Abstract
Sitosterolemia is a rare autosomal recessive disorder characterized by (a) intestinal hyperabsorption of all sterols, including cholesterol and plant and shellfish sterols, and (b) impaired ability to excrete sterols into bile. Patients with this disease have expanded body pools of cholesterol and very elevated plasma plant-sterol species and frequently develop tendon and tuberous xanthomas, accelerated atherosclerosis, and premature coronary artery disease. In previous studies, we have mapped the STSL locus to human chromosome 2p21. Recently, we reported that a novel member of the ABC-transporter family, named “sterolin-1” and encoded by ABCG5, is mutated in 9 unrelated families with sitosterolemia; in the remaining 25 families, no mutations in sterolin-1 could be identified. We identified another ABC transporter, located <400 bp upstream of sterolin-1, in the opposite orientation. Mutational analyses revealed that this highly homologous protein, termed “sterolin-2” and encoded by ABCG8, is mutated in the remaining pedigrees. Thus, two highly homologous genes, located in a head-to-head configuration on chromosome 2p21, are involved as causes of sitosterolemia. These studies indicate that both sterolin-1 and sterolin-2 are indispensable for the regulation of sterol absorption and excretion. Identification of sterolin-1 and sterolin-2 as critical players in the regulation of dietary-sterol absorption and excretion identifies a new pathway of sterol transport.
Introduction
The hypothesis that dietary-cholesterol absorption may be regulated by a complex molecular mechanism was first given credence with the seminal publication of a clinical study >25 years ago; Bhattacharyya and Connor (1974) studied two affected sisters who had presented with arthralgias and tendon xanthomas but who did not show the classic features of familial hypercholesterolemia on biochemical testing. Those authors reported a new disorder, characterized instead by very elevated plant sterols, and named the disorder “β–sitosterolemia” (MIM 210250), after the major species of plant sterol that was elevated in the plasma of the two sisters. Although this condition appears to be relatively rare, >40 families that have it have been reported (Bjorkhem and Boberg 1995; Patel et al. 1998b). Affected individuals may present with tendon and/or tuberous xanthomas, arthralgias, hemolytic anemia, and premature atherosclerotic disease (Bhattacharyya and Connor 1974; Hidaka et al. 1990; Salen et al. 1992; Bjorkhem and Boberg 1995). Affected individuals have significantly elevated plasma levels of noncholesterol sterols of dietary origin, which are mainly plant sterols, but with normal to mildly elevated plasma cholesterol levels; shellfish sterols also have been reported to be elevated (Gregg et al. 1986). Phytosterols are normally almost undetectable in plasma from normal individuals (Bjorkhem and Boberg 1995; Patel et al. 1998b). Indeed, previous studies had established that plant sterols were excluded from the body, despite being present, in the normal diet, in equal amounts almost to those of cholesterol (Gould et al. 1969). Almost no sitosterol was absorbed in normal individuals, and this finding has been reproduced in other mammals, suggesting that the exclusion of noncholesterol sterols may be more universal (Kuksis and Huang 1962; Ikeda et al. 1988). Studies >50 years ago had shown that sitosterol, when administered orally, interfered with dietary-cholesterol absorption; and sitosterol was used as the first therapeutic modality for hypercholesterolemia, prior to the advent of potent hypolipidemic agents (Pollock 1953; Best et al. 1955; Farquhar et al. 1956). That a single gene, when disrupted, led to the loss of sterol selectivity in dietary-sterol absorption and was associated with accelerated atherosclerosis suggested that specific molecular mechanisms regulate this pathway (Salen et al. 1992; Patel et al. 1998a). Clinical and laboratory studies suggested that the liver normally rapidly excretes into bile any noncholesterol sterols that are introduced into the bloodstream and that this process is also deficient in patients with sitosterolemia (Salen et al. 1992). Bile from affected individuals was found to be very cholesterol poor, suggesting that the same gene that is responsible for selective sterol absorption from the intestine may also mediate the excretion of cholesterol into bile. Recently, the STSL locus has been mapped to chromosome 2p21 (Patel et al. 1998b) and has been further localized to a <2-cM region bounded by markers D2S2294 and D2S2291 (Lee et al. 2001a).
