Abstract
We completed the mapping of a cutaneous CRH signaling system in two species with widely different determinants of skin functions, humans and mice. In human skin, the CRH receptor (CRH-R) 1 was expressed in all major cellular populations of epidermis, dermis, and subcutis with CRH-R1α being the most prevalent isoform. The CRH-R2 gene was expressed solely in hair follicle keratinocytes and papilla fibroblasts, whereas CRH-R2 antigen was localized predominantly in hair follicles, sebaceous and eccrine glands, muscle and blood vessels. In mouse skin, the CRH-R2 gene and protein were widely expressed in all cutaneous compartments and in cultured normal and malignant melanocytes. CRH-binding protein mRNA was present in dermal fibroblasts, melanoma cells, and sc fat of human skin and undetectable in mouse skin. The urocortin II gene was expressed equally in mouse and human skin. Taken together with our previous investigations, the present studies document the preferential expression of CRH-R1 in human skin, which mirrors CRH-R2 expression patterns in human and mouse skin. They are likely reflecting different functional activities of human and mouse skin. The adnexal location of CRH-R2 suggests a role for the receptor in hair growth. The differential interspecies CRH signaling expression pattern probably reflects adaptation to species-specific skin function determinants.
Abbreviations: CRH-R, CRH receptor; hCRH, human CRH; rh, recombinant human; TPA, phorbol 12-myristate 13-acetate; Ucn, urocortin
CRH IS THE MOST proximal element in the hypothalamic-pituitary-adrenal axis, an organization that co-ordinates the response to systemic stress (1–4). At the central level, CRH together with related urocortin (Ucn) I–III peptides also regulate a complex array of behavioral, autonomic, endocrine, reproductive, cardiovascular, gastro-intestinal, metabolic, and immune systemic functions (1–4). Moreover, the same CRH and related peptides can act in peripheral organs as modulators of local immune, cardiovascular, gastrointestinal, and vascular functions, and regulate cell proliferation (2–7). The general mechanism of action of these peptides in mammalian systems involves interactions with membrane-bound CRH receptors type 1 (CRH-R1) and type 2 (CRH-R2) (1–4, 8). Both receptor types belong to the group II subfamily of G protein-coupled receptors. CRH-R1 binds CRH and Ucn I with high affinity but does not bind Ucn II (strescopin-related protein) (2–4, 9, 10). CRH-R2 shows preferential activation after binding Ucn II (2, 3, 9, 10). CRH-R2 also binds CRH, however, with lower affinity than CRH-R1 (2, 3). Signal transduction through CRH-Rs is coupled to the activation of adenylate cyclase, phospholipase C, and calcium channels (1–7). Bioactivity of CRH and related peptides is also dependent upon CRH-binding proteins (CRH-BP), with their interaction resulting in either inhibitory effects (i.e. by decreasing ligand concentration available to CRH-Rs) or enhancing effects by increasing the peptide bioactivity (11, 12). The latter is probably mediated by CRH-BP, preventing CRH degradation or by facilitating the delivery of bound peptides to distant sites (11).
The gene coding for human CRH-R1 contains 14 exons and can generate seven alternatively spliced CRH-R1 transcripts (3, 4, 7, 13–15). These include CRH-R1β (containing all 14 exons), CRH-R1α (exon 6 is spliced out), CRH-R1c (exons 3 and 6 are spliced out), CRH-R1d (exons 13 is absent), CRH-R1e (exons 3, 4 and 6 are absent), CRH-R1f (exon 12 is absent), CRH-R1 g (exons 11, 27 bp of exon 10 and 28 bp of exon 12 are absent), and CRH-R1 h (insertion of cryptic exon between exons 4 and 5). The CRH-R2 gene contains 15 exons, as predicted from the analysis of human genomic DNA and generates at least three major isoforms (CRH-R2α, β, and γ) (3, 7, 16–19). CRH-R2α is expressed predominantly in the periphery in humans, whereas in rodents CRH-R2β is the major variant found in peripheral tissues (3). CRH-R2γ detection has been restricted to the brain (2, 3, 18). Additional alternatively spliced forms of CRH-R2, as well as multiple promoters for this gene, have also been described (16).
CRH, Ucn, and functional CRH-R1 are all produced in human and rodent skin supporting the local functional significance of this system (6, 7, 20–25). Within this context, the diversity of skin cell sub-populations and the known pleio-tropic effects of CRH and Ucn, may require selectivity of signal transduction pathway systems; for example, via differential expression of CRH-R1 isoforms (7, 15, 26). Elucidation of CRH/Ucn’s role in skin physiology and pathology would therefore require the full definition of the expression pattern of CRH-R2 and CRH-BP in human and rodent skin. Thus, in the present work, we tested various sub-populations of skin cells of human and mouse origin for the expression of CRH-R2 and CRH-BP genes and also examined the expression of CRH-R2 antigens in situ. These studies were complemented by the mapping of CRH-R1 protein expression in human scalp, and with testing of the expression of gene coding for the recently described Ucn II and III peptides.
Materials and Methods
Tissues
Human tissue samples included skin, pituitary, adrenal gland, gestational tissues, and skeletal muscle. Pituitaries were obtained from the National Hormone and Pituitary Program, NIDDK, and placenta and fetal membranes were collected after normal delivery. Skeletal muscle was obtained after autopsy, whereas skin and adrenal gland were removed during surgery. Total RNA from human brain and sc adipose tissue was obtained commercially (Ambion, Austin, TX). The tissues were stored at −80 C until the time of molecular analyses. Immunohistochemistry protocols are as described below. The University of Tennessee Health Science Center (UTHSC) Committee on Research Involving Human Subjects approved the use of human tissues.
Murine tissue samples consisted of brain, pituitary, spleen, and of skin isolated at telogen and anagen IV, V, and VI stages of hair cycle (27). Mice, C57BL/6 strain female, 8 weeks old were purchased from Taconic Farms (Taconic, NY) and housed in community cages at the animal facilities of the Albany Medical College (Albany, NY). The animals were killed under pentobarbital anesthesia and selected organs, including back skin, were collected following protocols routinely used in our laboratory (27). Tissue specimens were frozen rapidly in liquid nitrogen and stored at –80 C until further analysis or processed for immunohistochemistry analyses as described below. The Institutional Animal Care and Use Committee at Albany Medical College approved the original experimental protocol, and a similar protocol for mice was approved at UTHSC.
Cell culture
The normal, immortalized and malignant human and rodent cells or cell lines used in this study are listed in Table 1. Proliferating normal epidermal human keratinocytes, melanocytes, and dermal fibroblasts were purchased from Cascade Biologicals (Portland, OR). The keratinocytes were propagated in Epilife medium supplemented with 0.06 mm calcium chloride and EpiLife Defined Growth Supplement containing purified BSA, purified 5 μg/ml bovine transferrin, 0.18 μg/ml hydrocortisone, recombinant human (rh) IGF-I, prostaglandin E2, 0.2 ng/ml rh epidermal growth factor and antibiotics (Cascade Biologicals). Normal adult epidermal melanocytes and immortalized human epidermal melanocytes line PIG-1 (gift of Dr. Catherine LePoole, Loyola Medical Center, Maywood, IL) were cultured in Medium 154 (Cascade Biologicals) supplemented with 0.2% vol/vol bovine pituitary extract, 0.5% vol/vol fetal bovine serum, 5 μg/ml bovine insulin, 5 μg/ml bovine transferrin, 3 ng/ml basic fibroblast growth factor, 0.18 μg/ml hydrocortisone, 3 μg/ml heparin, 10 ng/ml phorbol 12-myristate 13-acetate. Dermal adult fibroblasts were purchased commercially (Cascade Biologics, Inc.) and cultured in Cascade 106 medium plus Cascade Low Serum Growth Supplement (containing fetal bovine serum, 2% vol/vol; hydrocortisone, 1 μg/ml; human epidermal growth factor, 10 ng/ml; basic fibroblast growth factor, 3 ng/ml; and heparin, 10 μg/ml) and 1% penicillin/streptomycin/amphotericin B solution (Sigma, St. Louis, MO).
TABLE 1.
Characteristics of cell lines tested
| Origin | Cell type | Citation/source |
|---|---|---|
| HUMAN | ||
| a) Primary cultures | ||
| Foreskin | Neonatal keratinocytes | (21) |
| Corporal skin | Adult epidermal keratinocytes | (28) or (Cascade) |
| Corporal skin | Adult epidermal melanocytes | (28) or (Cascade) |
| Corporal skin | Adult dermal fibroblasts | (28) or (Cascade) |
| Scalp skin | Hair follicle keratinocytes | (29) |
| Scalp skin | Hair follicle melanocytes | (29) |
| Scalp skin | Hair follicle papilla fibroblasts | (29) |
| b) Cell lines | ||
| Epidermis | Immortalized (PIG-1) melanocytes | (48) |
| Epidermis | Immortalized (HaCaT) keratinocytes | (49) |
| Cervix | Squamous cell carcinoma (C4-1) | (30) |
| RGP melanoma | WM 35 melanoma (amelanotic line) | (32) |
| VGP melanoma | WM 98 melanoma (amelanotic line) | (32) |
| Metastatic melanoma | WM 164 melanoma (amelanotic line) | (32) |
| Metastatic melanoma | WM 852 melanoma (amelanotic line) | (32) |
| Melanoma | SKMEL-188 melanoma (hypomelanotic line) | (32) |
| HAMSTER | ||
| Skin | Bomirski MI transplantable hypomelanotic melanoma | (35) |
| Transplantable melanoma | AbC-1 amelanotic melanoma line | (34) |
| MOUSE | ||
| DBA2J mouse | Cloudman S91 melanoma | (50) |
| C57BL/6 mouse | Melan-a immortalized follicular melanocytes | (51) |
RGP, Radial growth phase; VGP, vertical growth phase.
