Abstract
In our continued studies on corticotropin releasing factor receptor (CRFR1) signaling in the skin, we tested functional activity of CRFR1α, e, f, g and h isoforms after transfection to COS cells. Both membrane-bound and soluble variants are translated in vivo into final protein products that undergo further post-translational modifications. CRFR1 α was the only isoform coupled directly to adenylate cyclase with the exception of an artificial isoform (CRFR1h2) with the insertion of 37 amino acids between the ligand binding domain and the first extracellular loop that was capable of producing detectable levels of cyclic AMP (cAMP). Soluble isoforms could modulate cell response with CRFR1e attenuating and CRFR1h amplifying CRFR1α-coupled cAMP production stimulated by urocortin. Testing with plasmids containing the luciferase reporter gene, and inducible cis-elements (CRE, CaRE, SRE, AP1 or NF-κB) demonstrated that only CRFR1α was involved directly in the transcriptional regulation, while CRFR1g inhibited CRE activity. Significantly higher reporter gene expression by CRF was observed than that mediated by 4β-phorbol 12-myristate 13-acetate and forskolin alone, being compatible with the concomitant treatment by phorbol 12-myristate 13-acetate and forskolin. This suggests that both protein kinase A and C can be involved in CRF-dependent signal transduction.
Keywords: skin, CRFR1, CRF, urocortin, COS cells
Abbreviations: AEBSF, amino-ethyl benzene sulfonyl fluoride; cAMP, cyclic AMP; CaRE, calcium responsive element; CMV, cytomegalovirus; CRE, cyclic AMP responsive element; CREB, cyclic AMP responsive element binding protein; CRF, corticotropin releasing factor; CRFR1, corticotropin releasing factor receptor; IP3, inositol-1,4,5-triphosphate; NF-κB, nuclear factor-kappa B; PKA, protein kinase A; PKC, protein kinase C; PMA, 4β-phorbol 12-myristate 13-acetate; SRE, serum responsive element; TSH, thyroid stimulating hormone
Corticotropin releasing factor receptors (CRFRs) are recognized as main central regulators in the humoral and behavioral responses to systemic stress [1–4]. They also play an important role in the regulation of peripheral organ functions [2,3,5,6]. In the skin they may serve as coordinators of its homeostatic response to external stress [6–8]. CRFRs represent a family with at least three distinct members (CRFR1, CRFR2 and CRFR3) encoded by separate genes, which share high-sequence homology (≈ 70%) and belong to the seven transmembrane segment receptor proteins coupled to the Gs signaling system [1–3,9]. After binding of CRF or related peptides, CRFR1 interacts with the cellular effectors system via activation of adenylate cyclase with production of cAMP and subsequent activation of protein kinase A (PKA)-dependent pathways; or activation of phospholipase C with production of inositol-1,4,5-triphosphate (IP3); this in turn, activates protein kinase C (PKC)-dependent and calcium-activated pathways [2,9,10]. In addition, CRFR1 signal transduction is coupled directly to calcium channels [11,12]. Some authors also demonstrated that CRF receptors can activate MAP kinase-dependent signaling pathways [13] and nitric oxide production [14]. These CRFR1-activated signal transduction pathways can regulate cellular phenotype both on the central and peripheral levels.
The established genomic structures for the human CRFR1 and CRFR2 (GenBank accession number AF039510-AF3523; L24096) contained at least 14 and 15 exons, respectively. Eight alternatively spliced transcripts have been identified in humans (GenBank accession numbers are in parentheses); CRFR1α in which exon 6 is spliced (L23332); a longer variant CRFR1β (variant) that contains all 14 exons (L23333); CRFR1c isoform where exon 3 and exon 6 are spliced out (U16273); CRFR1d isoform where exons 6 and 13 are missing (AF180301); CRH-R1e with deletion of exons 3 and 4 (AF369651); CRFR1f with deletion of exon 12 (AF369652); CRFR1g with deletion of exon 11, 27 basepairs of exon 10 and 28 basepairs of exon 12 (AF369653); and CRFR1h with addition of a cryptic exon (AF374231) [15–18]. In rodents, three CRFR1 isoforms have already been identified in rats [19], four in mice [18] and nine in hamsters [20]. It was proposed that differential and tissue-specific expression of alternatively spliced CRFR forms are linked to the functional activity of placenta, decidua, fetal membranes, endometrium, myometrium, uterine vasculature and the immune system [2,16,21,22]. In skin, such expression is defined by anatomic or histological location, physiological status, coexisting pathology, or hair cycle phase [18,23]. In addition, we have demonstrated that alternative splicing of CRFR1 is modulated by external factors such as ultraviolet radiation or exposure to forskolin or 4β-phorbol 12-myristate 13-acetate (PMA) [18]. The above findings raise the question about the significance of generation of alternatively spliced CRFR1 mRNA forms. In general, the importance of alternative splicing is emphasized by the fact that up to 50% of human genes may be alternatively spliced, that this mechanism is frequently deregulated in cancer cells and that environmental factors can modulate the splicing process [18,24].
CRFR1 α, β, c and d isoforms differ in their ability to bind ligands and activate G proteins [10,16,25]. CRFR1 α is the most efficient in the stimulation of cAMP production, CRFR1c and CRFR1β have a decreased CRF binding capacity [10,25], while CRFR1d is poorly coupled to G proteins [16].
Recently we have described four new human CRFR1 isoforms, which included messages with internal deletions and unusual isoforms composed of soluble extracellular (ligand-binding) domains [18]. As the skin shows polymorphism in CRFR1 expression and its functional diversity may require differential expression of isoforms of CRFR1 to precisely couple selectively activated phenotypic targets, we performed molecular characterization of newly described CRFR1 isoforms. First, we tested whether these messages are translatable. In the second step, we characterized their coupling to different signaling pathways and their modulatory role on the CRFR1α activity.
Materials and methods
CRFR1 constructs preparations
Full-length sequences of human CRFR1 isoforms were constructed by PCR. Plasmid phCRFR82 (generous gift of Dr N. Vita, Sano. Recherche, Labege, France) containing human CRFR1α cDNA was used as an initial template. The reaction mixture (25 μL) contained 2 mm MgCl2, 2.5 of each dNTP, 0.4 μm of each primer, 20 mm Tris/HCl (pH 8.8), 10 mm KCl, 10 mm (NH4)2SO4, 0.1% Triton X-100, 0.1 mg·mL−1 bovine serum albumin and 1.25 μ of Pfu DNA polymerase (Stratagen, La Jolla, CA, USA). The mixture was heated to 95 °C for 2.5 min and then amplified for 25 cycles: 94 °C for 30 s (denaturation), 56 °C for 40 s (annealing) and 72 °C for 1.5 min (extension).