We isolated the entire sitosterolemia critical region in a sequence-ready bacterial artificial chromosome (BAC) contig and built a transcript map of this region (Lu et al. 2001). A number of expressed sequence tags (ESTs) and genes were mapped in the region of interest. One of these was found to encode a “half-ABC” transporter, ABCG5/sterolin-1 (MIM 605459), and also was found to be mutated in nine families with sitosterolemia (Lee et al. 2001b ; Shulenin et al., in press). Examination of two other ESTs (GenBank accession numbers AA034046 and AA700586) showed they were derived from a single cDNA and encoded a highly homologous protein, sterolin-2. The gene for sterolin-2 has been assigned the name “ABCG8” (MIM 605460). Mutational analyses showed that sterolin-2 was mutated in the remaining 25 probands. Thus, defects in either of two related genes are responsible for causing sitosterolemia. Berge et al. (2000) also have reported that mutations in ABCG8 cause sitosterolemia. We report here the characterization of the genomic structure of this region, a compendium of mutations in one of the largest collections of affected individuals known worldwide. We also provide genetic evidence that the products of ABCG5 and ABCG8—sterolin-1 and sterolin–2, respectively—either may function as heterodimers or are tightly coupled in a pathway regulating dietary-sterol absorption and excretion.
Families and Methods
Pedigrees
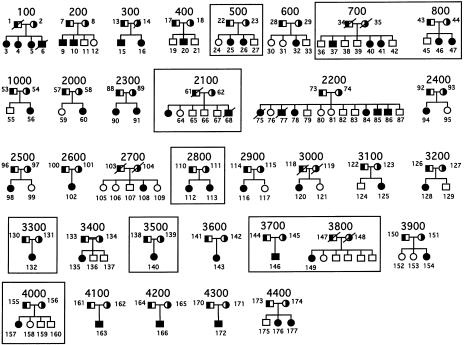
All pedigrees (fig. 1) were recruited on the basis of criteria defined elsewhere (Patel et al. 1998a, 1998b). Clinical features of some of the probands and their family members also have been described elsewhere (Patel et al. 1998a, 1998b). In brief, all probands had (a) clinical features compatible with a diagnosis of sitosterolemia and (b) diagnostically elevated plasma sitosterol levels. The pedigrees include 6 Japanese families (700, 800, 2100, 2800, 3300, 3500, and 3700), 2 South African families of Asian origin (500 and 3600) and 1 South African family of white origin (family 600), 16 U.S. white families (100, 200, 300, 1000, 2000, 2200, 2300, 2400, 2600, 2700, 3000, 3100, 3200, 4000, 4200, and 4300), 1 U.S. African-American family (3800), 1 Dutch family (2500), 1 Norwegian family (2900), 2 Finnish families (400 and 3900), 1 Swedish family (4100), 1 French family (4400), and 1 Colombian family (3400). The only known consanguineous family is family 3400. Many of the probands reported by Berge et al. (2000) are also part of the present study. Informed consent was obtained from all participants, in accordance with local institutional-review-board guidelines. These families include the original index cases (in pedigree 2300) as well as two Amish-Mennonite families (pedigrees 2200 and 2700).
Figure 1.
Pedigrees with sitosterolemia. The parents, who are depicted as obligate carriers (half-black symbols), and the affected individuals (black symbols) are indicated. Only one of these pedigrees, 3400, is consanguineous. The boxed pedigrees are those that are mutant for sterolin-1 (see the Results section).