In addition, normal adult epidermal keratinocytes, epidermal melanocytes and dermal fibroblasts were isolated from skin explants and cultured as described previously (28). Primary cell cultures of neonatal keratinocytes were established from foreskin as previously described (21). The cells were propagated in low-calcium (0.15 mm) serum-free keratinocyte growth medium containing bovine pituitary extract and antibiotics (Clonetics Corp., San Diego, CA). Normal human hair follicle-derived cells included hair follicle keratinocytes, hair follicle melanocytes and hair follicle papilla fibroblasts. Cells were isolated from normal human adult scalp and cultured as previously described (29).
Human cell lines were cultured according to standard protocols and media were changed every second day (15, 30, 31). CO2 was routinely used at a concentration of 5% with the exception of normal mouse melanocytes that were grown in 10% CO2. Human immortalized keratinocytes (HaCaT) and squamous cell carcinoma (C4 –1), were propagated in DMEM (GIBCO, Gaithersburg, MD), whereas human SKMEL188 melanoma cells were grown in Ham’s F10 medium supplemented with 10% fetal bovine serum and antibiotics (GIBCO) (30, 31). Other human melanoma lines included those established from radial growth phase (WM 35 and SBCE2), vertical growth phase (WM 98 and WM 1341D) and metastatic (WM 164) melanomas (gift of Dr. M. Herlyn, Wistar Institute, Philadelphia, PA); they were cultured in DMEM supplemented with 10% fetal bovine serum, insulin (5 μg/ml) and antibiotics (32).
Immortalized normal mouse skin melanocyte line melan-a from C57BL/6J strain (gift of Dr. D. Bennett, Saint George Hospital, London, UK) was cultured in RPMI 1640 media supplemented with 10% of bovine serum and 200 nm TPA (phorbol 12-myristate 13-acetate) in the presence of 10% CO2. Mouse Cloudman S91 and hamster AbC-1 melanoma cells were grown in Ham’s F10 medium supplemented with 10% fetal bovine serum and antibiotics (GIBCO) (33, 34) Bomirski hamster MI melanoma was serially transplanted in Syrian hamsters as described previously (35).
After washing with PBS melanoma cells were detached with Tyrode’s solution containing 1 mm EDTA (30, 34), whereas keratinocytes, squamous cell carcinoma cells, fibroblasts, and normal melanocytes were trypsinized (15, 30). The cells were centrifuged and suspended in RNA isolation solution (Trizol reagent). Total RNA was extracted using Trizol isolation kit (Life Technologies, Inc.-BRL, Gaithersburg, MD).
cDNA preparation
Poly (A+) mRNA was isolated using an Oligotex mRNA Mini Kit (QIAGEN, Valencia, CA). The synthesis of first-strand cDNA was performed using the Superscript pre-amplification system (Invitrogen, Carlsbad, CA). An aliquot of 5 μg of total RNA or 0.1 μg mRNA per reaction was reverse transcribed using oligo(deoxythymidine) as the primer. The expression of genes having only one coding exon was evaluated using cDNA synthesized from deoxyribonuclease-treated poly(A) mRNA. Lack of DNA contamination was confirmed by negative amplification of RNA without prior RT.
PCRs
All samples were standardized by the amplification of housekeeping gene glyceraldehyde-3-phosphate dehydrogenase as described previously (15). Primers were synthesized by Integrated DNA Technology Inc. (Coralville, IA). Nested PCR was used to detect different CRH-R1 isoforms in scalp skin cells. The amplification conditions were as previously described (15).
Detection of human CRH-R2 was obtained by nested PCR amplification of corresponding cDNAs. First, we tested expression of CRH-R2 by amplifying the fragment common to CRH-R2α, β, and γ; and second, we tested for expression of CRH-R2α and β in selected cells lines. Sequences of the primers used for PCR amplifications are listed in Table 2, whereas their exonal allocation together with gene structure is presented in Fig. 1.
TABLE 2.
Nucleotide sequence of primers used for RT-PCR amplification
| Gene | Primer | Primer location | PCR product size (bp) |
|---|---|---|---|
| Human genes | |||
| CRH-R2 (common fragment) | First pair of primers | ||
| P152: GCCATCGGCAAGCTCTACTATG (sense) | Exon 10 | ||
| P153: TCTCCATTGAAGAAGCAGTAGAAG (antisense) | Exon 15 | ||
| Nested primers | |||
| P154: CCCCATCATTCTCGTGCTCCTG (sense) | Exon 10 | ||
| P155: TGACGAAGAAGAGCATGTAGGTG (antisense) | Exon 15 | 176 | |
| CRH-R2α | First pair of primers | ||
| P333: ATGGACGCGGCACTGCTCCA (sense) | Exon 4 | ||
| P339: GGTCATACTTCCTCTGCTTGTC (antisense) | Exon 7 | ||
| Nested primers | |||
| P334: AGAGCTGCTCTTGGACGGCT (sense) | Exon 4 | ||
| P340: GTCATCCAAAATGGGCTCACAC (antisense) | Exon 7 | 253 | |
| CRH-R2β | First pair of primers | ||
| P335: ACCATCATGACCCTCACCAAC (sense) | Exon 2 | ||
| P339: GGTCATACTTCCTCTGCTTGTC (antisense) | Exon 7 | ||
| Nested primers | |||
| P336: CATGACCCTCACCAACCTCTC (sense) | Exon 2 | ||
| P340: GTCATCCAAAATGGGCTCACAC (antisense) | Exon 7 | 232 | |
| CRHBP | P354: CCTGGGACACGTAAATGGTC (sense) | Exon 3 | |
| P355: GCGCACCACAGTGTTGTCAC (antisense) | Exon 4 | 187 | |
| URC II | P573: TGTGCTCTGCTGTTGCTGATG (sense) | Exon 1 | |
| P574: GCGATAGGACAATGCGCGAG (antisense) | Exon 1 | 217 | |
| Mouse genes | |||
| CRH-R2 | P236: TGCCTGAGGAATGTGATCCACTG (sense) | Exon 4 | |
| P237: CGTGGTGGATGCTCGTAACTTCG (antisense) | Exon 6 | 462 | |
| CRH-BP | P579: TCTTACCAGAAGGAGCATCAG (sense) | Exon 5 | |
| P580: GTGTCCGAGGGTAAGATCAG (antisense) | Exon 6 | 241 | |
| URC II | P571: GTGGTGTTCGTGGTCCTGATG (sense) | Exon 1 | |
| P572: GGAACATCCAGGGAGAGTATG (antisense) | Exon 1 | 217 | |
URC, Urocortin.
Fig. 1.
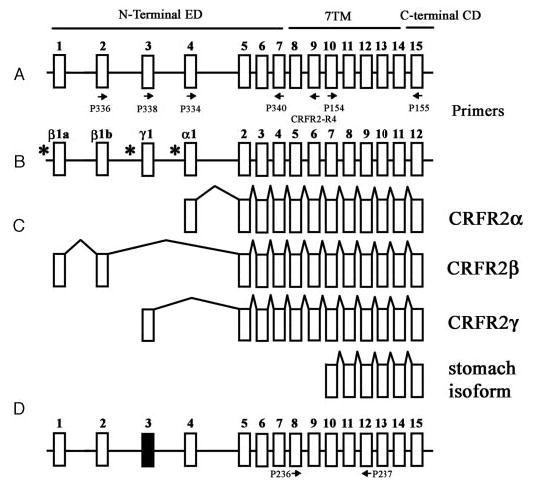
Schematic representation of mammalian CRH-R2 gene structure. A, Human CRH-R2 according to Slominski et al., 2000 (7). B, Human CRH-R2 according to Catalona et al., 2003 (16). C, Alternatively spliced CRH-R2 isoforms α, β, γ (7, 16), and the isoform from stomach (GenBank accession No. E12750; patent no. JP199707289-A). Arrows represent position of PCR primers. Putative promoters sites are shown by * (16). D, Predicted structure of mouse CRH-R2 gene. The structure of mouse gene has been obtained by comparing mouse genomic DNA with published mouse CRH-R2 cDNA and by comparing mouse genomic DNA with human CRHR2 exons (Blast program at http://www.ncbi.nlm.nih.gov/blast). Black box represents mouse exon 3 corresponding to human exon 3 (65% homology). Murine exon 3 contains multiple stop codons and its insertion into the final CRH-R2 mRNA would not produce a functional CRH-R2 receptor. Thus, the equivalent of the human CRH-R2γ isoform will not be expressed in the mouse. Exon 4 has 77% homology at the nucleotide level and 91.3% at the protein level with its human counterpart. Arrows represent position of PCR primers.