CRFR1α was amplified from phCRFR82 plasmid by primers E3 (5′-AAAAGCTTAGGACCCGGGCATTC AGGA-3′) and E11 (5′-AAGAATTCTCAGACTGCTGTGGACTGCT-3′).
Full-length CRFR1g DNA was obtained in three PCR reactions. First, a fragment spanning 5′ untranslated sequence and exons 1 through 10 was amplified by primers E3 and E9 (5′-GAAGGAGTTGAAGTAGATGTAGTCGGTGTACA-3′). Second fragment (exons 12–14) was amplified by primers E12 (5′-CATCTACTTCAACTCCTTCCTG-3′) and E11. Finally, the first two fragments were assembled together by primers E3 and E11. This was possible because primer E9 contained a sequence homologous to primer E12.
Similarly, for CRFR1f construct exons 1–11 of CRF receptor were amplified by primers E3 and E18 (5′-ACAAAGAAGCCCTGTACTGAATGGTCTCAG-3′), and exons 13 and 14 by primers E16 (5′-CATTCAGTACAGGGCTTCTTTGTGTCTGTG-3′) and E17 (5′-AAGAATTCTCATCCCCCCAGCCACAG-3′). The full sequence was obtained by combining those two fragments together by primers E3 and E17.
CRFR1e DNA was constructed in a slightly different manner. Fragments spanning exons 1–2 and 5–14 were amplified by primers E3, E26 (5′-CTTGCTTTTTTTGAGATGTTGCTGGCCAGGGA-3′) and E25 (5′-AAAAAAAGCAAGGTGCACTACC-3′), E11, respectively. The first fragment was slightly extended in nested PCR by primers E3 and E28 (5′-TGGTAGTGCACCTTGCTTTTTTTGAGATGTTGC-3′). Finally, full-length CRFR1e DNA was assembled by PCR of these two fragments with primers E3 and E11.
Two different constructs were produced for CRFR1h isoform. The first contained exons 1–4 and a fragment of the cryptic exon up to the translation terminator. These constructs were also assembled in three steps. In the first PCR we amplified exons 1–4 by primers E3 and E 24 (5′-CTCCTCATTGAGGATCTCCT-3′). The second PCR amplified the cryptic exon by primers E21 (5′-GTG CCAGGAGATCCTCAATG-3′) and E19 (5′-AAGAATTCTTTGTCCCACCACGGTGTGCTC-3′). The third PCR assembled the CRFR1h DNA. Another construct (CRFR1h2) was designed to contain an in-frame insertion of the cryptic exon. It was produced by six separate amplifications. First PCR amplified exons 1–4 by primers E3 and E24. The second one produced a fragment spanning exons 5–14 (primers E25, E11). The first half of the cryptic exon was amplified by primers E21 and E20 (5′-TGATGTCCCACCACGGTGTG-3′). The second half was amplified by primers E22 (5′-GTGGGACATCAAAACGGATTCTGGGGGTCTG-3′) and E23 (5′-CTTGCTTTTTTTCTCTCCCCACACGGTGAAC-3′). Primers E20 and E22 contained two mutations eliminating translation terminator and introducing additional nucleotide to preserve translation frame of CRF receptor. The mutated fragment was reassembled by primers E21 and E23 and connected to CRFR1 (exons 1–4) by primers E3 and E23. This fragment was slightly extended by primer E27 (5′-GGTAGTGCACCTTGCTTTTTTTCTCTCCCCA-3′) and attached to another fragment of CRF receptor in the final PCR by primers E3 and E11.
To attach V5 epitope to the CRFR1α, g, h2 and e2 isoforms we amplified the corresponding DNA fragments with primers E3 and primer E29 (5′-AAGAATTCTTGACTGCTGTGGACTGCT-3′). Isoform CRFR1f was amplified with primers E3 and primer E30 (5′-AAGAATTCTTTCCCCCCAGCCACAG-3′) and CRFR1e with primers E3 and primer E31 (5′-AAGAATTCTTGCTGGACCACGAACCAGGT-3′).
Final PCR fragments were purified by GFX gel band purification kit (Amersham-Pharmacia-Biotech), digested by HindIII and EcoRI enzymes and cloned in expression vector pcDNA6/V5-His version B (Invitrogen, Carlstand, CA, USA).
Luciferase constructs
The starting vector to construct luciferase (luc) reporter gene plasmids was pGL3-basic (Promega). We had to modify the promoter region to insert TATA box and convenient restriction sites. Thus, we deleted the luciferase gene by amplification pGL3-basic with P762 (5′-TCGAATTCCC TAGGGCCGCTTCGAGCAGACATGA-3′) and P763 (5′-TTCTCGAGACGCGTTATCGATAGAGAAATGTTCTGGC-3′) and digested the PCR product with EcoRI and XhoI. The insert was synthesized with primers P764 (5′-AACTCGAGGCTAGTCTGCAGGAGCTCAAGCTTTCTAGAGAATTCA-3′) and P765 (5′-TGAATTCTCTAGAAAGCTTGAGCTCCTGCAGACTAGCCTCGAGTT-3′). It was also digested with EcoRI and XhoI, ligated with the vector and cloned in JM109 Escherichia coli.
Luciferase gene was amplified from pGL3-basic vector by primers P766 (5′-AAAAGCTTCCCGGGCATTCCGGTACTGTTGGTAAA-3′), P767 (5′-GGGAATTCGACTCTAGAATTACACGGCGA-3′), digested with HindIII and EcoRI and inserted in the vector described above. This plasmid was named pLuc.
The minimal promoter containing TATA box was amplified from pcDNA6/V5-HisA vector (Invitrogen) by primers P768 (5′-AACTGCAGGAGCTCCCCATTGACGCAAATGGGCG-3′), P769 (5′-GGAAGCTTTTCGATAAGCCAGTAAGCAGTG-3′), digested with PstI and HindIII and inserted in pLuc. This plasmid was named pP1-Luc.
pP1-Luc was used to construct the reporter plasmids containing CRE, CaRe, NF-κB, AP1, SRE sequences. These sequences were synthesized as 45 basepair-long oligonucleotides and assembled in 158 basepair-long fragments according the reported protocols [26]. Assembled fragments were digested by XhoI, PstI and inserted in pP1-Luc.
