of fusion protein expression is a major consideration in the interpretation of live cell imaging experiments, it is important to emphasize that the cells at the lower end of this range contained GFP signals that would be difficult or impossible to detect by eye. Therefore, the population of cells analyzed contained the cells expressing low levels of GFP-GRIP that would not have been captured using conventional qualitative imaging approaches. Although absolute quantification of the relationship between FP concentration and fluorescence intensity in living cells requires specialized techniques,28 in vitro characterization of GFP over a 1000-fold concentration range revealed a linear relationship when measured using an epifluorescence microscope and CCD detector that was similar to the instrument used in this study.29 Because the dimmest cells that we observe are at the limit of detection, it seems likely that the differences in fluorescence intensity that we measure here linearly represent the relative changes in the nuclear concentration of the coregulatory protein.
3.3 Normalizing Protein Organization Measurements for Differences in Fusion Protein Expression Levels
The interactions of coregulatory proteins such as GRIP with their nuclear receptor partners have been shown to affect the subnuclear distribution of the coactivators. For example, the distribution of GRIP in the living cell nucleus is affected by the expression of the estrogen receptor alpha (ERα).8,10 However, the variability of GRIP distribution within the cell population makes the analysis of the interaction with coexpressed nuclear receptors very difficult to quantify without a rigorous cell population analysis based on an unbiased cell selection. Here, we illustrate this problem by sorting the cell population quantified in Fig. 3 into two subpopulations, expressing low and high levels of GFP-GRIP [Figs. 4(a) and 4(b)]. Based on differences in fluorescence intensity per nucleus, the expression level in these two subpopulations varies by more than three-fold [Fig. 4(a)]. As expected from the earlier analysis [Fig. 3(a)], these subpopulations had significant differences in OF values because of the relationship of GFP-GRIP expression level to its subnuclear organization [Fig. 4(b)]. If a second variable were added, such as the expression of an interacting protein partner, then the accurate interpretation of the results would require that the GRIP expression level effects be statistically separated from the effects of the coexpressed protein. Therefore, it is important to normalize the measurements of subnuclear organization for any differences in fusion protein expression level.
Fig. 4.
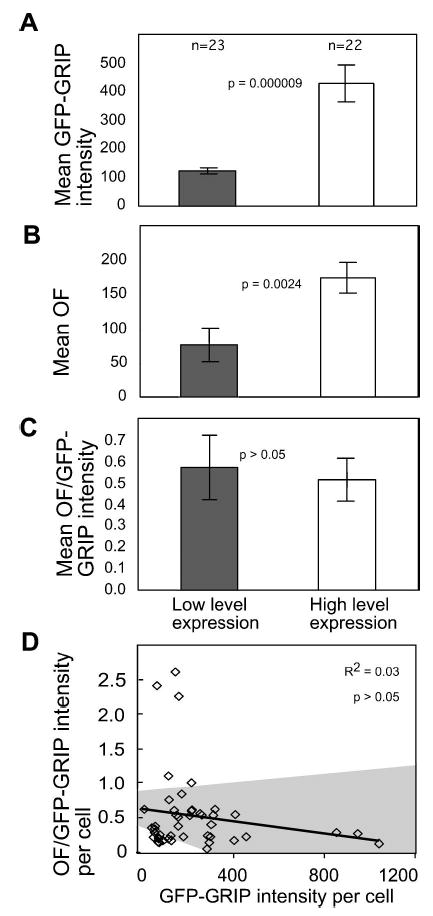
Normalization of GFP-GRIP morphometric data for differences in fusion-protein expression level. (a), (b), and (c) The morphometric data from 45 cells shown in Fig. 3 were divided into two subpopulations based on expression of low levels (gray bars) and high levels (white bars) of GFP-GRIP. The graphs display the mean values for the morphometric data representing the two cell subpopulations. The displayed p-values estimate the significance of the difference between the two subpopulations as calculated by ANOVA and post hoc t-test. Error bars denote standard error of the mean. (d) For each cell, the OF value quantifying GFP-GRIP organization was normalized for the relative level of GFP-GRIP fusion protein expression. In the plot, each square represents the normalized data from a single cell. The gray areas define the 95% confidence intervals for the best-fit line. The R2 value and the ANOVA F test p-value indicate that there is no significant correlation between the normalized morphometric data and GFP-GRIP expression level in each cell.