Mouse CRH-R2, CRH-BP, Ucn II (strescopin-related protein) and human CRHBP, Ucn II (strescopin-related protein) were amplified by one round of PCR (30 cycles). Primer sequences for these genes are listed in Table 2 and conditions of amplification are described below. Hamster CRH-R2 was amplified by nested PCR using primers designed for mouse gene. Primers P210 from exon 4 (P210: GAGACCGTGC-CCCGAGTA) and P211 from exon 6 (CGTGGTGGATGCTCGTAACT-TCG) were used in the first round of amplification and primers P236 and P237 (listed in the Table 2) in the second round.
The reaction mixture (25 μl) contained 2.5 mm MgCl2, 2.5 mm of each deoxynucleotide triphosphate, 0.4 μm of each primer, 75 mm Tris-HCl (pH 8.8), 20 mm (NH4)2SO4, 0.01% Tween 20, and 0.25 U of Taq polymerase (Promega). The mixture was heated to 94 C for 2.5 min and then amplified for 30 cycles as specified: 94 C for 30 sec (denaturation), 55 C for 30 sec (annealing) and 72 C for 40 sec (extension). Amplification products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining according to standard protocols used in our laboratory (15).
Sequencing
The identified PCR products were excised from the agarose gel and purified by GFX PCR DNA and gel band purification kit (Amersham-Pharmacia-Biotech, Piscataway, NJ). PCR products were sequenced from both ends. Sequencing was performed in the Molecular Resource Center at the University of Tennessee HSC (Memphis, TN) using the Applied Biosystems 3100 Genetic Analyzer and BigDye Terminator Kit.
Immunohistochemistry
1. CRH-R1.
The rabbit polyclonal antibody against the CRH-R1 (named JR2A) was obtained from Dr. E. Linton, Oxford University (Oxford, UK). The characteristics and specificity of the antibody have been fully described (36). In brief, the antibody was raised against the peptide sequence 335–346 of the human CRH-R1 receptor. This sequence, present in the extracellular loop of the seven-transmembrane domains protein, is unique to the CRH-R1 receptor and does not occur within any of the membrane spanning regions (36). The resulting anti-CRH-R1 (JR2A) antibody does not cross-react in RIAs with ACTH, Metenkephalin, angiotensin I, αMSH, arginine vasopressin, hproCRH (125–151), human CRH (hCRH) (1–41), ovine CRH (1–41), hCRH (36 –41), hCRH (1–20), bovine thyroglobulin, human serum albumin, or urotensin I, sauvagine. Moreover, the specificity of our immunohistochemistry tests was confirmed by substituting the preimmune serum for the same JR2A antiserum as negative control for the immunostaining.
Deparaffinized sections of human scalp skin were incubated in blocking buffer (PBS/5% BSA/0.2% Fish Skin Gelatin) for 30 min at RT. Sections were incubated with rabbit anti-CRH-R1 antibody (JR2A) or preimmune sera overnight at 4 C. Sections were washed three times in PBS, and incubated with biotinylated goat antirabbit IgG (1:250 dilution in PBS; Vector Laboratories, Burlingame, CA) for 1 h at RT. After three washes in PBS, sections were incubated with ABC solution for 30 min at RT. Following a wash in PBS, peroxidase activity was detected with Vector VIP peroxidase substrate kit, following the manufacturer’s instructions. Sections were washed in H2O, dehydrated, and then mounted with Permount (Fisher Scientific, Fair Lawn, NJ).
2. CRH-R2.
Immunocytochemistry was performed with a commercially available antibody against CRH-R2 (CRF-RII [N-20]; Santa Cruz Bio-technology, Inc., Santa Cruz, CA). As reported by the manufacturer (Santa Cruz Biotechnology data sheet) CRF-RII (N-20) is an affinity purified goat polyclonal antibody raised against a peptide mapping at the amino terminus of corticotropin-releasing factor (hormone) receptor 2 (CRH-R2) of mouse origin. CRF-RII (N-20) reacts with CRH-R2 of mouse, rat and, to a lesser extent, human origin as determined by Western blotting and immunohistochemistry. The reactivity with the human CRH-R2 protein is due to the 91% homology between mouse and human sequences in the 12 amino acid epitope used to raise this antibody. This antibody does not cross-react with CRH-R1, being specific for CRH-R2, as per the manufacturer’s data sheet.
Frozen sections and formalin-fixed, paraffin-embedded biopsies of normal human scalp and mouse pelage skin were studied by immunohistohemistry. Normal human scalp skin was obtained after elective plastic surgery with informed consent from a 35-yr-old male and embedded in OCT medium (RA Lamb, East Sussex, UK). Murine anagen skin was obtained from the dorsum of normal C57Bl/6 mice. Seven-micrometer cryosections of murine and human skin were air-dried, fixed in ice-cold acetone, hydrated in PBS, and blocked in 10% donkey serum. The sections were incubated with an affinity-purified goat antimouse CRF-RII polyclonal antibody (N-20; Santa Cruz Biotechnology, Inc.) diluted 1:10 for 1 h. After removal of the primary antibody the sections were washed in PBS and incubated with horseradish peroxidase-labeled donkey antigoat IgG secondary antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). After removal of the secondary antibody, the sections were washed in PBS and developed with the LSAB2 HRP/AEC kit (Dako Ltd., Cambridgeshire, UK). Negative controls included incubation of sections with primary antibody depleted with blocking peptide (sc-1826P; Santa Cruz) and also by replacing the primary antibody with PBS.
In the latter protocol, deparaffinized sections of mouse skin were incubated with goat anti-CRF-R2 or primary antibody preadsorbed with blocking peptide, followed by washes in PBS and incubation with biotinylated donkey antigoat IgG (Santa Cruz Biotechnology, Inc.). Following washes in PBS and incubation in ABC solution, peroxidase activity was detected with substrate solution containing 0.05% 3,3′-diaminobenzidine-4HCl in PBS (pH 7.6), containing 0.01% H2O2. Sections were washed in H2O, dehydrated, and mounted with Permount. Sections were examined on an Olympus BX 50 light microscope (Olympus America, Inc., Melville, NY) and images captured with an Optronics MagnaFire-SP digital camera (Optronics, Goleta, CA).
Results and Discussion
1. CRH-R1 is predominantly expressed in human skin, whereas CRH-R2 expression is restricted to adnexal cells
CRH-R1.
As previously documented, expression of CRH-R1 or, of alternatively spliced products of the CRH-R1 gene was found to be widespread throughout the skin, both human and mouse skin, and also in cultured normal and malignant epidermal melanocytes and keratinocytes. CRH-R1 α was the prevalent isoform throughout (Table 3) (7, 15). Similarly, the current investigation, using skin cells derived from the human scalp, also found that the predominant isoform expressed was CRH-R1α; CRH-R1α was the sole isoform expressed by adult epidermal and follicular keratinocytes and melanocytes and by hair follicle dermal papilla fibroblasts (Table 3 and Fig. 2).
TABLE 3.
Expression of CRH-R genes in human skin cells
| Human skin cell type | CRH-R2α | CRH-R2β | CRH-R2 (common fragment) | CRH-R1 (isoform) |
|---|---|---|---|---|
| Neonatal epidermal keratinocytes | − | − | − | α, d, f, g3 |
| Adult epidermal keratinocytes | − | − | − | α |
| Adult hair follicle keratinocytes | + | − | + | α |
| Immortalized HaCaT keratinocytes | − | − | − | α,1,3 |
| Squamous cell carcinoma C1–4 | − | − | − | α, f 3 |
| C1–4 cells after UV treatment | ND | ND | + | α, c, g3 |
| Epidermal melanocytes | − | ND | − | α3 |
| Hair follicle melanocytes | − | − | − | α |
| Immortalized PIG-1 melanocytes | − | − | − | α, c, e |
| Melanoma SKMEL188 | − | − | − | d2,3 |
| Melanoma SBCE2 | − | ND | − | α, d, e, f, g3 |
| Melanoma WM35 | − | ND | − | α, d, e, f, g3 |
| Melanoma WM98 | − | ND | − | α, e, g3 |
| Melanoma WM164 | − | ND | − | α, e, g3 |
| Melanoma WM1341D | − | − | − | α, d, f, g3 |
| Dermal fibroblasts | − | − | − | α, e, f |
| Hair follicle papilla fibroblasts | + | − | + | α |
| sc Fat | − | − | − | α, e, f |
Fig. 2.
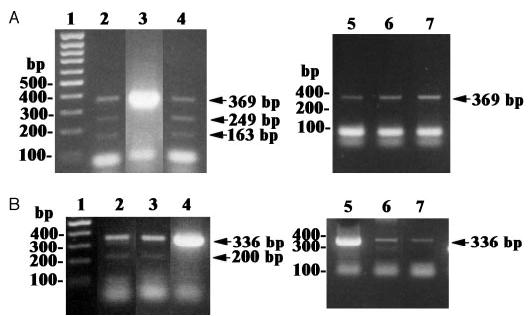
RT-PCR amplification of isoforms of human CRH-R1 mRNA. A, Amplification of the fragment spanning exons 2 through 7. B, Amplification of the fragment spanning exons 9 through 14. DNA ladder, 1; sc adipose tissue, 2; dermal fibroblasts, 3; immortalized epidermal melanocytes (Pig 1), 4; hair follicle dermal papilla fibroblasts, 5; hair follicle keratinocytes, 6; hair follicle melanocytes, 7. Arrows, Amplified CRH-R1 transcripts.