In summary, the newly produced constructs were as follows: pCRE-Luc (contained four CRE elements); pCaRe-Luc (four CaRe elements); pAP1-Luc (fiive AP1 elements); pSRE-luc (two SRE elements); pNF-κB-Luc (four pNF-κB elements) pNF-κB-Luc2 (two pNF-κB sequences). pL-L uc served as negative control. It contained 158 basepair-long random sequence. The positive control was pCMV-luc. It contained CMV promoter. The sequences of the cis elements were as follow: CRE (5′-TGACGTCA-3′), CaRE (5′-TGACGTTT-3′), NF-κB (5′-GGGGACTTTCC-3′), AP1 (5′-TGACTAA-3′), SRE (5′-CCATATTAGG-3′).
Transfections of COS cells with the plasmids
For transfection we used 4000 cells per well of 96-well plate. Cells were washed with antibiotic-free Dulbecco’s modified Eagle’s media (DMEM) and transfected by constructs using Lipophectamine Plus reagent (Invitrogen, Carlstand, CA, USA) according to the manufacturers’ protocol. We always used equal amount of plasmid DNA in each experiment (0.1 μg·well−1). Plasmid pcDNA6/V5-His version B (further named as pcDNA) was used as an empty vector. Four hours after transfection an equal volume of DMEM media containing 10% fetal bovine serum was added and cells were incubated overnight. Next morning, the cells were washed by DMEM and incubated in DMEM media containing 5% fetal bovine serum and antibiotics for 24 h. After that cells were stimulated by CRF or urocortin.
Western blotting
Transfected cells were detached by trypsin, centrifuged at 1000 g for 10 min at 4 °C. The cell pellets were then washed with NaCl/Pi and frozen in −70 C. For protein isolation frozen cell pellets were solubilized by pipetting into an iced buffer containing 20 mm Tris, pH 7.5, 150 mm NaCl, 15% glycerol, 1% Triton X100, 120 μg·mL−1 leupeptin, 3 μM pepstatin and 3 mm amino-ethyl benzene sulfonyl fluoride (AEBSF). Cellular homogenates were centrifuged at 16 000 g for 10 min at 4 °C, and the supernatants were removed and stored at −80 °C for further analysis. Separate aliquots of 5 μL were used for protein determination by Micro protein Kit (Sigma). Fifty micrograms of protein were loaded on 12% SDS-PAGE, transferred to immobilion-p poly(vinylidene difluoride) membrane (Millipore Corp, Bedford, MA, USA) for 3 h at 4 °C and blocked for 4 h at room temperature in 5% non fat powdered milk in TBST (50 mm Tris, pH 7.5, 150 mm NaCl, 0.01% Tween-20). Immunodetection of the V5-tagged proteins was performed after 1-h incubation with anti-V5 mouse antibodies (dilution 1 : 10 000) (Invitrogen). After that membranes were washed twice in TBST for 10 min and incubated 1 h with antimouse antibodies coupled to horse-radish peroxidase (dilution 1 : 4000, 1 h) (Santa Cruz Biotechnology). Membranes were washed twice in TBST and once in TBS. Bands were visualized by ECL reagent (Amersham Pharmacia Biotech) according to the manufacturers’ instructions (Amersham Pharmacia Biotech).
CRF and urocortin treatment and cAMP assays
Serial dilutions of CRF and urocortin peptides were added to DMEM containing 5% fetal bovine serum, antibiotics and 0.5 mm 3-isobutyl-1-methylxanthine (IBMX), and transfected COS cells were incubated with the ligand for 1 h at 37 °C and 5% CO2 in the incubator.
Cyclic AMP concentration was measured by cAMP functional assay kit (Packard BioScience, Meriden, CT, USA). Stimulated cells were washed three times by NaCl/Pi and incubated for 1 h in 25 μL of lysis buffer at room temperature. Lysis buffer contained 0.4 × Hank’s balanced salt solution (Gibco BRL), pH 7.4, 50 mm Hepes, 2 g·L−1 MgCl2, 0. 01mm IBMX, 0.05% Triton X100, 0.01 μM biotinilated cAMP, 4 μL·mL−1 of donor beads and 4 μL·mL−1 of acceptor beads. The signal was measured by Fusion α instrument (Packard BioScience, Meriden, CT, USA). cAMP concentration was recalculated from the standard curve according to the manufacture’s protocol (Packard BioScience, Meriden, CT, USA).
Luciferase expression assays
Luciferase expression was measured by dual-luciferase reporter assay system (Promega) according to the manufacturers’ protocol. Cells were cotransfected with the experimental constructs and phRL-TK plasmid containing Renilla luciferase. Experimental constructs were pCRFR1α and plasmids containing firefly luciferase under control of different cis-elements. Renilla luciferase was used to normalize the data (see below). Transfected cells were exposed to CRF or urocortin peptides for 12 h, lysed and the luminescence was measured. The luminescence background represented by untransfected COS cells was subtracted, the firefly luciferase counts were divided by Renilla luciferase counts and the relative luciferase expression was calculated. It was determined as a ratio of experimental sample vs. positive control. Firefly luciferase driven by the CMV promoter (pCMV-Luc construct) was used as a positive control.
In some experiments, PMA (200 nM), forskolin (10 μM) or H89 inhibitor of PKA (10 μM) were added directly to the experimental media (alone or in combination) to measure reporter gene response.
Statistical analysis
Data was presented as mean ± SEM, and analyzed using one-way analysis of variance and appropriate post hoc test or by Student’s t-test using prism 4.00 software (GraphPad Software, San Diego, CA, USA). Significant differences are denoted with asterisks: *P<0.05 or P<0.001; for the details see figure legends.
Results and discussion
Figure 1A shows alternatively spliced CRFR1 isoforms including CRFR1e, CRFR1f, CRFR1g and CRFR1h, which were recently characterized by us [18]. Together with CRFR1α, they were cloned into the expression vector pcDNA6/V5-HisB (Fig. 1B). This vector contains cytomegalovirus (CMV) immediate-early promoter that drives high-level transcription in wide range of mammalian cells. The constructs were named according to the isoform they contained. For example, pCRFR1a corresponds to the plasmid containing CRFR1α isoform. We also constructed an artificial CRFR1h2 isoform by introduction of two point mutations that restore the original reading frame (Fig. 1B). Thus, the CRFR1h2 protein is similar to CRFR1α except that it contains an insertion between the ligand binding domain and the first transmembrane domain (Fig. 1B) .
Fig. 1. The structure of CRFR1 isoforms.