In the method employed here, this normalization is accomplished for each cell in the sampled population by dividing the GFP-GRIP OF by the relative GFP-GRIP fluorescence intensity for each cell. The resulting normalized value is indicated by the abbreviation OF/VFP. If the OF value is linearly related to the fusion protein fluorescence intensity, then this simple arithmetic process will quantitatively cancel the effect of changing expression levels from the OF values. In contrast, more complicated curve fitting methods would be required if the relationship between expression level and organization exhibited a strong nonlinear component. However, a linear relationship was suggested by the correlation of OF values with GFP-GRIP expression [Fig. 3(a)]. To test the feasibility of this simple linear approach, we normalized the two different GFP-GRIP expressing subpopulations described in Figs. 4(a) and 4(b). As expected, the mean OF/VFP values were statistically identical for the two cell groups with different expression levels [Fig. 4(c)]. The accurate normalization for the GFP-GRIP expression levels was further confirmed by demonstrating that there was no significant correlation between the OF/GFP values and the GFP-GRIP expression level in each cell in the population [Fig. 4(d)]. This statistical analysis showed that the expression level normalization method could effectively remove the influence of GRIP expression level from the morphometric data measuring GRIP subnuclear organization, allowing other experimental factors to be examined in isolation.
3.4 Comparison of Coactivator and Corepressor Subnuclear Organization
Similarly to the coactivator GRIP, the transcriptional corepressor SMRT has also been reported to form spherical subnuclear bodies.13 Here, we directly compare the nuclear focal bodies formed by GRIP and SMRT. When GRIP and SMRT were coexpressed in the same cells, each protein formed its own distinct population of nuclear focal bodies [Fig. 5(a)]. Considering this observation, it seems likely that protein-specific interactions function to concentrate these divergent proteins into distinct subnuclear domains. Therefore, although focal body formation is a behavior common to these divergent protein families, these coregulators are not simply sequestered via widely utilized protein interactions, such as those targeting general proteasome-mediated protein degradation.30
Fig. 5.
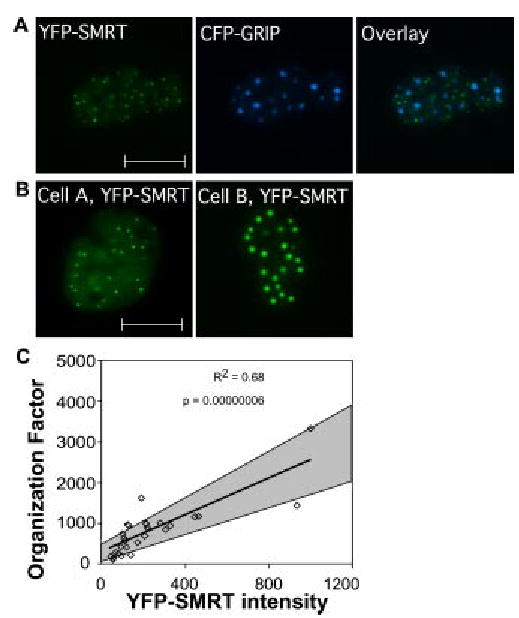
YFP-SMRT and CFP-GRIP form spatially distinct subnuclear bodies. (a) Pituitary GHFT1-5 cells were cotransfected with vectors encoding YFP-SMRT and CFP-GRIP. Images of a living cell show the fluorophores separately as labeled, and together in the overlay. (b) GHFT1-5 cells were cotransfected with vectors encoding mRFP and YFP-SMRT. The living cells expressing the fusion proteins were selected for imaging using the RFP signal. The YFP fluorescence channel images are displayed for two example cells with differing levels of YFP-SMRT expression. The scale bar denotes 10 μm. The morphometric data that were automatically extracted from these cells are summarized in Table 2. (c) Images of 28 cells coexpressing YFP-SMRT and mRFP were acquired and analyzed. In the plots, each square represents data from a single cell. The best-fit line shows the relationship for the cell population between YFP-SMRT subnuclear organization and relative YFP-SMRT expression level. The gray areas define the 95% confidence intervals for the best-fit line. The R2 value and the ANOVA F test p-value estimate the correlation between the parameters as calculated by linear regression analysis.