Dermal fibroblasts and sc fat cells additionally expressed the e and f isoforms of CRH-R1, whereas an immortalized line of human melanocytes derived from a vitiligo patient also expressed the c and e isoforms (Fig. 2 and Table 3). Previously, we have shown that squamous cell carcinoma and melanomas coexpressed other isoforms (15). Thus, the CRH-R1 gene was consistently expressed in the main cellular compartments of the skin and in sc fat. Of the multiple CRH-R1 isoforms, CRH-R1α was the most prevalent or the solely expressed isoform, leading us to propose that CRH-R1 α is the main regulator or coordinator of CRH-dependent skin functions in the adult organism; other CRH-R1 isoforms may play secondary roles. This expanded pattern of CRH-R1 isoforms in malignant or abnormal cells may then reflect cellular heterogeneity in cell lines that could play a role in stabilizing tumoral phenotype and inducing resistance to external manipulation. Alternately, the same pattern of isoform heterogeneity seen in normal dermal fibroblasts or fat cells (both of mesenchymal origin) could signal a potential for functional transdifferentiation that has already been lost or suppressed in the more highly specialized hair follicle papilla fibroblasts, or reflect a certain level of cellular immaturity (neonatal foreskin keratinocytes) or indeed defective function (immortalized melanocytes from the vitiligo patient).
Immunohistochemical analysis of adult human scalp biopsies confirmed a functional expression of the CRH-R1 gene by demonstrating the CRH-R1 antigen in almost all skin compartments (Fig. 3) and is similar to the pattern reported by Kono, 2001 (20). In addition to our detection of CRH-R1 in the epidermis, hair follicle (Fig. 3, A–D) and sebaceous glands, we also detected the CRH-R1 antigen in eccrine glands and their ducts, smooth muscle of the media of blood vessels, endothelial cells and in erector pili muscle cells (Fig. 3). This finding is in agreement with the reported detection of CRH-R1 in smooth muscle of the myometrium (37), the vasodilatory action of CRH on isolated blood vessels, the presence of specific CRH binding sites on the endothelium and expression of functional CRH-R1 in sebaceous gland and cultured sebocytes (38 –41). Taken together, the data on ubiquitous expression of CRH-R1 is strongly suggestive of an important role in skin physiology and pathology, most likely through its regulation of local pigmentary, epidermal, ad-nexal, secretory, vascular, and dermal activity. Moreover, the molecular data emphasize that this regulation would be mediated predominantly by CRH-R1α isoform.
Fig. 3.
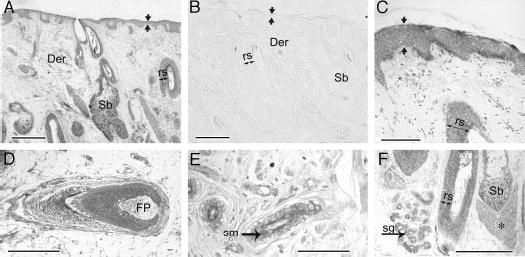
Expression of CRH-R1 in human scalp A and C–F (formalin-fixed, paraffin-embedded sections) reacted with monospecific rabbit antiserum to CRH-R1; B is the preimmune control. Panels A and B are survey images to show large region of scalp, and panels C–F are higher magnification images to demonstrate expression in subcompartments. Der, Dermis; Sb, sebaceous unit; rs over the double-ended arrow, outer root sheath; FP, follicular papilla; sm, smooth muscle; sg, sweat gland; asterisk, erector pili muscle; apposing arrows, epidermis. Bar in panels A and B, 500 μm; bar in C, 100 μm; bar in D–F, 250 μm.
CRH-R2.
We tested the expression of the CRH-R2 gene in human skin performing extensive RT-PCR analyses in a large panel of samples. Control and test samples were standardized by amplification of the common housekeeping gene glyceraldehyde-3-phosphate dehydrogenase. Positive controls run for each set of experiments were represented by cDNA obtained from CRH-R2 expressing tissues such as brain, placenta and other gestational tissues or pituitary. Because the human CRH-R2 gene is composed of 15 exons, contains multiple promoter sites and generates several alternatively spliced isoforms, we performed extensive analyses using primers at different locations that included the N-and C-terminal ends (Fig. 1). Of note, the C terminus is less prone to alternative splicing (16) and locating the primers at exons 10 and 15 would cover all the insofar-described CRH-R2 isoforms. Therefore, negative results using the latter primers should indicate a true lack of CRH-R2 gene expression. As expected, RT-PCR products, corresponding to either CRH-R2 fragments common to α, β, or γ isoforms or to fragments corresponding specifically to CRH-R2 α or β, were readily detected in positive control tissue (Fig. 4A). However, none of those products was detected in epidermal normal and malignant keratinocytes, normal and malignant melanocytes, dermal fibroblasts, sc adipose tissue, or biopsies of human corporal skin (Fig. 4A and Table 3). By contrast, CRH-R2 α was readily detectable in cells derived from human scalp adnexal structures (hair follicle keratinocytes and hair follicle papilla fibroblasts) (Fig. 4B and Table 3). The amplified fragment showed 100% sequence homology with database for human CRH-R2 α cDNA. Thus, at the molecular level CRH-R2 that was absent in the main cell populations of the epidermis (melanocytes and keratinocytes), was nevertheless detected in epithelial (follicular keratinocytes) and mesenchymal (hair follicle papilla fibroblasts) cells of the adnexal structures, but absent in dermal fibroblasts and sc fat. In agreement with previous studies in human peripheral tissues, (3) the prevailing isoform detected in adnexal structures was CRH-R2α with absence of expression of the α isoform (Fig. 4C).
Fig. 4.
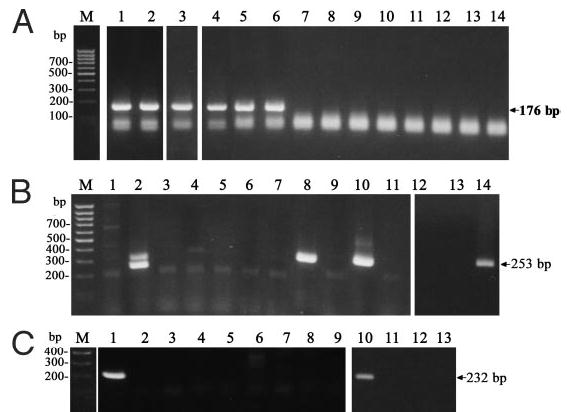
Expression of CRH-R2 in human tissues. A, Amplification of common CRH-R2 fragment: DNA ladder, M; placenta, 1; fetal membrane, 2; brain, 3; adrenal gland, 4; hair follicle papilla fibroblasts, 5; follicular keratinocytes, 6; epidermal keratinocytes, 7; hair follicle melanocytes, 8; dermal fibroblasts, 9; immortalized (HaCaT) keratinocytes, 10; C1–4, 11; skin, 12; melanoma lines SKMEL-188, 13; and WM35, 14. B, Amplification of CRH-R2α isoform: DNA ladder, M; neonatal keratinocytes, 1; adrenal gland, 2; immortalized (HaCaT) keratinocytes, 3; SKMEL-188 melanoma, 4; C1–4 squamous cell carcinoma cells, 5; immortalized (Pig 1) epidermal melanocytes, 6; WM1341 melanoma, 7; follicular keratinocytes, 8; adult epidermal keratinocytes, 9; brain, 10; skin biopsy, 11; dermal fibroblasts, 12; hair follicle melanocytes, 13; hair follicle papilla fibroblasts, 14. C, Amplification of CRH-R2α isoform: DNA ladder, M; fetal membrane, 1; epidermal keratinocytes, 2; follicular keratinocytes, 3; melanoma WM1341, 4; immortalized (Pig 1) epidermal melanocytes, 5; C4 –1 squamous cell carcinoma cells, 6; SKMEL-188 melanoma, 7; immortalized (HaCaT) keratinocytes, 8; neonatal keratinocytes, 9; muscle, 10; follicular melanocytes, 11; hair follicle papilla fibroblasts, 12; skin biopsy, 13.