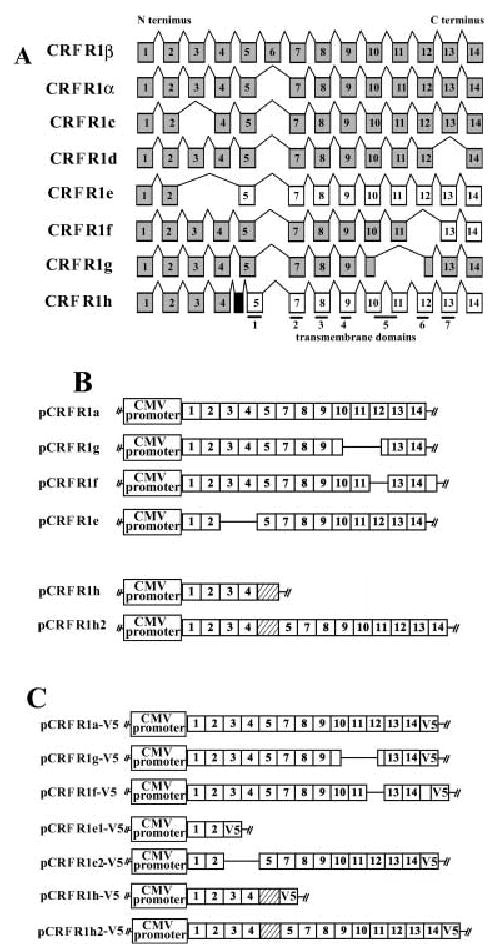
(A) Alternatively spliced isoforms of CRFR1 . Shaded boxes, translated exons; open boxes, exons located after a frame-shift; black boxes, insertion of a cryptic exon. (B) The structure of constructs used for functional assays.(C) The structure of constructs used for Western blotting.
Protein expression
To verify that the constructs produce proteins of the expected masses, we attached the V5 epitope to the C terminus of the CRFR1 isoforms (Fig. 1C). The predicted masses of the isoforms without/with V5 tag are as follows: CRFR1α (47.7/52 kDa), CRFR1e1 (10.8/15.1 kDa), CRFR1e2 (28.1/32.4 kDa), CRFR1f (43.1/47.4 kDa), CRFR1g (39.2/43.5 kDa), CRFR1h (13.5/18 kDa), CRFR1h2 (52.9/57.4 kDa). Western blotting experiments of extracts from COS cells transfected with CRFR1 isoforms identified specific proteins that were common or specific for a tested isoform and absent in control COS cells transfected with empty plasmid (Fig. 2). The molecular mass (including tag) of these isoforms is listed in Table 1. Thus, the mRNA from the alternatively spliced CRFR1 forms is translated into final protein products, which are the subject for further post-translational modifications (Fig. 2). The sole exception was pCRFR1e2, which did not produce any band, indicating that this putative open reading frame was not translated.
Fig. 2. Levels cAMP accumulation in transiently transfected COS cells with different CRFR1 isoforms after stimulation by CRF (A, C, E, G, I) or urocortin (B, D, F, H, J).
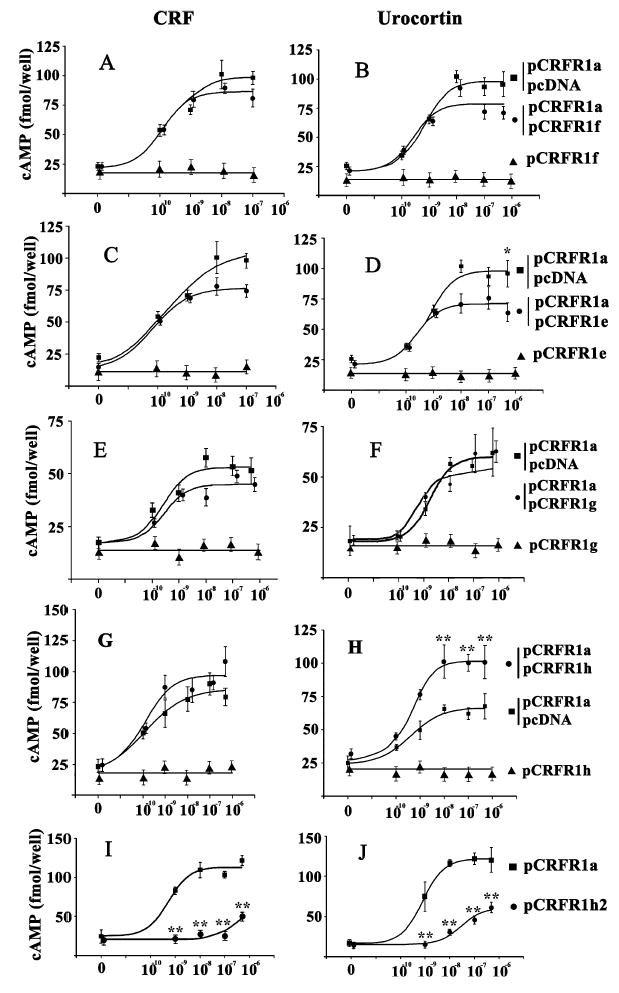
Cells were transiently transfected by the constructs alone or together with pCRFR1a. Significant differences between controls and ligand-stimulated cells are denoted as follows *P < 0.05 and **P < 0. 01.
Table 1. Molecular mass of the CRFR1 proteins expressed in COS cells.
The data represent estimated molecular mass of the proteins detected by anti-V5 Igs.
| Band number | Molecular mass (kDa) |
|---|---|
| 1 | 85–90 |
| 2 | 55–60 |
| 3 | 50–55 |
| 4 | 48 |
| 5 | 43 |
| 6 | 39 |
| 7 | 34 |
| 8 | 30 |
| 9 | 27 |
| 10 | 20 |
| 11 | 16 |
In general the majority of our isoforms were translated into proteins (Fig. 2) with the predicted size (Table 1). For example, band 4 (48 kDa) corresponds to the expected 47.4 kDa for pCRFR1f-V5; band 5 (43 kDa) to 43.5 kDa molecular mass for pCRFR1g-V5; band 11 (16 kDa) to 15.1 kDa molecular mass for CRHR1e1. The exception was isoform pCRHR1h producing protein with molecular mass 27 kDa (band 9; Fig. 2) vs. the expected 18 kDa (AF374231). The most likely explanation for the latter difference is that the synthesized protein undergoes rapid post-translational modification, e.g. glycosylation. Similar explanation is proposed for artificial construct pCRFR1h2, where instead of a band with 53 kDa a smear ranging from 50 to 60 kDa was noted (bands 3, Fig. 2 ) .
Proteins with different than expected molecular mass included bands 1, 2, 6–8 and 10. Protein with molecular mass 85–90 kDa (band 1) was seen in all isoforms containing transmembrane domains (Fig. 2) and therefore it may represent dimmer or fully glycosylated receptor form. Broad band 2 seen in CRFR1α has an apparent molecular mass of 55–65 kDa and most likely represents glycosylated receptor. We note that others have also reported detection of CRFR1 proteins with molecular mass at a similar range [27]. Protein glycosylation is also the most likely explanation for detection of an additional CRFR1e1 protein (band 10) with molecular mass of 20 kDa (Fig. 2). Proteins with lower molecular mass than expected included band 6 (39 kDa) for CRFR1f, band 7 (34 kDa) for CRFR1g; and band 8 (30 kDa) for CRFR1h2 (Fig. 2, Table 1) may represent products of post-translational proteolytic processing and/or degradation.