When YFP-SMRT was expressed in cells, we observed that it had similar heterogeneity in its subnuclear distribution when compared to GRIP. Some cells had a more diffuse pattern containing very small focal bodies, while other cells had SMRT organized in larger, more concentrated focal bodies [Fig. 5(b) and Table 2]. Next, YFP-SMRT expressing cells were randomly selected (n = 28 cells) for image acquisition using coexpressed mRFP, and morphometric data describing the subnuclear organization were consistently extracted using the automated algorithm. Statistical analysis of the morphometric data revealed a linear correlation between YFP-SMRT expression levels and focal body organization [Fig. 5(c)]. These results paralleled our previous studies of the coactivator GRIP (Fig. 3) and corepressor NCoR,15 and suggested that the organization of both coactivators and corepressors is highly sensitive to changes in the concentrations of these divergent transcriptional coregulatory proteins.
Table 2.
Summary of example cell morphometric data coexpressing mRFP and YFP-SMRT. All intensity data are relative fluorescence intensity with gray level per second camera exposure time. EF is the mean foci intensity/surrounding intensity. OF is the mean foci size ×mean enrichment factor.
| Morphometric data | Cell A | Cell B |
|---|---|---|
| Mean whole nucleus YFP intensity | 54 | 216 |
| Mean whole cell mRFP intensity | 38 | 65 |
| Mean foci area (pixels) | 47 | 231 |
| Mean foci intensity | 183 | 1335 |
| Mean surrounding intensity | 88 | 315 |
| Mean enrichment factor (EF) | 2.1 | 4.2 |
| Mean organization factor (OF) | 98 | 974 |
To extend these observations and place them in context with our analysis of coactivator protein subnuclear distribution, the morphometric data for YFP-SMRT expressing cells [Fig. 5(c)] were divided into two subpopulations based on expression level. The results of this statistical analysis supported the linear correlation between expression level and focal body organization [Figs. 6(a) and 6(b)]. When the two different YFP-SMRT subpopulations were normalized for differences in expression level, the OF/YFP values were statistically the same [Fig. 6(c)], indicating the accurate normalization of the morphometric data. In addition, linear regression analysis confirmed that there was no significant correlation between OF/YFP values and YFP-SMRT expression in each cell within the population [Fig. 6(d)]. These results are very similar to those characterizing the organization of GFP-GRIP in the cell population (compare Figs. 4 and 6), suggesting that this expression level normalization method will be useful in the study of many different subnuclear features.
Fig. 6.