Isolation of Full-Length cDNA and Gene-Structure Characterization
After fine mapping and construction of a BAC-and-YAC contig across the region of interest, we mapped a number of genes and cDNAs onto this contig (Lee et al. 2001a; Lu et al. 2001). Three ESTs were identified (GenBank accession numbers AA700586, AA034046, and T99836). One of these, T99836, encoded the ABCG5/sterolin. ABCG5 was found to be mutated in nine families (Lee et al. 2001b). The two other ESTs were used to screen cDNA libraries from the human liver and intestine, and nine clones were identified and sequenced. On the basis of sequence analyses, we were able to infer that both ESTs were part of a single cDNA and that they encoded another ABC-family member. This gene, encoding sterolin-2, was assigned the name “ABCG8, according to the HUGO nomenclature.
Image clones corresponding to ESTs that mapped to the region of interest were obtained from Research Genetics and were fully sequenced. Probes were generated and used to screen both phage lambda and plasmid human liver and intestine cDNA libraries (Clontech and Stratagene). All positive clones were screened for insert sizes, and the longest clones were fully sequenced. A cDNA clone of size 2.4 kb was identified that contained exons 4–13 of sterolin-2 cDNA. To identify the 5′ end of the cDNA, RACE was undertaken, with total RNA obtained from the human liver, as well as with commercially available cDNA (Origene). Additionally, we screened a genomic library made from HindIII-digested BAC clone R-328I4 with a probe containing exon 1 of ABCG5 sequences. A 4.9-kb fragment was identified that contained exons 1 of both ABCG5 and ABCG8 and that was completely sequenced from both ends. Using oligonucleotides based on this information, we were able to amplify exons 1–3 from cDNAs and RACE experiments and, thereby, were able to determine the full-length cDNA for human sterolin-2. The gene structure was characterized by PCR with BAC genomic DNA as the template. A list of the oligonucleotides used for exon amplification and of the approximate intron sizes is presented in table 1. Intron 6 acceptor-site boundary sequence is currently not available.
Table 1.
Intron/Exon Boundaries for ABCG8
|
Exon |
Intron |
Exon |
|||||||
| GenBankAccessionNumber | Number | Size(bp) | 3′-End Sequence | Splice-Donor Site | Number | Size(bp) | Splice-Acceptor Site | 5′-End Sequence | Number |
| AF351812 | 1 | 153 | CCCCAGGATACCTCG | gtgagtgagcaatgg | 1 | 5,486 | tcctgtctcccacag | GGCCTCCAGGATAGA | 2 |
| AF351813 | 2 | 102 | GACCTCAACTGCCAG | gtagaggcacgcctg | 2 | 1,546 | gatatctccccacag | GTGGACCTGGCCTCT | 3 |
| AF351814 | 3 | 157 | TCATAGGGAGCTCAG | gtaccggaaaggcaa | 3 | ∼6,000 | ctggtggctttgcag | GTTGTGGGAGAGCCT | 4 |
| AF351815 | 4 | 239 | CAGCGTGACAAAAGG | gtaactaactggccc | 4 | 531 | ccttctggcccacag | GTGGAAGACGTGATC | 5 |
| AF351816 | 5 | 133 | TCCTGTGGAACCCAG | gtgagggcctggggg | 5 | 112 | ctctgtgttggaaag | GAATCCTTATTGTCG | 6 |
| AF351817 | 6 | 270 | CTGCTGACTTCTATG | gtgagtccccaaggc | 6 | >40 kb? | TGGACCTGACCAGCA | 7 | |
| AF351818 | 7 | 163 | CACCTGTGTGGAAAG | gtaaggtggcaggcg | 7 | 87 | tctgcctcccagcag | CGTGACCCCACTAGA | 8 |
| AF351819 | 8 | 81 | TAAGACGCTGATCCG | gtaattatctgtcat | 8 | ∼1,000 | cttttggtttttaag | TCGTCAGATTTCCAA | 9 |