Coupling to cAMP production
Figure 3 shows the effect of CRF and urocortin on cAMP production in COS cells transfected by single construct or cotransfected by several plasmids. As expected [2,3,28] cAMP increases mediated by alpha isoform were similar for CRF and urocortin (Fig. 3, Table 2). None of the other isoforms had any effect on cAMP accumulation when transfected alone with the exception of CRFR1h2 (Fig. 3) . In the latter, a significantly lower cAMP response (Fig. 3, Table 2) demonstrates that an insertion of 37 amino acid peptide segment between the ligand binding domain and the first transmembrane domain attenuates coupling of CRFR1h2 to cAMP transduction system. Nevertheless , the ability to produce cAMP in the latter system suggests that the CRFR1 receptor structure is relatively stable and it can survive such major changes as insertions or deletions without loosing its function.
Fig. 3. Expression of CRFR1 proteins in transiently transfected COS cells with plasmids containing receptor isoforms.
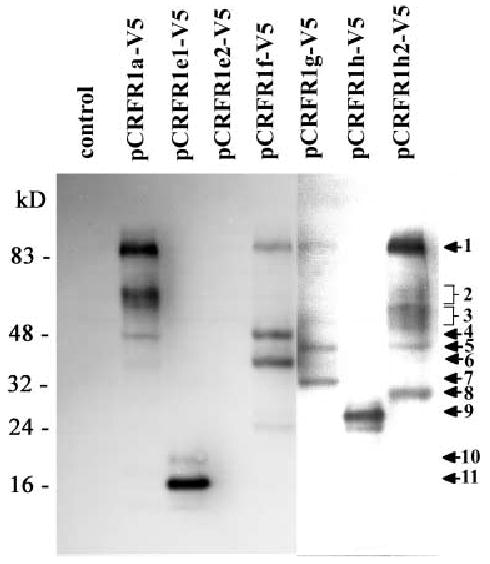
Data represents detection of the receptor proteins in extracts from COS cells transfected by V5-tagged constructs: pCRHR1a-V5; pCRFR1e1-V5; pCRFR1e2-V5; pCRFR1f-V5; pCRFR1g-V5; pCRFR1h-V5 and pCRFR1h2-V5. Ne gative control was represented by untransfected COS cells. Primary antibody: mouse anti-V5; secondary antibody: goat anti-mouse HRP-conjugated Ig.
Table 2. EC50 values for cAMP accumulation in COS cells expressing CRFR1 receptors.
Cells were transfected with CRFR1α with empty vector (pcDNA6/V5-His version B) or isoforms listed. The values are from the representative experiment presented in Fig. 3.
| EC50 |
||
|---|---|---|
| Isoform | CRF | Urocortin |
| A | 3.80 × 10−10 ± 2.50 × 10−10 | 7.33 × 10−10 ± 3.05 × 10−10 |
| A + empty vector | 2.97 × 10−10 ± 1.17 × 10−10 | 6.00 × 10−10 ± 8.56 × 10−10 |
| A + E | 8.01 × 10−11 | 2.42 × 10−10 |
| A + F | 1.17 × 10−10 | 2.65 × 10−10 |
| A + G | 2.81 × 10−10 | 1.68 × 10−9 |
| A + H | 3.56 × 10−10 | 8.41 × 10−10 |
| H2 | 5.43 × 10−6 | 2.88 × 10−8 |
| A + H2 | 7.39 × 10−10 | 1.72 × 10−9 |
The inability of the isoforms e–h to induce accumulation of cAMP suggests that functionally important domain(s) are missing in the final proteins. For example, CRFR1e encodes soluble protein of 11 kDa (Fig. 2) containing first 40 amino acids of distal N-terminal sequence with a remaining sequence different from the CRFR1α receptor due to the frame shift [18]. Similarly, CRFR1h isoform encodes truncated protein having only CRF-binding domain coded by exons 1–4, because of the translation terminator in the cryptic exon 4 [18]. With regard to membrane bound isoforms, CRFR1f lacks exon 12 and has C-terminus different from CRFR1α [18], which most likely will diminish its efficient coupling to Gs. Although CRFR1g preserves the original reading frame (the message is translated in a protein only 74 amino acids shorter than alpha isoform); it does not accumulate cAMP in response to CRF of urocortin. This suggests that the fifth and sixth transmembrane regions corresponding to the missing fragments in this isoform (Fig. 1A) are vitally important for the receptor coupling to adenylate cyclase.
To find possible interactions between the fully active alpha isoform and other variants, we conducted a series of cotransfection experiments and compared ligand-induced accumulation of cAMP(Fig. 3). Although the level of cAMP accumulated in COS cells cotransfected by CRFR1e, CRFR1f or CRFR1g and pCRFR1a was slightly lower than in the cells transfected by pCRFR1α and empty vector (Fig. 3), none of these differences were statistically significant with the exception of pCRFR1e after stimulation by urocortin (Fig. 3). In the latter the cotransfection with pCRFR1e inhibited significantly (P < 0.05) the maximal response (accumulation of cAMP) to urocortin but not CRF (Fig. 3F). EC50 values for the representative experiments shown in Table 2 were in a similar range to the alpha isoform, indicating that the affinities of the ligands for receptors had not changed significantly. The only exception was the CRFR1h2 isoform, which had a much lower affinity for CRF or urocortin in comparison to the control (Table 1).
A different pattern was observed for the CRFR1h isoform. When this construct was transfected together with the pCRFR1α, it dramatically amplified its cAMP responsiveness to urocortin (P < 0.01), with CRF having a statistically insignificant effect (P > 0.05) (Fig. 3H). This observation is reflected in the data of Perrin et al.[29]. They showed a higher binding potency for urocortin than CRF in the soluble form of the N-terminal domain (coded by the first four exons) that had been proteolytically removed from human CRFR1 [29]. Thus , the affect we have observed may result from the higher affinity of urocortin to the ligand binding domain. Nevertheless, it is unclear how a soluble protein can amplify cellular responsiveness. A possible explanation may be offered by experiments performed with thyroid stimulating hormone (TSH) receptors, where the activity of wild-type TSH receptor is higher when it is coexpressed together with the extracellular (TSH-binding) domain; the proposed mechanism included dimerizaton of the extracellular domains [30]. However, a satisfactory explanation for CRFR1h-associated enhancement of cAMP accumulation requires further extensive experimentation.