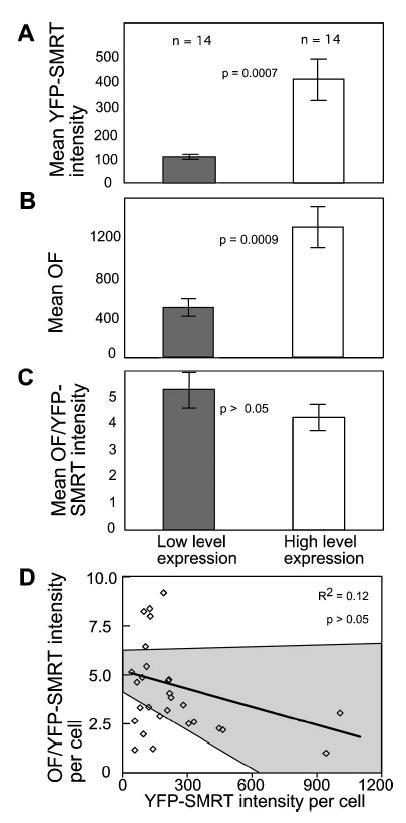
Normalization of YFP-SMRT morphometric data for differences in fusion-protein expression level. (a), (b), and (c) The morphometric data from 28 cells shown in Fig. 5(c) were divided into two subpopulations based on expression of low levels (gray bars) and high levels (white bars) of YFP-SMRT. The graphs show the mean values for the morphometric data representing the two cell subpopulations. The displayed p-values estimate the significance of the difference between the two subpopulations as calculated by ANOVA and post hoc t-test. Error bars denote standard error of the mean. (d) For each cell, the OF value quantifying YFP-SMRT organization was normalized for the relative level of YFP-SMRT fusion protein expression. In the plot, each square represents the normalized data from a single cell. The gray areas define the 95% confidence intervals for the best-fit line. The R2 value and the ANOVA F test p-value indicate that there is no significant correlation between the normalized morphometric data and YFP-SMRT expression level in each cell.
4 Discussion
The assembly of transcriptional coregulatory proteins into highly ordered complexes within the cell nucleus is well documented, but little is known of biological mechanisms that regulate this organization.8–13,22,31 Fluorescence microscopy of proteins labeled with the VFPs provides a way to directly visualize the assembly of coregulatory proteins into these complexes, but this approach is complicated by the cell-to-cell heterogeneity in the subnuclear distribution of these proteins within the cell population. We showed that the coactivator GRIP is found in patterns ranging from a diffuse nucleoplasmic distribution to an arrangement of highly concentrated focal bodies (Fig. 1). This heterogeneity prevents the accurate analysis of GRIP subnuclear morphology under different experimental conditions using qualitative image analysis, since no single image can adequately represent the cell population.
To address this issue, we developed an integrated imaging method for the rigorous analysis of subnuclear protein morphology in heterogeneous cell populations. The approach combines unbiased image acquisition, automated morphometric data extraction, and statistical modeling to minimize subjectivity and error in the quantitative results. Each component of this method was designed to extract reliable biological information from complex digital imaging datasets. The statistical modeling of the quantitative imaging data was then used to characterize the complex behavior of GRIP morphology in cell populations, establishing significant differences between distinct subpopulations. By correlating measurements of multiple cellular features, we demonstrated that a concentration-dependent mechanism accounts for a large component of heterogeneity between cells for GRIP subnuclear organization.
The application of statistical modeling to digital imaging datasets requires an unbiased selection method to acquire the images. This is critical because unintentional user bias during the selection of cells for image acquisition will prevent the sampled cells from accurately representing the population. Although various fluorescent dyes have been used to select cells for automated image analysis,32 these staining methods would not preferentially identify cells that exogenously express the VFP-labeled fusion protein of interest. In this study, we used the diffuse fluorescence signal from the cotransfected mRFP to select cells for analysis of GRIP subnuclear distribution with no prior knowledge of the localization of GFP-GRIP. This analysis revealed a weak correlation between the mRFP and GFP-GRIP expression levels (Fig. 3). Importantly, this weak correlation allowed the selection of a wide range of GFP-GRIP expression in the sampled cells, including cells that contained very low levels of GFP-GRIP that would not have been detected by eye. The application of statistical analysis to the selected populations of cells indicated that the level of mRFP expression was not correlated with the formation of GRIP subnuclear bodies. Taken together with our earlier population analysis of cells expressing YFP-labeled corepressor proteins,15 these results confirm that the mRFP can be used to select cells during image acquisition without affecting the subnuclear organization of coexpressed transcription factors.
User bias in the selection and measurement of specific subcellular features also introduces inaccuracies into the morphometric data. This problem becomes more pronounced when multiple features within sets of digital images are subjectively measured, and the resulting error will hinder the analysis.33 However, multiple morphometric measurements must be made for each cell within the sampled population to reveal the biological mechanisms that underlie the subcellular distribution of proteins. To resolve these issues, we applied a customized image analysis algorithm that automatically selected and measured multiple cellular features for each image within the dataset. The results demonstrated that the automated algorithm was robust, appropriately selecting subnuclear GRIP structures even in cells with a high degree of diffuse nucleoplasmic signal (Fig. 2). The important advantage of this approach is that all the cell images were reproducibly measured using the same rule-based system, with no user intervention.