| AF351820 | 9 | 201 | ATGTCATCTCCAAAT | gtgagtgtggcccac | 9 | 419 | tcccttactttttag | GTTACTCAGAGAGGG | 10 |
| AF351821 | 10 | 77 | TATTTCTTTGCCAAG | gtgactgggcagggt | 10 | 665 | ggctgttctttgcag | ATCCTCGGGGAGCTT | 11 |
| AF351822 | 11 | 268 | GCAGCCTGTGGACAG | gtaaggcctgccccc | 11 | ∼1,600 | gctgtctgtctccag | TGCCCGCGTGGATTT | 12 |
| AF351823 | 12 | 128 | GTCTCAGGAGATAAA | gtaagcggggaaggc | 12 | 87 | tgtctgtgtctccag | ATCCTCAGTGCCATG | 13 |
| AF351824 | 13 | 672 | ATGTATTGAGCATCT | ||||||
Exon Amplification and DNA Sequencing
Exons were amplified by PCR using oligonucleotide primers that are located within the flanking intronic areas (table 1) and that have been described elsewhere (Lee et al. 2001b). SSCP variants were identified by incorporation PCR-SSCP (IPS), as described elsewhere (Sossey-Alaoui et al. 1999). Direct sequencing of PCR products was performed by an Amplicycle sequencing kit (Perkin Elmer), and the products were analyzed by an ABIPRISM 377 Genetical Analyzer (Perkin Elmer). Both strands were sequenced, to confirm the identified mutations. Sequence alignment was aided by the use of MacVector software (Oxford Molecular) running on an Apple iMac. PCR products were digested with appropriate restriction enzymes and were analyzed by agarose- or acrylamide-gel electrophoresis (Yu et al. 2000).
Expression Analyses
Northern analysis was performed as described elsewhere (Wu et al. 1999). A ready-made blot (Clontech) was utilized for northern analyses of the human ABCG8 gene, under conditions specified by the manufacturer. Additionally, reverse transcriptase–PCR (RT-PCR) analyses were performed with commercially available genomic DNA-free cDNAs (Origene) and forward (MF2 [5′-ATGAACTGGAAGACGGGC]) and reverse (MR5 [5′-TGAAGGGTCTGCTCAG]) oligonucleotides located in exons 10 and 13, respectively, yielding a cDNA PCR product of 637 bp. Amplification of a β-actin cDNA segment, by oligonucleotides provided by the manufacturer (Origene), was used as a positive control.
Sequence Alignments and Phylogenetic Analyses
Sterolin protein sequences were aligned with other ABC-transporter proteins, by CLUSTAL W (version 1.7) (Thompson et al. 1994). ABC-protein sequences included in the alignment were based on a variety BLAST and TFASTX (Genetics Computer Group, Madison) searches of databases. To ensure comparison of ABC proteins that included half-transporters (i.e., proteins such as sterolins) to ABC proteins that contain two ATP-binding sites and ABC motifs, alignment was performed by identification of the ATP-binding site, selection of ∼10 amino acids upstream through 120 amino acids downstream of this site, and analysis of each such segment independently. Thus, ABC1, which has two such segments, is represented twice, the two segments designated by the suffixes “-1” and “-2.” The alignment was assessed with 1,000 bootstrap resamplings by the integral options of CLUSTAL W. In addition, the alignment was subjected to the heuristic bootstrap-analysis option of the maximum-parsimony program PAUP (version 4.0) (Swofford and Olsen 1996). A further bootstrap resampling was performed by the protein-distance programs of the PHYLIP package (version 3.5c) (Felsenstein 1996). The trees produced by the programs were transferred to TreeView version 1.5 (Page 1996), for manipulation and printing.