Immunohistochemistry on frozen sections of skin scalp confirmed expression of the CRH-R2 antigen, concentrated to the outer layer of the outer root sheath of anagen hair follicles (Fig. 5A), being associated to the cell membrane proximally (Fig. 5A, lower inset), and to the cytoplasm distally (Fig. 5A, upper inset). Moderate expression was found also in the hair bulb matrix, inner root sheath and pre-cortex, the mesenchymal follicular papilla and along the glassy membrane separating the outer root sheath epithelium from the connective tissue sheath. During the apoptosis-driven regression phase of the hair follicle cycle, increased CRH-R2 expression was detected in the regressing hair bulb, follicular papilla, and the papilla-associated collapsing vascular capillary loop (Fig. 5B). Of note, in formalin-fixed, paraffin-embedded sections CRH-R2 was below detectability in any of the compartments listed above, except for sebaceous glands where sebocytes showed CRH-R2 immunoreactivity (not shown). Using frozen tissues, CRH-R2 immunostaining was absent in epidermal keratinocytes; only scattered cells of the basal layer with distribution similar to that of melanocytes (Fig. 5C) and dendritic cells of the mid epidermis that exhibited Langerhans cell-type distribution (Fig. 5D) showed positive CRH-R2 immunoreactivity. In adnexal structures, CRH-R2 antigen was detected in scattered cells in the basal or suprabasal layers of the sebaceous gland (Fig. 5E), and in epithelial cells of the eccrine sweat gland duct (Fig. 5F). Other skin components expressing the CRH-R2 antigen included muscle cells (Fig. 5G) and blood vessel endothelium. No CRH-R2 immunoreactivity was observed when using the antibody-containing solution that had been treated with CRH-R2 peptide (Fig. 5H). Thus, the in situ analysis was consistent with the molecular data with the exception of detection by immunocytochemistry of scattered CRH-R2 antigen positive cells at histological locations specific for epidermal melanocytes. The latter discrepancy may be explained by the biological properties of the cells in situ and in culture. Epidermal melanocytes do not proliferate in situ (proliferation is seen only in atypical pigmented lesions), whereas this is a primary property of cultured cells, and this may be the reason for a possible change in gene expression pattern. Another potential explanation is a false immunopositive signal, e.g. due to cross-reactivity with melanocyte-specific antigens. These questions may only be addressed using laser dissection methodology applied by others to analyze methodically CRH/CRH-R axis in the skin in situ (20). Detection of CRH-R2 in other cutaneous cells such as sebaceous glands, smooth muscle, blood vessels, and Langerhans cells is in agreement with reports showing the presence of CRH-R2 in vascular endothelium or smooth muscle of peripheral organs (5) in cultured sebocytes (41) or in immune cells (5) of which Langerhans cells are an example.
Fig. 5.
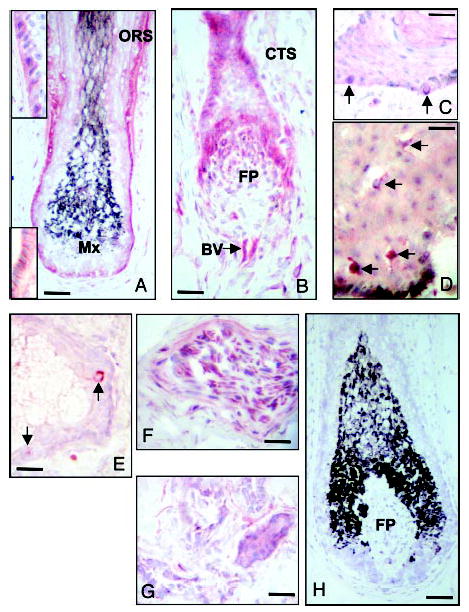
Expression of CRH R2 in adult human scalp. Anagen VI hair follicle (A); catagen hair follicle (B); epidermis (C and D); sebaceous gland (E); muscle (F); eccrine gland (G); negative control in which the antiserum was absorbed with the CRH R2 peptide (H). Outer root sheath (ORS), matrix (MX), follicular papilla fibroblast population (FP), proximal collapsing capillary loop (BV). Arrows show singly scattered basal cells in the epidermis (C) dendritic cells of the mid-epidermis (D), and sebaceous gland epithelium (E). Scale bars: A, 45 μm; B, 40 μm; C, 30 μm; D, 30 μm; E, 30 μm; F, 35 μm; G, 30 μm; and H, 45 μm.
Thus, both molecular and in situ studies document that the predominant form of the CRH receptor expressed in human skin is the CRH-R1. This is present in all the main cellular compartments of epidermis, dermis, and in sc fat. CRH-R2 expression was fully documented only in cells of adnexal structures, smooth muscle, blood vessels, and selected cells of immune origin. Of great interest is the relation between these findings and our proposed cutaneous neuroendocrine system that would coordinate responses to external stress in analogy to the central response where the CRH-R1 plays a major role (42, 43). Within this context, the differential expression of CRH-Rs in human skin correlates with data suggesting that CRH-R1 is the main coordinator of the stress response at the central level and CRH-R2 plays only a modulatory role (42–44). Therefore, we suggest that the central position of CRH-R1 in coordinating the systemic stress response may be analogous to its role in the human skin.
2. CRH-R2 is widely expressed in mouse skin
In contrast to its scarcity in human skin, mouse skin showed a wide expression of the CRH-R2 gene detected during telogen and different phases of anagen, and also in normal and malignant melanocytes (Fig. 6 and Table 4). The CRH-R2 message was indeed detected by both direct PCR (30 cycles; for primers location see Fig. 1) and by nested PCR (see Table 2) with the amplified fragment showing 100% sequence homology with the database for mouse CRH-R2 cDNA. We also tested CRH-R2 expression in skin cells derived from another rodent species, e.g. hamster melanomas (tumors of cutaneous origin). Nested PCR amplification using mouse primers amplified a cDNA corresponding to a common fragment of CRH-R2 showed expression in hamster pituitary and eye, and in transplantable Bomirski hypomelanotic MI melanoma line, with 94% homology with the published mouse sequence (GenBank accession no. AY300753) (Fig. 6B). In addition, we detected an aberrant CRH-R2 isoform of 564 bp (GenBank accession no. AY300754) in amelanotic hamster AbC-1 melanoma line. The 564-bp cDNA contains an insertion of a new fragment of 102 bp into the 462-bp cDNA fragment. Because the sequence of the insertion fragment is without a stop codon or frame-shift, it would code for the functional CRH-R2 with an additional peptide segment of 94 amino acids.
Fig. 6.
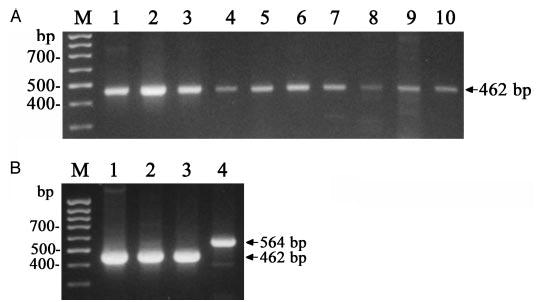
Expression of CRH-R2 gene in rodent skin cells. A, Expression of CRH-R2 in the C57BL/6 mouse (arrow): molecular markers, M; brain, 1; skin at anagen IV (2), V (3), mid anagen VI (4), late anagen (VI) (5) and telogen (6 –7); immortalized melanocytes melan-a, 8; Cloudman S91 melanoma, 9; spleen, 10. B, Expression of CRH-R2 in hamster tissues (arrows): molecular markers, M; eye, 1; pituitary, 2; transplantable MI hypomelanotic melanoma, 3; amelanotic AbC1 melanoma, 4.
TABLE 4.
Relative expression of CRH-R2 mRNA in mouse skin samples in relation to brain, pituitary and spleen
| Mouse tissue | CRH-R2 |
|---|---|
| Brain | +++ |
| Pituitary | +++ |
| Spleen | +++ |
| Telogen skin | +++ |
| Anagen IV skin | ++ |
| Anagen V skin | ++ |
| Early anagen VI skin | + |
| Mid anagen VI skin | + |
| Late anagen VI skin | + |
| Immortalized melanocytes | + |
| Melanoma (Cloudman S91) | + |
Immunohistochemistry studies were performed on either formalin-fixed, paraffin-embedded sections or on frozen sections; both techniques demonstrated that CRH-R2 was present in all compartments of mouse skin (Figs. 7 and 8). Thus, CRH-R2 antigen was expressed in epidermal and follicular (outer and inner root sheath as well as matrix kera-tinocytes), compartments, follicular papilla fibroblasts, dermal fibroblasts, blood vessels and sebaceous glands and eccrine glands. CRH-R2 antigen expression was most prominent in the follicular papilla and peripheral hair follicle matrix epithelium and outer root sheath keratinocytes (Fig. 8A and inset). Within murine epidermis, moderate CRH-R2 immunoreactivity was seen in keratinocytes, although concentrated staining occurred in scattered cells throughout the basal layer (Fig. 8C). The same receptor was also observed in a subpopulation of mesenchymal dermal cells. No immu-noreactivity was observed in the negative controls where the antibody had been depleted with CRH-RII peptides (Figs. 7B and 8D).
Fig. 7.
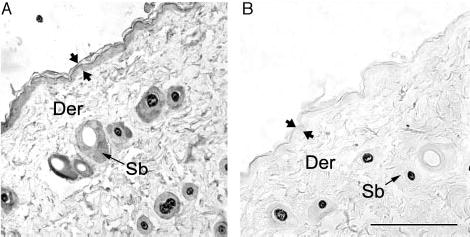
Expression of CRH R2 in adult mouse dorsal skin (formalin-fixed, paraffin-embedded sections). A, CRH-R2 antigen is detected in the epidermis, hair follicle and sebaceous glands. B, Negative control incubated with antibodies preabsorbed with CRH-R2 antigen. Der, Dermis; Sb, sebaceous unit; apposing arrows delineate the epidermis. Scale bar, 250 μm.