In addition to the coactivator and corepressor proteins, many other nuclear proteins also organize in focal bodies or “speckles,” including splicesome subunits, p80 coilin, and promelocytic leukemia protein (PML).4–6 These structures form by a process of self-assembly, and the proteins within these structures are in dynamic equilibrium with proteins in the surrounding nucleoplasm.7,16,31 Moreover, the interactions between the coregulatory proteins and the nuclear hormone receptors, which bind to DNA and function to control transcription in response to specific ligands, regulate the fraction of proteins localized in focal bodies.8,9,16,34,35 For example, GRIP is recruited from focal bodies by its interactions with both the estrogen and androgen receptors.8,34 Further, the subnuclear organization and function of SRC-1, a related coactivator protein, are also regulated by these nuclear steroid hormone receptors in a ligand-dependent manner.35,36 Similarly, the transcriptional corepressor RIP140 forms spherical focal bodies that are redistributed by the glucocorticoid receptor.9 In light of the heterogeneous distribution of the coregulatory proteins, however, the analysis of how the nuclear receptors affect the localization of the coregulatory proteins requires the use of an unbiased and quantitative imaging approach. Importantly, we show here that it is possible to normalize coregulatory protein subnuclear organization for the effects of expression level in each cell (Figs. 4 and 6). This normalization method allows the effects of DNA-binding factors to be statistically verified even in cell populations with different levels of coregulator concentration. Since differences in expression levels would interfere with the comparison of different experimental cell populations, these quantitative techniques will be critical for understanding how DNA-binding transcription factors regulate the subnuclear organization and function of coregulatory proteins.
Recent studies indicate that the transcriptional activity of the nuclear receptor-coregulatory protein complex is coupled to protein degradation,37 and GRIP foci have been shown to contain some proteasome components.11 Because several pathologies are linked to the formation of nuclear focal bodies by aberrant proteins that recruit proteasomes, focal bodies have been regarded as generalized sites of protein degradation.38,39 However, several lines of evidence argue against applying this point of view to all subnuclear bodies. First, the coregulatory proteins are not irreversibly trapped in focal bodies, but are in dynamic equilibrium with the surrounding nucleoplasm.36,40 Additionally, the coregulatory proteins associated with focal bodies maintain interactions with other functional protein partners, and the nuclear receptors regulate the fraction of proteins localized in focal bodies.8,9,13,34,36 We showed for the first time that the focal bodies formed by a coactivator protein (GRIP) were separate and distinct from those bodies formed by a corepressor protein (SMRT) [Fig. 5(a)]. This novel finding suggests that specific functional interactions must target coactivators and corepressors to the different subnuclear regions, supporting the previous evidence that subnuclear bodies are not merely sites of degradation for abberant proteins. We also used the quantitative image analysis approach to compare the formation of nuclear focal bodies by GRIP and SMRT. Despite their distinct subnuclear localization, statistical analysis revealed that the formation of both GRIP and SMRT focal bodies was directly related to the expression level of the coregulatory proteins [Figs. 3(a) and 5(c)]. In addition to the effects we have shown for coregulatory proteins, similar concentration-dependent behavior has been observed for other distinct spherical subnuclear structures, such as Cajal bodies.41 It has been hypothesized that the exchange of proteins between subnuclear structures and the nucleoplasm is intricately regulated,31 and changing protein expression levels may contribute to this process. Taken together, these results support a regulated mechanism of self assembly of like proteins into distinct multimeric complexes, potentially serving as a reservoir for those proteins.42–44
The broad goal of many cellular imaging studies is to understand how different experimental conditions regulate subcellular structure and function. In these studies, it is critical that the analytical techniques rigorously account for variability in the cell population. Here, the subnuclear organization of transcriptional coregulatory proteins was used to exemplify the complexity of imaging data that accurately represent the cell population. We have shown that rigorous quantitative imaging techniques are necessary to simplify the analysis of these complex datasets. The integrated methods that were validated in this report could be easily adapted to study the organization of other transcriptional regulators, or other subnuclear structures such as Cajal bodies or PML bodies.4,42 Based on these results, we propose that this novel combination of unbiased image acquisition, automated morphometric data extraction, and statistical modeling techniques will be essential for rigorously addressing many critical questions in cell biology.