Results
Gene Structure and Expression Pattern of Sterolin-2
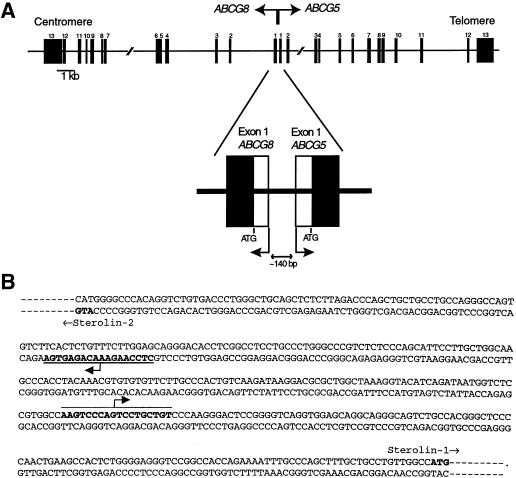
The gene structure for sterolin-1 and sterolin-2 was characterized (fig. 2). Exons 1 and 2 of ABCG5 and ABCG8 are located on a single BAC, C-569J16, that is ⩽∼80 kb, and thus these two genes are located in close proximity but are transcribed in opposite directions. Note that the two initiator ATGs of the two respective genes are separated by 340 bp only, and we estimate that the start-transcription sites are ⩽140 bp apart (data not shown). The exact start-transcription sites have not been accurately determined. When reverse oligonucleotides located in either exon 3 or exon 4 were used, RT-PCR yielded specific products. Overlapping oligonucleotides only 10 bp upstream of these failed to yield a PCR product (data not shown), suggesting that the start-transcription sites are very close to these areas. Although the mouse homologous sequence contains one TATA box (K. Lu, M.-H. Lee, and S. B. Patel, unpublished data), none was identified in the human “promoter” sequence.
Figure 2.
A, Structure and organization of ABCG5 and ABCG8. The intronic sizes (see table 1) were estimated on the basis of either long-range PCR products or, when available, genomic sequence. B, Promoter-region DNA sequence between the two first exons of each gene. The minimum start-transcription sites were estimated on the basis of both the presence of particular sequences in cDNA and the ability of an oligonucleotide located in such sequences to amplify spliced cDNA sequences. We estimate that the maximum distance between the two genes' start-transcription sites is ⩽140 bp. No canonical TATA sequence can be identified in this minimal region.
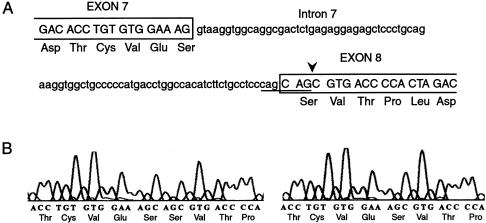
Sequence analyses of several sterolin-2 cDNA clones showed that a few clones had a CAG codon deleted, with the loss of serine 376. This position corresponds to the boundary of exons 7 and 8. Examination of the genomic sequence both showed that the splice-acceptor site at exon 8 contained two CAGs in tandem and suggested alternative splicing as a source of this apparent “polymorphism” (fig. 3). To confirm this, we isolated RNA from the human liver and, after RT-PCR and cloning of cDNA fragments, sequenced 20 clones at random. Approximately 10% of the cDNAs contained a deletion of the CAG and would result in the deletion of serine 376. The functional consequences of this deletion in a minority of ABCG8 transcripts are not known.
Figure 3.
Alternative splicing of ABCG8 mRNA transcripts. The genomic sequence of exon 7–exon 8 region is shown (A), with the splice-acceptor site for exon 8, which contains a CAG repeat (underlined) that results in two different processed transcripts being made. The majority of clones are spliced by the first AG (B, left), but ∼10% of transcripts are spliced by the second AG (A, downward-facing arrowhead).