Fig. 8.
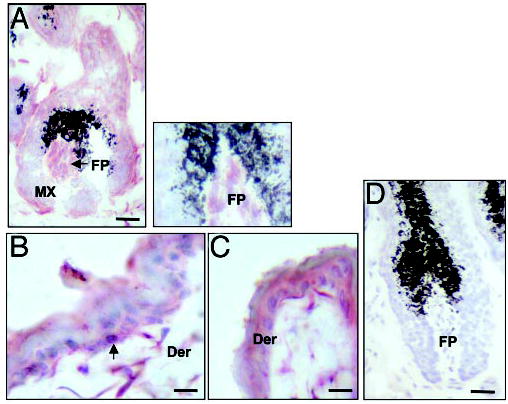
Expression of CRH R2 in C57BL/6 mouse dorsal skin (frozen sections). Anagen hair follicle (A), epidermis with adjacent dermis (B and C), negative control in which the antiserum was absorbed with the CRH R2 peptide (D). Follicular papilla (FP), arrow (epidermis), dendritic cells in the dermis (Der). The gap is the hair bulb (A) reflects a tear in the tissue that occurred upon sectioning. Scale bar: A, 40 μm; B, 20 μm; C, 20 μm; D, 40 μm.
Thus, both molecular and immunocytochemistry data document wide expression of CRH-R2 in all the main cellular compartments of mouse skin (epidermis and dermis). We previously demonstrated that mouse skin expressed CRH-R1 gene, albeit at very low levels during telogen and high levels at anagen VI phases of hair cycle (22, 45). However, immunocytochemistry demonstrated restricted localization of the CRH-R1 antigen to the follicular keratinocytes and hair follicle papilla fibroblasts and its absence in the epidermis (46). It is striking therefore, that the in situ expression patterns of CRH-R1 and 2 show opposite (mirror) compartmental localization in murine and human skin. It is becoming progressively clear that in mouse skin, in opposition to human skin, CRH-R2 may play an equal if not a major role in the CRH-mediated regulation of cutaneous functions. Still, for both species CRH-R2 may at least be an important regulator of adnexal (hair follicle) functions.
3. Expression of CRH-related genes in mammalian skin
CRH-BP.
The biological effects of CRH and related peptides can be modified by CRH-BP on the central, local or systemic levels (11). Expression of the CRH-BP gene was therefore examined in human and mouse skin cells. Expression of the gene coding for the CRH-BP showed heterogeneous expression in human skin (Fig. 9A). Thus, CRH-RB message was expressed in human dermal fibroblasts, in three of six mel-anoma lines, and in sc fat cells. CRH-RB message was not expressed in human normal epidermal melanocytes, kera-tinocytes, or squamous cell carcinoma cells. In mouse skin and in normal and malignant melanocytes, CRH-BP was below detectability (Fig. 9C). As expected, CRH-BP was expressed in mouse pituitary that served as positive control (Fig. 9C). Production of CRH-BP by human cutaneous fibro-blasts or fat cells may serve as an additional mechanism to modulate CRH signaling in dermal or sc compartments. In melanoma CRH-BP expression could seal the tumor from host (CRH) regulation. The expression of CRH-R1e and f isoforms in dermal fibroblasts and fat cells may serve as an additional line of regulation by sequestering locally produced CRH-related peptides and thus preventing their entry into systemic circulation via the dermal or sc vascular plexuses. Although CRH-BP is not produced by normal human epidermal or adnexal cells, the regulatory functions could be exerted by products of alternatively spliced CRH-R1e, f, and h isoforms (15, 26). Lack of expression of CRH-BP in mouse skin is consistent with its reported restricted expression in this species, to the brain and pituitary (11). Thus, modulation of the CRH signaling in the mouse skin could be performed by CRH-R1e and f isoforms.
Fig. 9.
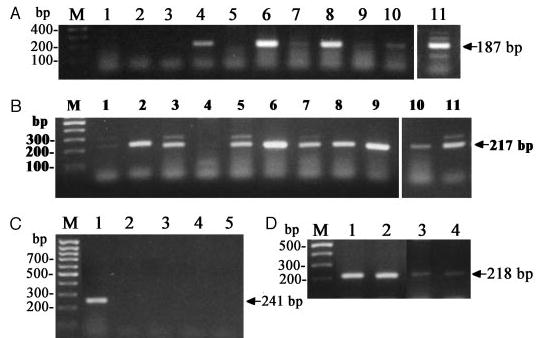
Expression of CRH-BP and Ucn II. A, Amplification of human CRH-BP: immortalized (HaCaT) keratinocytes, 1; immortalized (Pig 1) epidermal melanocytes, 2; adult epidermal keratinocytes, 3; dermal fibroblasts, 4; melanoma lines SKMEL-188 (5) SBCE2 (6), WM35 (7), WM98 (8), WM164 (9), WM1341 (10); sc adipose tissue, 11. B, Amplification of human stresscopin-related gene (Ucn II): dermal fibro-blasts, 1; adult epidermal melanocytes, 2; C1–4 squamous cell carcinoma, 3; melanoma lines SKMEL-188 (4), SBCE2 (5), WM35 (6), WM98 (7), WM164 (8), WM1341 (9), immortalized (HaCaT) keratin-ocytes (10); adult epidermal keratinocytes, 11. C, Amplification of mouse gene similar to CRH binding protein: pituitary, 1; anagen skin, 2; telogen skin, 3; Cloudman S91 melanoma, 5; immortalized normal (melan-a) melanocytes. D, Amplification of mouse Ucn II gene: ana-gen skin, 1; telogen skin, 2; anagen skin, 3; Cloudman S91 melanoma, 4. Total RNA was used for 3 and 4 and mRNA for 1 and 2.
Ucn II.
Human skin reportedly expresses both CRH and Ucn mRNA and protein (20, 22–25, 47), whereas mouse skin expresses CRH protein but not CRH mRNA (suggesting neural delivery of the peptide) and both Ucn mRNA and protein (7, 22, 25, 45, 46). Presence of CRH and Ucn-1 peptides in human skin was documented both by RIA coupled with RP-HPLC separation and by liquid-chromatography mass spectrometry (LC/MS) (23–25). These peptides were additionally localized by immunocytochemistry to epidermal and dermal cells of the skin (22, 24, 46, 49). Similarly, mouse skin expresses both Ucn-1 mRNA and protein, although in the mouse, intracutaneous concentrations vary in tight linkage with phase of hair cycle, in a pattern opposite to that observed for CRH (24).
In the present study, we demonstrated expression of Ucn II (strescopin-related peptide) mRNA with direct RT-PCR (30 cycles) in all tested human skin cells. Thus, Ucn II mRNA was found in normal epidermal adult melanocytes and keratin-ocytes, immortalized HaCaT keratinocytes, normal dermal fibroblasts, sc adipose tissue, squamous cell carcinoma (C4 –1) line and in 6 (all) human melanoma lines (Fig. 9B). Ucn II mRNA was also detected in mouse telogen and anagen skin and cultured malignant melanocytes (Fig. 9D). Similar findings have been noted in rat skin where Ucn II mRNA has been detected (9). Because Ucn II may activate CRH-R2 but not CRH-R1, we suggest that Ucn II expression increase specificity and sensitivity of cutaneous signaling by targeting only one specific receptor expressed on selected cells.
4. Concluding remarks
We have conclusively found marked differences in the expression of a local CRH signaling system in human vs. mouse (rodent) skin. The most contrasting features were the opposite or mirrored tissue-restricted nature of CRH-R1 and CRH-R2 expression patterns in human and mouse skin, the preferential expression of CRH-R1 in human skin, and the lack of expression of CRH-BP in mouse skin. These differences are likely to reflect evolutionary traits resulting in different functional activities of human and mouse skin. In humans, skin is continuously exposed to solar radiation, whereas mouse skin (nocturnal animals) is shielded from solar radiation by fur, which also effectively buffers other physical factors (temperature). Thus, in human skin, the stress-reaction may be centered on the CRH-R1, also a major mediator in activation of the hypothalamic-pituitary-adrenal axis. Expression of CRH-BP would serve as a secondary buffering system that could negatively regulate the local availability of CRH and CRH-related peptides and thus prevent them from entering systemic circulation. In mice, the presence of fur and nocturnal behavior significantly reduce exposure of their skin to biohazardous environmental factors which consequently may have reduced evolutionary pressure on, or need for, the CRH system. This may explain why CRH-R2 is more prevalent than CRH-R1, CRH-BP expression is absent and CRH is not produced by murine skin cells but delivered via neural pathways, whereas Ucn and Ucn II are expressed in their skin cells. The similar location of CRH-R2 in adnexal structures in both humans and mice suggest that at this site this receptor type plays a predominant role in the regulation of hair growth. In conclusion, we mapped the expression of the CRH signaling system in the skin. We found differential, spatiotemporal selectivity and species restricted expression of CRH-Rs and CRH-BP. We suggest that the differences observed may be evolutionary driven to adapt to the species predominant determinant of skin function: human, solar radiation and thermal energy; mice, hair cycle and production of chemical messengers.