Acknowledgments
We thank R. Tsien for the plasmid encoding the optimized mRFP and F. Schaufele for the expression vectors encoding GFP-GRIP and the human SMRT cDNA. We also wish to thank A. Periasamy, Director of the W. M. Keck Center for Cellular Imaging, for his help with these studies. This work was supported by NIH DK47301 (RND) and F32 DK60315 (TCV).
References
- 1.Andrews PD, Harper IS, Swedlow JR. “To 5D and beyond: quantitative fluorescence microscopy in the postgenomic era,”. Traffic. 2002;3:29. doi: 10.1034/j.1600-0854.2002.30105.x. [DOI] [PubMed] [Google Scholar]
- 2.Swedlow JR, Goldberg I, Brauner E, Sorger PK. “Informatics and quantitative analysis in biological imaging,”. Science. 2003;300:100–102. doi: 10.1126/science.1082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eils R, Athale C. “Computational imaging in cell biology,”. J Cell Biol. 2003;161:477–481. doi: 10.1083/jcb.200302097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spector DL. “Nuclear domains,”. J Cell Sci. 2001;114:2891–2893. doi: 10.1242/jcs.114.16.2891. [DOI] [PubMed] [Google Scholar]
- 5.Carmo-Fonseca M. “The contribution of nuclear compartmentalization to gene regulation,”. Cell. 2002;108:513–521. doi: 10.1016/s0092-8674(02)00650-5. [DOI] [PubMed] [Google Scholar]
- 6.Lamond AI, Earnshaw WC. “Structure and function in the nucleus,”. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 7.Misteli T. “The concept of self-organization in cellular architecture,”. J Cell Biol. 2001;155:181–185. doi: 10.1083/jcb.200108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaufele F, Chang CY, Liu W, Baxter JD, Nordeen SK, Wan Y, Day RN, McDonnell DP. “Temporally distinct and ligand-specific recruitment of nuclear receptor-interacting peptides and cofactors to subnuclear domains containing the estrogen receptor,”. Mol Endocrinol. 2000;14:2024–2039. doi: 10.1210/mend.14.12.0572. [DOI] [PubMed] [Google Scholar]
- 9.Tazawa H, Osman W, Shoji Y, Treuter E, Gustafsson JA, Zilliacus J. “Regulation of subnuclear localization is associated with a mechanism for nuclear receptor corepression by RIP140,”. Mol Cell Biol. 2003;23:4187–4198. doi: 10.1128/MCB.23.12.4187-4198.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Day RN, Nordeen SK, Wan Y. “Visualizing protein-protein interactions in the nucleus of the living cell,”. Mol Endocrinol. 1999;13:517–526. doi: 10.1210/mend.13.4.0259. [DOI] [PubMed] [Google Scholar]
- 11.Baumann CT, Ma H, Wolford R, Reyes JC, Maruvada P, Lim CP, Yen PM, Stallcup MR, Hager GL. “The glucocorticoid receptor interacting protein 1 (GRIP1) localizes in discrete nuclear foci that associate with ND10 bodies and are enriched in components of the 26S proteasome,”. Mol Endocrinol. 2001;15:485–500. doi: 10.1210/mend.15.4.0618. [DOI] [PubMed] [Google Scholar]
- 12.Soderstrom M, Vo A, Heinzel T, Lavinsky RM, Yang WM, Seto E, Peterson DA, Rosenfeld MG, Glass CK. “Differential effects of nuclear receptor corepressor (N-CoR) expression levels on retinoic acid receptor-mediated repression support the existence of dynamically regulated corepressor complexes,”. Mol Endocrinol. 1997;11:682–692. doi: 10.1210/mend.11.6.0018. [DOI] [PubMed] [Google Scholar]