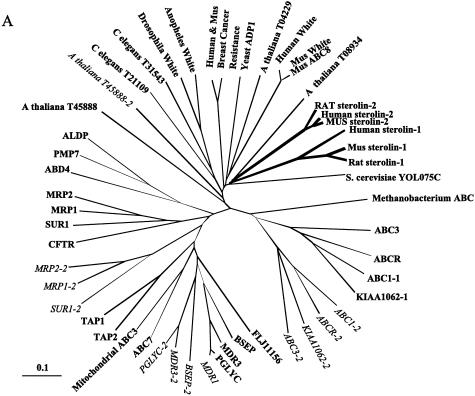
Phylogenetic analyses of sterolin-2 showed it to be highly homologous to sterolin-1 and placed it firmly within the ABCG family (fig. 4A). Comparison of the mouse, rat, and human proteins showed a very high degree of conservation, with >85% similarity at the peptide level (fig. 4B). Both sterolins contain the characteristic ABC motifs, located at the N-terminal half, with predicted six transmembrane domains located at the carboxyl-terminal end (fig. 4B). Although sterolin-1 contains two potential N-glycosylation signals in the largest lumenal facing loop, only one such potential site is present in sterolin 2 (fig. 4B). A number of mutations were identified in both ABCG5 and ABCG8 (see below), and the positions of the missense mutations are as indicated in fig. 4B.
Figure 4.
Homology of human, mouse, and rat ABC proteins. A, Phylogenetic analyses of sterolins and other ABC proteins, performed as described in the text. Each ABC-family member was included on the basis of identification based on its ATP-binding domain and the inclusion of ∼10 amino acids upstream and 120 amino acid downstream of this site. For ABC proteins that contain two ATP-binding sites, each site was analyzed independently; the first site is represented by the thinner unbroken lines, the second site by broken lines. Reassuringly, all such proteins clustered, indicating that such “full” ABC proteins are closely related and probably evolved after being organized as full ABC proteins. Sterolin, which forms a distinct family, is represented by the thicker unbroken lines; its nearest neighbor is the WHITE gene family, the mammalian homologues of which include ABC8-family members. B, Human, mouse, and rat sterolin proteins, aligned by CLUSTALW. The Walker A (i.e., ATP-binding domain) and Walker C motifs are indicated by the series of downward-pointing black triangles and the series upward-pointing black triangles, respectively; the Walker B motif is indicated by the sequence of alternating upward- and downward-pointing black triangles. The thick horizontal lines indicate the predicted transmembrane domains. Residues with the highest homology are denoted by the black portions of the boxes, those with moderate homology by the unblackened boxes and portions of boxes. Missense mutational changes are indicated by black ovals and diamonds, and polymorphic coding changes are indicated by the unblackened ovals and diamonds; changes pertaining to sterolin-1 (ABCG5) are indicated, above the alignments, by diamonds, and those pertaining to sterolin-2 (ABCG8) are indicated below the alignments, by ovals. In sterolin-2, serine 376, which is deleted by alternative splicing, is indicated by the solitary upward-pointing triangle; phenylalanine 570, which is deleted by a 3-bp deletion, is indicated by the arrow (see the text).
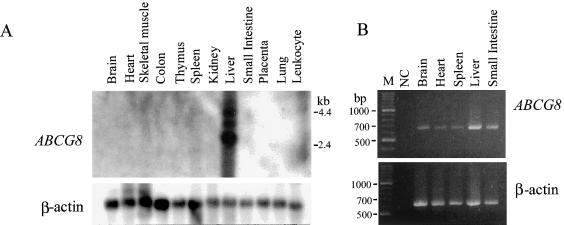
We have previously reported the expression pattern for murine ABCG5 mRNA (Lee et al. 2001b). A human full-length ABCG8 (sterolin-2) cDNA was used to probe a multiple-tissue northern filter, and the results of this are shown in figure 5. Northern analyses show robust expression in the human liver only, although, in RT-PCR analyses (fig. 5B), expression is detectable in a wide variety of human tissues. Since the forward and reverse primers used for RT-PCR are located in exons 10 and 13, respectively, genomic contamination is unlikely. Since RT-PCR is considerably more sensitive than are northern analyses, the implication is that sterolin-2 is more widely expressed, albeit at low levels. Confirmation of this wider expression at the protein level is pending development of antibody reagents. This is in contrast to murine sterolin-1 whose mRNA expression, by both northern and RT-PCR analyses, appears to be confined to the liver and intestine (Berge et al. 2000; Lee et al. 2001b).