Coupling to signal transduction pathways distant from the cell membrane
As activation of CRF receptors has been shown to be coupled to different second messengers [2,6,10], we designed a set of constructs allowing assessment of the in vivo activation of different signal transduction pathways. These constructs contained the luciferase reporter gene, which was controlled by basic promoter element (TATA box) and inducible cis-element (Fig. 4). The cis-elements contained direct repeats of the cAMP response element (CRE), calcium response element (CaRE), serum response element (SRE), activator protein 1 (AP1) or binding sites for nuclear factor-kappa B (NF-κB). The control vector with a random sequence instead of the cis-element was also constructed. These constructs were transfected to COS cells together with different CRFR1 isoforms. COS cells were stimulated by CRF or urocortin and the luciferase expression was measured.
Fig. 4. Relative expression of luciferase in COS cells cotransformed by constructs containing CRE, CaRe, AP1, NF-κB and SRE elements and different CRFR1 isoforms.
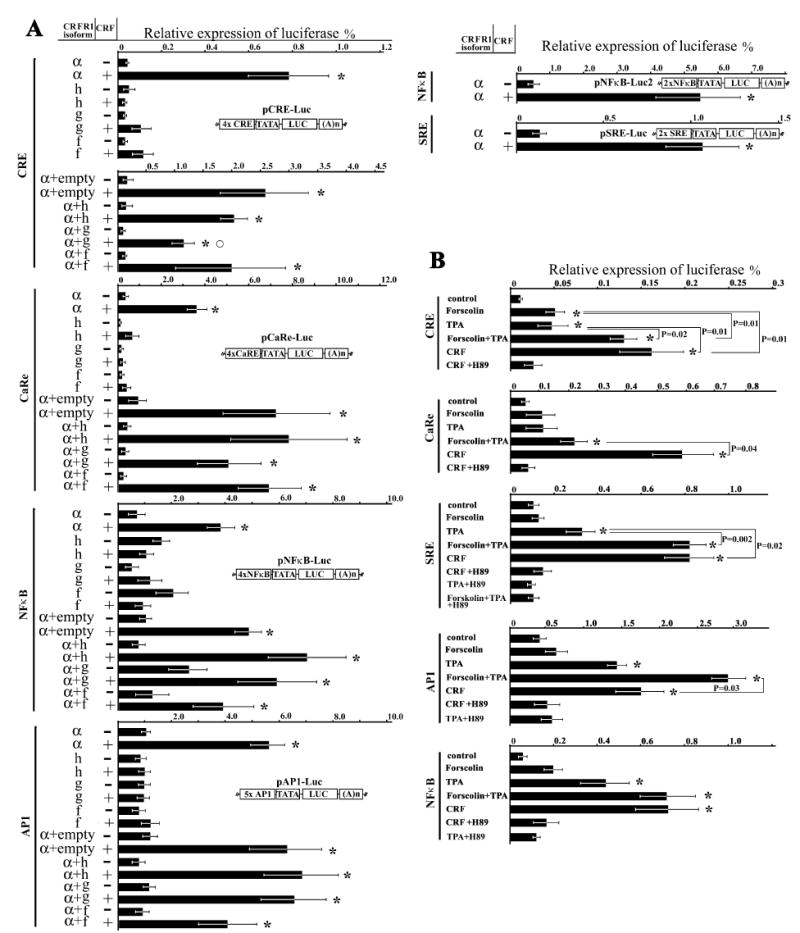
(A) Stimulation by CRF.(B) Stimulation by CRF, forskolin and PMA (TPA) and inhibition by PKA inhibitor (H89). Significant differences between controls and CRF-stimulated cells (P < 0.05) are denoted with an asterisk (*). Open circles denote significant differences between CRF-stimulated cells (pCRFR1a + empty vector and pCRFR1a + pCRFR1g) (P < 0.05).
cis-Elements containing CRE or CaRE should stimulate reporter gene expression in response to cAMP and calcium. The CaRE sequence is highly homologous to CRE. It was first identified as an element required for the induction of c-fos transcription in response to membrane depolarization and calcium influx [26]. CREB was subsequently identified as the c-fos promoter calcium-response element binding protein and shown to mediate both cAMP and calcium induction of c-fos expression through the CRE/CaRE sequence [31]. Thus CRE and CaRE can function as regulatory elements that integrate both calcium and cAMP signals in the control of gene expression. The SRE, AP1 or NF-κB binding sites should also report activation of protein kinase C and the MAP kinase pathways [32].
CRFR1α stimulated luciferase expression through all cis-elements (Fig. 4). Reporter gene expression induced by CaRE was always higher than for CRE, although both elements should bind with CREB. A possible explanation is that either CREB binds to CaRE more efficiently then to CRE or CaRE, or that it may bind some other factors. Thus, higher reporter gene expression induced by CaRE could result from additive effects of PKA and other factors including those induced by calcium. This is in agreement with our previous demonstration that in skin cells, activation of CRFR1 is coupled with the membrane-associated calcium channels through a mechanism independent of cAMP and IP3 [11,12,33].
Neither CRFR1f, g or h isoforms were able to stimulate any of the cis-elements. Instead the reporter gene expression decreased when these isoforms were cotransfected together with the CRFR1α (Fig. 4). For CRFR1g the inhibition of CRE-dependent luciferase expression was statistically significant (Fig. 4). Thus, only the α-isoform is directly coupled to tested signal transduction systems. Activation of different cis-elements by the α-isoform indicates that it is coupled to several different signal transduction pathways, either directly or through a cross-talk mechanism between different pathways. To test this hypothesis we induced cAMP accumulation by forskolin or stimulated protein kinase C with PMA. As expected, forskolin stimulated CRE and CaRE, which is characteristic of the cAMP-dependent pathway (Fig. 4B). SRE-, AP1-and NF-κB-dependent reporter expression was stimulated by PMA but not forskolin. The highest response was detected when forskolin and PMA were used together (Fig. 4). In this case the expression level of the reporter gene was similar to the expression induced by CRF. Thus, CRF induced the same level of response as simultaneous activation of PKA and PKC together. We attempted to separate these effects by the addition of PKA inhibitor (H89). Unfortunately, these compounds inhibited reporter expression induced not only by CRF and forskolin but also by PMA (Fig. 4B), not allowing proper distinction between those two pathways.
In conclusion, we suggest that the CRF/CRFR1α signaling system can stimulate gene expression through CRE, CaRE, SRE, AP1 and NF-κB elements and that PKA, PKC and MAP kinase pathways are involved in the regulation of transcriptional activity. This hypothesis is in agreement with a recent demonstration that CRFR can activate multiple G proteins with the subsequent activation of diverse signal transduction pathways [34–36].