Acknowledgments
We thank Dr. Elizabeth Linton (Oxford University) for the antibody against CRH-R1, Dr. Dorothy Bennett (Saint George Hospital, London, UK) and Dr. Vincent Hearing (NIH, Bethesda, MD) for immortalized mouse melanocytes line melan-a, Dr. Catherine LePoole (Loyola Medical Center, Maywood, IL) for immortalized human epidermal melanocytes line PIG-1 and Dr. Meenhard Herlyn (Wistar Institute, Philadelphia, PA) for WM melanoma lines. We also thank Ms. Christine Crawford for skillful secretarial assistance.
Footnotes
This project was supported by NIH Grant 1R01-AR047079-01A2 (to A.S. and D.T.) and by a grant from the Center of Excellence in Genomics and Bioinformatics, University of Tennessee Health Science Center (to A.P. and A.S.).
References
- 1.Aguilera G. Corticotropin releasing hormone, receptor regulation and the stress response. Trends Endocrinol Metab. 1998;9:329 –336. doi: 10.1016/s1043-2760(98)00079-4. [DOI] [PubMed] [Google Scholar]
- 2.Dautzenberg FM, Hauger RL. The CRF peptide family and their receptors: yet more partners discovered. Trends Pharmacol Sci. 2002;23:71–77. doi: 10.1016/s0165-6147(02)01946-6. [DOI] [PubMed] [Google Scholar]
- 3.Grammatopoulos DK, Chrousos GP. Functional characteristics of CRH receptors and potential clinical applications of CRH-receptor antagonists. Trends Endocrinol Metab. 2002;13:436 –444. doi: 10.1016/s1043-2760(02)00670-7. [DOI] [PubMed] [Google Scholar]
- 4.Perrin MH, Vale WW. Corticotropin releasing factor receptors and their ligand family. Ann NY Acad Sci. 1999;885:312–328. doi: 10.1111/j.1749-6632.1999.tb08687.x. [DOI] [PubMed] [Google Scholar]
- 5.Linton EA, Woodman JR, Asboth G, Glynn BP, Plested CP, Bernal AL. Corticotrophin releasing hormone: its potential for a role in human myome-trium. Exp Physiol. 2001;86:273–281. doi: 10.1113/eph8602183. [DOI] [PubMed] [Google Scholar]
- 6.Slominski A, Wortsman J, Luger T, Paus R, Salomon S. Corticotropin releasing hormone and proopiomelanocortin involvement in the cutaneous response to stress. Physiol Rev. 2000;80:979 –1020. doi: 10.1152/physrev.2000.80.3.979. [DOI] [PubMed] [Google Scholar]
- 7.Slominski A, Wortsman J, Pisarchik A, Zbytek B, Linton EA, Mazurkiewicz JE, Wei ET. Cutaneous expression of corticotropin-releasing hormone (CRH), urocortin, and CRH receptors. FASEB J. 2001;15:1678 –1693. doi: 10.1096/fj.00-0850rev. [DOI] [PubMed] [Google Scholar]
- 8.Chalmers DT, Lovenberg TW, Grigoriadis DE, Behan DP, De Souza EB. Corticotropin-releasing factor receptors: from molecular biology to drug design. Trends Pharmacol Sci. 1996;17:166 –172. doi: 10.1016/0165-6147(96)81594-x. [DOI] [PubMed] [Google Scholar]
- 9.Reyes TM, Lewis K, Perrin MH, Kunitake KS, Vaughan J, Arias CA, Ho-genesch JB, Gulyas J, Rivier J, Vale WW, Sawchenko PE. Urocortin II: a member of the corticotropin-releasing factor (CRF) neuropeptide family that is selectively bound by type 2 CRF receptors. Proc Natl Acad Sci USA. 2001;98:2843–2848. doi: 10.1073/pnas.051626398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu SY, Hsueh AJW. Human stresscopin and stresscopin-related peptide are selective ligands for the corticotropin-releasing hormone receptor. Nat Med. 2001;7:605–611. doi: 10.1038/87936. [DOI] [PubMed] [Google Scholar]
- 11.Seasholtz AF, Valverde RA, Denver RJ. Corticotropin-releasing hormone-binding protein: biochemistry and function from fishes to mammals. J Endocrinol. 2002;175:89 –97. doi: 10.1677/joe.0.1750089. [DOI] [PubMed] [Google Scholar]
- 12.Potter E, Behan DP, Fischer WH, Linton EA, Lowry PJ, Vale WW. Cloning and characterization of the cDNAs for human and rat corticotropin releasing factor-binding proteins. Nature. 1991;349:423–426. doi: 10.1038/349423a0. [DOI] [PubMed] [Google Scholar]
- 13.Sakai K, Yamada M, Horiba N, Wakui M, Demura H, Suda T. The genomic organization of the human corticotropin-releasing factor type-1 receptor. Gene. 1998;219:125–130. doi: 10.1016/s0378-1119(98)00322-9. [DOI] [PubMed] [Google Scholar]
- 14.Grammatopoulos DK, Dai Y, Randeva HS, Levine M, Karteris E, Easton A, Hillhouse EW. A novel spliced variant of the type 1 corticotropin-releasing hormone receptor with a deletion in the seventh transmembrane domain present in the human pregnant term myometrium and fetal membranes. Mol Endocrinol. 1999;13:2189 –2202. doi: 10.1210/mend.13.12.0391. [DOI] [PubMed] [Google Scholar]
- 15.Pisarchik A, Slominski A. Alternative splicing of CRH-R1 receptors in human and mouse skin: identification of new variants and their differential expression. FASEB J. 2001;15:2754 –2756. doi: 10.1096/fj.01-0487fje. [DOI] [PubMed] [Google Scholar]
- 16.Catalano RD, Kyriakou T, Chen J, Easton A, Hillhouse EW. Regulation of corticotropin-releasing hormone type 2 receptors by multiple promoters and alternative splicing: identification of multiple splice variants. Mol Endocrinol. 2003;17:395–410. doi: 10.1210/me.2002-0302. [DOI] [PubMed] [Google Scholar]
- 17.Liaw CW, Lovenberg TW, Barry G, Oltersdorf T, Grogoriadis EE, DeSouza EB. Cloning and characterization of he human corticotropin-releasing factor-2 receptor complementary deoxyrubonucleic acid. Endocrinology. 1996;137:72 –77. doi: 10.1210/endo.137.1.8536644. [DOI] [PubMed] [Google Scholar]
- 18.Kostich WA, Chen A, Sperle K, Largent BL. Molecular identification and analysis of a novel human corticotropin-releasing factor (CRF) receptor: The CRF2g receptor. Mol Endocrinol. 1998;12:1077–1085. doi: 10.1210/mend.12.8.0145. [DOI] [PubMed] [Google Scholar]
- 19.Valdenaire O, GIller T, Breu V, Gottowick J, Kilpatrick G. A new functional isoform of the human CRF2 receptor for corticotropin-releasing factor. Biochim Biophys Acta. 1997;1352:129 –132. doi: 10.1016/s0167-4781(97)00047-x. [DOI] [PubMed] [Google Scholar]
- 20.Kono M, Nagata H, Umemura S, Kawana S, Osamura RY. In situ expression of corticotropin-releasing hormone (CRH) and proopiomelanocortin (POMC) genes in human skin. FASEB J. 2001;15:2759 –2766. doi: 10.1096/fj.01-0254fje. [DOI] [PubMed] [Google Scholar]
- 21.Quevedo MA, Slominski A, Pinto W, Wei E, Wortsman J. Pleiotropic effects of corticotropin releasing hormone (CRH) on normal human keratin-ocytes. In Vitro Cell Dev Biol. 2001;37:50 –54. doi: 10.1290/1071-2690(2001)037<0050:peocrh>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 22.Slominski A, Botchkarev V, Choudhry M, Fazal N, Fechner K, Furkert J, Krause E, Roloff B, Sayeed M, Wei E. Cutaneous expression of CRH and CRH-R: is there a “skin stress system? Ann NY Acad Sci. 1999;885:287–311. doi: 10.1111/j.1749-6632.1999.tb08686.x. [DOI] [PubMed] [Google Scholar]
- 23.Slominski A, Ermak G, Mazurkiewicz JE, Baker J, Wortsman J. Characterization of corticotropin releasing hormone (CRH) in human skin. J Clin Endocrinol Metab. 1998;83:1020 –1024. doi: 10.1210/jcem.83.3.4650. [DOI] [PubMed] [Google Scholar]
- 24.Slominski A, Roloff B, Curry J, Dahiya M, Szczesniewski A, Wortsman J. The skin produces urocortin. J Clin Endocrinol Metab. 2000;85:815–823. doi: 10.1210/jcem.85.2.6381. [DOI] [PubMed] [Google Scholar]
- 25.Slominski A, Szczesniewski A, Wortsman J. Liquid chromatography-mass spectrometry detection of corticotropin-releasing hormone and proop-iomelanocortin-derived peptides in human skin. J Clin Endocrinol Metab. 2000;85:3582–3588. doi: 10.1210/jcem.85.10.6863. [DOI] [PubMed] [Google Scholar]