- 13.Downes M, Ordentlich P, Kao HY, Alvarez JG, Evans RM. “Identification of a nuclear domain with deacetylase activity,”. ProcNatl Acad Sci USA. 2000;97:10330–10335. doi: 10.1073/pnas.97.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaufele F, Enwright JF, Wang X, Teoh C, Srihari R, Erickson R, MacDougald OA, Day RN. “CCAAT/enhancer binding protein alpha assembles essential cooperating factors in common subnuclear domains,”. Mol Endocrinol. 2001;15:1665–1676. doi: 10.1210/mend.15.10.0716. [DOI] [PubMed] [Google Scholar]
- 15.Voss TC, Demarco IA, Booker CF, Day RN. “A computer-assisted image analysis protocol that quantitatively measures subnuclear protein organization in cell populations,”. BioTechniques. 2004;36:240–247. doi: 10.2144/04362BI01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belmont A. “Dynamics of chromatin, proteins, and bodies within the cell nucleus,”. Curr Opin Cell Biol. 2003;15:304–310. doi: 10.1016/s0955-0674(03)00045-0. [DOI] [PubMed] [Google Scholar]
- 17.Hong H, Kohli K, Trivedi A, Johnson DL, Stallcup MR. “GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors,”. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen JD, Evans RM. “A transcriptional corepressor that interacts with nuclear hormone receptors [see comments],”. Nature (London) 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 19.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. “A monomeric red fluorescent protein,”. Proc Natl Acad Sci USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacGregor GR, Caskey CT. “Construction of plasmids that express E. coli beta-galactosidase in mammalian cells,”. Nucleic Acids Res. 1989;17:2365. doi: 10.1093/nar/17.6.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Day RN, Periasamy A, Schaufele F. “Fluorescence resonance energy transfer microscopy of localized protein interactions in the living cell nucleus,”. Methods. 2001;25:4–18. doi: 10.1006/meth.2001.1211. [DOI] [PubMed] [Google Scholar]
- 22.Day RN. “Visualization of Pit-1 transcription factor interactions in the living cell nucleus by fluorescence resonance energy transfer microscopy,”. Mol Endocrinol. 1998;12:1410–1419. doi: 10.1210/mend.12.9.0168. [DOI] [PubMed] [Google Scholar]
- 23.Tsien RY. “The green fluorescent protein,”. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 24.van Roessel P, Brand AH. “Imaging into the future: visualizing gene expression and protein interactions with fluorescent proteins,”. Nat Cell Biol. 2002;4:E15–20. doi: 10.1038/ncb0102-e15. [DOI] [PubMed] [Google Scholar]
- 25.Hadjantonakis AK, Nagy A. “The color of mice: in the light of GFP-variant reporters,”. Histochem Cell Biol. 2001;115:49–58. doi: 10.1007/s004180000233. [DOI] [PubMed] [Google Scholar]
- 26.Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. “Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP,”. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 27.Walsh MK, Lichtman JW. “In vivo time-lapse imaging of synaptic takeover associated with naturally occurring synapse elimination,”. Neuron. 2003;37:67–73. doi: 10.1016/s0896-6273(02)01142-x. [DOI] [PubMed] [Google Scholar]
- 28.Dundr M, McNally JG, Cohen J, Misteli T. “Quantitation of GFP-fusion proteins in single living cells,”. J Struct Biol. 2002;140:92–99. doi: 10.1016/s1047-8477(02)00521-x. [DOI] [PubMed] [Google Scholar]
- 29.Chiu CS, Kartalov E, Unger M, Quake S, Lester HA. “Single-molecule measurements calibrate green fluorescent protein surface densities on transparent beads for use with ‘knock-in’ animals and other expression systems,”. J Neurosci Methods. 2001;105:55–63. doi: 10.1016/s0165-0270(00)00354-x. [DOI] [PubMed] [Google Scholar]