Conclusions
We have conclusively demonstrated that messages from newly characterized CRFR1 isoforms, including membrane bound and soluble variants, were translated in vitro into final protein products that had undergone further post-translational modifications. Testing of cAMP production demonstrated that CRFR1α was the only isoform coupled to adenylate cyclase, whilst soluble isoforms modulated cell response to the agonist, e.g. CRFR1e attenuated while CRFR1h amplified CRFR1α coupled cAMP production stimulated by urocortin. The artificial isoform (CRFR1h2) with the insertion of 37 amino acids between ligand binding domain and the first extracellular loop was able to produce detectable levels of cAMP indicating that this region is not critical for the receptor function.
Testing with CRE, CaRE, SRE, AP1 and NF-κB elements demonstrated that only CRFR1α was directly involved in the transcriptional regulation. However , CRFR1g inhibited CRE activity suggesting that other isoforms might also play a modulatory role. Induction of CRE, CaRE, AP1, SRE and NF-κB-dependent luciferase reporter gene expression by CRF was higher than that mediated by PMA and forskolin alone and was compatible to the concomitant treatment by PMA and forskolin. Our data suggest that both protein kinase A and C can be involved in CRF-dependent signal transduction.
Acknowledgments
The project was supported by NIH grant number 1R01-AR047079–01A2 (A.S.), and by a grant from the Center of Excellence in Genomics and Bioinformatics, UTHSC (A.P. and A.S.). We also thank Ms Christine Crawford for skillful secretarial assistance.
References
- 1.A guilera G. Corticotropin releasing hormone, receptor regulation and the stress response. Trends Endocrinol Metab. 1998;9:329–336. doi: 10.1016/s1043-2760(98)00079-4. [DOI] [PubMed] [Google Scholar]
- 2.Grammatopoulos DK, Chrousos GP. Functional characteristics of CRH receptors and potential clinical applications of CRH-receptor antagonists. Trends Endocrinol. Metab. 2002;13:436–444. doi: 10.1016/s1043-2760(02)00670-7. [DOI] [PubMed] [Google Scholar]
- 3.Perrin MH, Vale, W.W Corticotropin releasing factor receptors and their ligand family. Ann NY Acad Sci. 1999;885:312–328. doi: 10.1111/j.1749-6632.1999.tb08687.x. [DOI] [PubMed] [Google Scholar]
- 4.Chrousos GP, Gold, P.W The concepts of stress and stress system disorders. JAMA. 1992;267:1244–1252. [PubMed] [Google Scholar]
- 5.Linton EA, Woodman JR, Asboth G, Glynn BP, Plested CP, Bernal, A.L Corticotrophin releasing hormone: its potential for a role in human myometrium. Exp Physiol. 2001;86:273–281. doi: 10.1113/eph8602183. [DOI] [PubMed] [Google Scholar]
- 6.Slominski A, Wortsman J, Pisarchik A, Zbytek B, Linton EA, Mazurkiewicz JE, Wei, E.T Cutaneous expression of corticotropin-releasing hormone (CRH), urocortin, and CRH receptors. FASEB J. 2001;15:1678–1693. doi: 10.1096/fj.00-0850rev. [DOI] [PubMed] [Google Scholar]
- 7.Slominski A, Wortsman J, Luger T, Paus R, Salomon, S Corticotropin releasing hormone and proopiomelanocortin involvement in the cutaneous response to stress. Physiol Rev. 2000;80:979–1020. doi: 10.1152/physrev.2000.80.3.979. [DOI] [PubMed] [Google Scholar]
- 8.Slominski A, Wortsman J. Neuroendocrinology of the skin. Endocrine Rev. 2000;21:457–487. doi: 10.1210/edrv.21.5.0410. [DOI] [PubMed] [Google Scholar]
- 9.Dautzenberg FM, Hauger, R.L The CRF peptide family and their receptors: yet more partners discovered. Trends Pharmacol Sci. 2002;23:71–77. doi: 10.1016/s0165-6147(02)01946-6. [DOI] [PubMed] [Google Scholar]
- 10.Nabhan C, Xiong Y, Xie LY, Abou-Samra, A. B. The alternatively spliced type II corticotropin-releasing factor receptor, stably expressed in LLCPK-1 cells, is not well coupled to the G protein(s) Biochem Biophys Res Commun. 1995;212:1015–1021. doi: 10.1006/bbrc.1995.2071. [DOI] [PubMed] [Google Scholar]
- 11.Fazal N, Slominski A, Choudhry MA, Wei ET, Sayeed, M.M Effect of CRF and related peptides on calcium signalling in human and rodent melanoma cells. FEBS Lett. 1998;435:187–190. doi: 10.1016/s0014-5793(98)01067-9. [DOI] [PubMed] [Google Scholar]
- 12.Wiesner B, Rolo. B, Fechner K, Slominski, A Intracellular calcium measurements of single human skin cells after stimulation with corticotropin-releasing factor and urocortin using confocal laser scanning microscopy. J Cell Sci. 2003;116:1261–1268. doi: 10.1242/jcs.00301. [DOI] [PubMed] [Google Scholar]
- 13.Rossant CJ, Pinnock RD, Hughes J, Hall MD, McNulty S. Corticotropin-releasing factor type 1 and type 2 alpha receptors regulate phosphorylation of calcium/cyclic adenosine 3′,5′-monophosphate response element-binding protein and activation of p42/p44 mitogen-activated protein kinase. Endocrinology. 1999;140:1525–1536. doi: 10.1210/endo.140.4.6656. [DOI] [PubMed] [Google Scholar]
- 14.Aggelidou E, Hillhouse EW, Grammatopoulos, D.K Up-regulation of nitric oxide synthase and modulation of the guanylate cyclase activity by corticotropin-releasing hormone but not urocortin II or urocortin III in cultured human pregnant myometrial cells. Proc Natl Acad Sci USA. 2002;99:3300–3305. doi: 10.1073/pnas.052296399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen R, Lewis KA, Perrin MH, Vale, W.W Expression cloning of human corticotropin-releasing factor receptor. Proc Natl Acad Sci USA. 1993;90:8967–8971. doi: 10.1073/pnas.90.19.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grammatopoulos DK, Dai Y, Randeva HS, Levine M, Karteris E, Easton A, Hillhouse, E.W A novel spliced variant of the type 1 corticotropin-releasing hormone receptor with a deletion in the seventh transmembrane domain present in the human pregnant term myometrium and fetal membranes. Mol Endocrinol. 1999;13:2189–2202. doi: 10.1210/mend.13.12.0391. [DOI] [PubMed] [Google Scholar]
- 17.Ross PC, Kostas CM, Ramabhadran, T.V A variant of the human corticotropin-releasing factor (CRF) receptor: cloning, expression and pharmacology. Biochem Biophys Res Commun. 1994;205:1836–1842. doi: 10.1006/bbrc.1994.2884. [DOI] [PubMed] [Google Scholar]
- 18.Pisarchik A, Slominski A. Alternative splicing of CRH-R1 receptors in human and mouse skin: identification of new variants and their differential expression. FASEB J. 2001;15:2754–2756. doi: 10.1096/fj.01-0487fje. [DOI] [PubMed] [Google Scholar]
- 19.Tsai-Morris CH, Buczko E, Geng Y, Gamboa-Pinto A, Dufau, M.L The genomic structure of the rat corticotropin releasing factor receptor. J Biol Chem. 1996;271:14519–14525. doi: 10.1074/jbc.271.24.14519. [DOI] [PubMed] [Google Scholar]
- 20.Pisarchik A, Slominski A. Corticotropin releasing hormone receptor type 1: molecular cloning and investigation of alternative splicing in the hamster skin. J. Invest. Derm. 2002;118:1065–1072. doi: 10.1046/j.1523-1747.2002.01770.x. [DOI] [PubMed] [Google Scholar]
- 21.Karteris E, Grammatopoulos DK, Randeva H.S.& Hillhouse, E.W The role of corticotropin-releasing hormone receptors in placenta and fetal membranes during human pregnancy. Mol Genet Metab. 2001;72:287–296. doi: 10.1006/mgme.2001.3159. [DOI] [PubMed] [Google Scholar]
- 22.Slominski, A., Wortsman, J., Linton, E.A., Pisarchik, A. & Zbytek, A.(2003) The skin as a model for the immunodulatory effects of corticoptropin-releasing hormone. In Mind Over Matter –Regulation of Peripheral Inflammation by the CNS (Schaffer, M. & Stein, C., eds), pp. 14–176, Birkahauser Verlag, Basel, Switzerland.