- 26.Pisarchik A, Slominski A. Corticotropin releasing hormone receptor type 1: molecular cloning and investigation of alternative splicing in the hamster skin. J Invest Derm. 2002;118:1065–1072. doi: 10.1046/j.1523-1747.2002.01770.x. [DOI] [PubMed] [Google Scholar]
- 27.Slominski A, Paus R, Plonka P, Maurer M, Chakraborty A, Pruski D, Luk-iewicz S. Melanogenesis during the anagen-catagen-telogen transformation of the murine hair cycle. J Invest Dermatol. 1994;102:862–869. doi: 10.1111/1523-1747.ep12382606. [DOI] [PubMed] [Google Scholar]
- 28.Kauser S, Schallreuter KU, Thody AJ, Gummer C, Tobin DJ. Regulation of human epidermal melanocye biology by β-endorphin. J Invest Dermatol. 2003;120:1073–1080. doi: 10.1046/j.1523-1747.2003.12242.x. [DOI] [PubMed] [Google Scholar]
- 29.Tobin DJ, Bystryn J-C. Different populations of melanocytes are present in hair follicles and epidermis. Pigment Cell Res. 1996;916:304 –310. doi: 10.1111/j.1600-0749.1996.tb00122.x. [DOI] [PubMed] [Google Scholar]
- 30.Slominski A. Identification of β-endorphin, α-MSH and ACTH peptides in cultured human melanocytes, melanoma and squamous cell carcinoma cells. Exp Dermatol. 1988;7:213–216. doi: 10.1111/j.1600-0625.1998.tb00326.x. [DOI] [PubMed] [Google Scholar]
- 31.Slominski A, Ermak G, Wortsman J. Modification of melanogenesis in cultured human melanoma cells. In Vitro Cell Dev Biol. 1999;35:564 –565. doi: 10.1007/s11626-999-0093-6. [DOI] [PubMed] [Google Scholar]
- 32.Coburn SP, Slominski A, Mahuren JD, Wortsman J, Hessle L, Millan JL. Cutaneous metabolism of vitamin B-6. J Invest Dermatol. 2003;120:292–300. doi: 10.1046/j.1523-1747.2003.12034.x. [DOI] [PubMed] [Google Scholar]
- 33.Slominski A, Moellman G, Kuklinska E, Bomirski A, Pawelek J. Positive regulation of melanin pigmentation by two key substrates of the melanogenic pathway: L-tyrosine and L-dopa. J Cell Sci. 1988;89:287–296. doi: 10.1242/jcs.89.3.287. [DOI] [PubMed] [Google Scholar]
- 34.Slominski A, Moellmann G, Kuklinska E. MSH inhibits growth in a line of amelanotic hamster melanoma cells and induces increases in cAMP levels and tyrosinase activity without inducing melanogenesis. J Cell Sci. 1989;92:551–559. doi: 10.1242/jcs.92.4.551. [DOI] [PubMed] [Google Scholar]
- 35.Bomirski A, Wrzolkowa T, Arendarczyk M, Bomirska M, Kuklinska E, Slominski A, Moellmann G. Pathology and ultrastructural characteristics of a hypomelanotic variant of transplantable hamster melanoma with high tyrosinase actiity. J Invest Derm. 1987;89:469 –473. doi: 10.1111/1523-1747.ep12460928. [DOI] [PubMed] [Google Scholar]
- 36.Castro MG, Morrison E, Perone MJ, Brown OA, Murray CA, Ahmed I, Perkins AV, Europe-Finner G, Lowenstein PR, Linton EA. Corticotro-phin-releasing hormone receptor type 1: generation and characterization of polyclonal antipeptide antibodies and their localization in pituitary cells and cortical neurones in vitro. J Neuroendocrinol. 1996;8:521–531. doi: 10.1046/j.1365-2826.1996.04866.x. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez-Linares B, Linton EA, Phaneuf S. Expression of corticotro-phin-releasing hormone receptor mRNA and protein in the human myome-trium. J Endocrinol. 1998;156:15–21. doi: 10.1677/joe.0.1560015. [DOI] [PubMed] [Google Scholar]
- 38.Lei S, Richter R, Bienert M, Mulvany MJ. Relaxing actions of cortico-tropin-releasing factor on rat resistance arteries. Br J Pharmacol. 1993;108:941–947. doi: 10.1111/j.1476-5381.1993.tb13490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dashwood MR, Andrews HE, Wei ET. Binding of [125I]Tyr-corticotropin-releasing factor to rabbit aorta is reduced by removal of the endothelium. Eur J Pharmacol. 1987;135:111–112. doi: 10.1016/0014-2999(87)90766-7. [DOI] [PubMed] [Google Scholar]
- 40.Barker DM, Corder R. Studies of the role of endothelium-dependent nitric oxide release in the sustained vasodilator effects of corticotrophin releasing factor and sauvagine. Br J Pharmacol. 1999;126:317–325. doi: 10.1038/sj.bjp.0702261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zouboulis CC, Seltmann H, Hiroi N, Chen W, Young M, Oeff M, Scherbaum WA, Orfanos CE, McCann SM, Bornstein SR. Corticotropin-releasing hormone: an autocrine hormone that promotes lipogenesis in human sebo-cytes. Proc Natl Acad Sci USA. 2002;99:7148 –7153. doi: 10.1073/pnas.102180999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Preil J, Muller MB, Gesing A, Reul JM, Sillaber I, van Gaalen MM, Land-grebe J, Holsboer F, Stenzel-Poore M, Wurst W. Regulation of the hypothalamic-pituitary-adrenocortical system in mice deficient for CRH receptors 1 and 2. Endocrinology. 2001;142:4946 –4955. doi: 10.1210/endo.142.11.8507. [DOI] [PubMed] [Google Scholar]
- 43.Smith GW, Aubry JM, Dellu F, Contarino A, Bilezikjian LM, Gold LH, Chen R, Marchuk Y, Hauser C, Bentley CA, Sawchenko PE, Koob GF, Vale W, Lee KF. Corticotropin releasing factor receptor 1-deficient mice display decreased anxiety, impaired stress response, and aberrant neuroendocrine development. Neuron. 1998;20:1093–1102. doi: 10.1016/s0896-6273(00)80491-2. [DOI] [PubMed] [Google Scholar]
- 44.Rivier CL, Grigoriadis DE, Rivier JE. Role of corticotropin-releasing factor receptors type 1 and 2 in modulating the rat adrenocorticotropin response to stressors. Endocrinology. 2003;144:2396 –2403. doi: 10.1210/en.2002-0117. [DOI] [PubMed] [Google Scholar]
- 45.Slominski A, Ermak, Hwang J, Mazurkiewicz J, Corliss, Eastman A. The expression of proopiomelanocortin (POMC) and of corticotropin releasing hormone receptor (CRH-R) genes in mouse skin. Biochim Biophys Acta. 1996;1289:247–251. doi: 10.1016/0304-4165(95)00159-x. [DOI] [PubMed] [Google Scholar]
- 46.Roloff B, Fechner K, Slominski A, Furkert J, Botchkarev VA, Bulfone-Paus S, Zipper J, Krause E, Paus R. Hair cycle-dependent expression of cor-ticotropin releasing factor (CRF) and CRF receptors (CRF-R) in murine skin. FASEB J. 1998;12:287–297. doi: 10.1096/fasebj.12.3.287. [DOI] [PubMed] [Google Scholar]
- 47.Slominski A, Ermak G, Hwang J, Chakraborty A, Mazurkiewicz JE, Mihm M. Proopiomelanocortin, corticotropin releasing hormone and cortico-tropin releasing hormone receptor genes are expressed in human skin. FEBS Lett. 1995;374:113–116. doi: 10.1016/0014-5793(95)01090-2. [DOI] [PubMed] [Google Scholar]
- 48.LePoole C, Boissy R, Sarangarajan R, Chen J, Forristal J, Sheth P, Westerhof W, Babcock G, Das P, Saelinger C. PIG3V, an immortalized human vitiligo melanocyte cell line, expresses dilated endoplasmic reticulum. In Vitro Cell Dev Biol. 2000;36:309 –319. doi: 10.1290/1071-2690(2000)036<0309:PAIHVM>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 49.Slominski A, Roloff B, Zbytek B, Wei ET, Fechner K, Curry J, Wortsman J. Corticotropin releasing hormone (CRH) and related peptides can act as bioregulatory factors in human keratinocytes. In Vitro Cell Dev Biol. 2000;36:211–216. doi: 10.1290/1071-2690(2000)036<0211:CRHARP>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 50.Pawelek JM. Factors regulating growth and pigmentation of melanoma cells. J Invest Dermatol. 1976;66:201–209. doi: 10.1111/1523-1747.ep12482134. [DOI] [PubMed] [Google Scholar]
- 51.Bennett DC. Colour genes, oncogenes and melanocyte differentiation. J Cell Sci. 1991;98:135–139. doi: 10.1242/jcs.98.2.135. [DOI] [PubMed] [Google Scholar]