- 30.Pickart CM, Cohen RE. “Proteasomes and their kin: proteases in the machine age,”. Nat Rev Mol Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- 31.Lamond AI, Spector DL. “Nuclear speckles: a model for nuclear organelles,”. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 32.Elliott JT, Tona A, Plant AL. “Comparison of reagents for shape analysis of fixed cells by automated fluorescence microscopy,”. Cytometry. 2003;52A:90–100. doi: 10.1002/cyto.a.10025. [DOI] [PubMed] [Google Scholar]
- 33.Webb D, Hamilton MA, Harkin GJ, Lawrence S, Camper AK, Lewandowski Z. “Assessing technician effects when extracting quantities from microscope images,”. J Microbiol Methods. 2003;53:97–106. doi: 10.1016/s0167-7012(02)00228-2. [DOI] [PubMed] [Google Scholar]
- 34.Black BE, Vitto MJ, Gioeli D, Spencer A, Afshar N, Conaway MR, Weber MJ, Paschal BM. “Transient, ligand-dependent arrest of the androgen receptor in subnuclear foci alters phosphorylation and coactivator interactions,”. Mol Endocrinol. 2004;18:834–850. doi: 10.1210/me.2003-0145. [DOI] [PubMed] [Google Scholar]
- 35.Stenoien DL, Mancini MG, Patel K, Allegretto EA, Smith CL, Mancini MA. “Subnuclear trafficking of estrogen receptor-alpha and steroid receptor coactivator-1,”. Mol Endocrinol. 2000;14:518–534. doi: 10.1210/mend.14.4.0436. [DOI] [PubMed] [Google Scholar]
- 36.Rivera OJ, Song CS, Centonze VE, Lechleiter JD, Chatterjee B, Roy AK. “Role of the promyelocytic leukemia body in the dynamic interaction between the androgen receptor and steroid receptor coactivator-1 in living cells,”. Mol Endocrinol. 2003;17:128–140. doi: 10.1210/me.2002-0165. [DOI] [PubMed] [Google Scholar]
- 37.Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. “A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors,”. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- 38.Taylor JP, Tanaka F, Robitschek J, Sandoval CM, Taye A, Markovic-Plese S, Fischbeck KH. “Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein,”. Hum Mol Genet. 2003;12:749–757. doi: 10.1093/hmg/ddg074. [DOI] [PubMed] [Google Scholar]
- 39.Tarlac V, Storey E. “Role of proteolysis in polyglutamine disorders,”. J Neurosci Res. 2003;74:406–416. doi: 10.1002/jnr.10746. [DOI] [PubMed] [Google Scholar]
- 40.Voss TC, DeMareo IA, Booker CF, Day RN. “Corepressor subnuclear organization is regulated by estrogen receptor via a mechanism that requires the DNA-binding domain,”. Mol Cell Endocrinol. 2005;231(1–2):33–47. doi: 10.1016/j.mce.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 41.Sleeman JE, Ajuh P, Lamond AI. “snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN,”. J Cell Sci. 2001;114:4407–4419. doi: 10.1242/jcs.114.24.4407. [DOI] [PubMed] [Google Scholar]
- 42.Matera AG. “Nuclear bodies: multifaceted subdomains of the interchromatin space,”. Trends Cell Biol. 1999;9:302–309. doi: 10.1016/s0962-8924(99)01606-2. [DOI] [PubMed] [Google Scholar]
- 43.Khochbin S, Kao HY. “Histone deacetylase complexes: functional entities or molecular reservoirs,”. FEBS Lett. 2001;494:141–144. doi: 10.1016/s0014-5793(01)02327-4. [DOI] [PubMed] [Google Scholar]
- 44.Hebert MD, Matera AG. “Self-association of coilin reveals a common theme in nuclear body localization,”. Mol Biol Cell. 2000;11:4159–4171. doi: 10.1091/mbc.11.12.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]