- 23.Slominski A, Pisarchik A, Tobin DJ, Mazurkiewicz JE, Wortsman J. Differential expression of a cutaneous corticotropin-releasing hormone system. Endocrinology. 2004;145:941–950. doi: 10.1210/en.2003-0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 25.Wille S, Sydow S, Palchaudhuri MR, Spiess J, Dautzenberg, F.M Identification of amino acids in the N-terminal domain of corticotropin-releasing factor receptor 1 that are important determinants of high-affinity ligand binding. J Neurochem. 1999;72:388–395. doi: 10.1046/j.1471-4159.1999.0720388.x. [DOI] [PubMed] [Google Scholar]
- 26.Sheng M, Dougan ST, McFadden G, Greenberg, M.E Calcium and growth factor pathways of c-fos transcriptional activation require distinct upstream regulatory sequences. Mol Cell Biol. 1988;8:2787–2796. doi: 10.1128/mcb.8.7.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castro MG, Morrison E, Perone MJ, Brown OA, Murray CA, Ahmed I, Perkins AV, Europe-Finner G, Lowenstein PR, Linton EA. Corticotrophin-releasing hormone receptor type 1: generation and characterization of polyclonal antipeptide antibodies and their localization in pituitary cells and cortical neurones in vitro. J. Neuroendocrinol. 1996;8:521–531. doi: 10.1046/j.1365-2826.1996.04866.x. [DOI] [PubMed] [Google Scholar]
- 28.Wei ET, Thomas HA, Christian HC, Buckingham JC, Kishimoto T. d-Amino acid-substituted analogs of corticotropin-releasing hormone (CRH) and urocortin with selective agonist activity at CRH1 and CRH2β receptors. Peptides. 1998;19:1183–1190. doi: 10.1016/s0196-9781(98)00085-0. [DOI] [PubMed] [Google Scholar]
- 29.Perrin MH, Sutton S, Bain DL, Berggren WT, Vale, W.W The first extracellular domain of corticotropin releasing factor-R1 contains major binding determinants for urocortin and astressin. Endocrinology. 1998;139:566–570. doi: 10.1210/endo.139.2.5757. [DOI] [PubMed] [Google Scholar]
- 30.Fremont, V., Tong, K.P., Weintraub, B.D. & S.M. (2001) Cell surface-anchored extracellular domain of the TSH Receptor modulates expression as well as basal and TSH-dependent activation. Paper Presented at the Annual Meeting of the American Thyroid Association, November 7–10, 2001, Washington DC.
- 31.Sheng M, McFadden G, Greenberg, M.E Membrane depolarization and calcium induce c-fos transcription via phosphorylation of transcription factor CREB. Neuron. 1990;4:571–582. doi: 10.1016/0896-6273(90)90115-v. [DOI] [PubMed] [Google Scholar]
- 32.Lodish, H., Berk, A., Zipursky, S.L., Matsudaira, P., Baltimore, D.& Darnell, J.(2000 ) Molecular Cell BiologyW. H. Freeman and Company, New York.
- 33.Slominski A, Botchkarev V, Choudhry M, Fazal N, Fechner K, Furkert J, Krause E, Rolo B, Sayeed M, Wei E. Cutaneous expression of CRH and CRH-R.: is there a ‘skin stress system?’. Ann. NY Acad. Sci. 1999;885:287–311. doi: 10.1111/j.1749-6632.1999.tb08686.x. [DOI] [PubMed] [Google Scholar]
- 34.Papadopoulou N, Chen J, Randeva HS, Levine MA, Hillhouse EW, Grammatopoulos, D.K Protein kinase A-induced negative regulation of the corticotropin-releasing hormone R1α receptor-extracellularly regulated kinase signal transduction pathway: the critical role of Ser301 for signaling switch and selectivity. Mol Endocrinol. 2004;18:624–639. doi: 10.1210/me.2003-0365. [DOI] [PubMed] [Google Scholar]
- 35.Grammatopoulos DK, Randeva HS, Levine MA, Kanellopoulou KA, Hillhouse, E.W Rat cerebral cortex corticotropin-releasing hormone receptors: evidence for receptor coupling to multiple G-proteins. J Neurochem. 2001;76:509–519. doi: 10.1046/j.1471-4159.2001.00067.x. [DOI] [PubMed] [Google Scholar]
- 36.Blank T, Nijholt I, Grammatopoulos DK, Randeva HS, Hillhouse EW, Spiess, J Corticotropin-releasing factor receptors couple to multiple G-proteins to activate diverse intracellular signaling pathways in mouse hippocampus: role in neuronal excitability and associative learning. J Neurosci. 2003;23:700–707. doi: 10.1523/JNEUROSCI.23-02-00700.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]