Figure 5.
Northern analyses of human sterolin-2 expression. A, Tissue expression of sterolin, analyzed by hybridization to poly (A)+ RNA from different human tissues. The upper panel shows that significant hybridization was detected for liver RNA only (lane 8) and not in small-intestine RNA (lane 9); the bottom panel shows hybridization to β-actin after the membrane has been stripped. B, Results of RT-PCR analyses of cDNAs from different human tissue types, suggesting that sterolin-2 transcripts can be detected in a number of different tissue types, including the small intestine (top panel). The PCR products are derived from amplification across exons. Amplification of a control transcript, β-actin, also is shown (β). NC = water-only control.
Mutations of Sterolin-2/ABCG8 as the Cause of Sitosterolemia
Information on the exon/intron boundaries was used to screen probands, including those known to be mutated for sterolin-1, and to compare them to normal controls. All SSCP variants were sequenced, and any nucleotide changes were compared for evidence of mutations. No proband previously found to be mutated for ABCG5/sterolin-1 (fig. 1, boxed pedigrees) was found to harbor nucleotide changes compatible with mutations in ABCG8/sterolin-2. To confirm that these changes are mutations, segregation analyses, using restriction-enzyme digestions when possible, were performed (data reviewed but not shown). All mutational changes were also screened in appropriate control samples of the respective populations, including individuals from the United States (n=69), Finland (n=95), and Japan (n=82). None of the mutational changes described were seen in the control samples. A compilation of all the mutations identified is given in table 2, including those reported elsewhere (Berge et al. 2000; Lee et al. 2001b). In three probands (102, 149 and 154), only a single mutation was identified. Table 3 shows a compilation of the frequency of each mutation identified thus far. Surprisingly, Trp361X is the most common mutation in sterolin-2, and Arg389His is the most common mutation in sterolin-1. The positions of the missense and coding polymorphic changes are indicated in the homology alignment (see fig. 4B). Of interest is a 3-bp deletion in the fourth transmembrane domain of sterolin-2, resulting in the loss of phenylalanine 570; this deletion is located next to two other missense mutations but also is adjacent to a missense polymorphism (Gly575Arg) that does not appear to be deleterious (fig. 4B).
Table 2.
Compilation of Mutations in ABCG5 and ABCG8
|
Mutations in |
||
| Patient (Nationality/Ethnicity) | ABCG5a | ABCG8b |
| 4 (U.S./white) | Trp361X (1173G→A) / Arg412X (1324C→T) | |
| 9 (U.S./white) | Arg543Ser (1719G→T) / Gln172X (604C→T) | |
| 56c (U.S./white) | Trp361X (1173G→A) / Tyr658X (2064C→G) | |
| 60 (U.S./white) | Trp361X (1173G→A) / IVS1 −2 A→G | |
| 90c (U.S./white) | Trp361X (1173G→A) / Trp361X (1173G→A) | |
| 94 (U.S./white) | Trp361X (1173G→A) / Arg184His (641G→A) | |
| 120 (U.S./white) | Trp361X (1173G→A) / Leu501Pro (1592T→C) | |
| 125 (U.S./white) | Trp361X (1173G→A) / Trp361X (1173G→A) | |
| 128 (U.S./white) | Leu596Arg (1877T→G) / IVS1 −2 A→G | |
| 172 (U.S./white) | Trp361X (1173G→A) / Tyr658Stop (2064C→G) | |
| 166c (U.S./white) | Trp361X (1173G→A) / Arg412X (1324C→T) | |
| 32 (SA/white) | Arg121X (451C→T) / Arg121X (451C→T) | |
| 98 (Dutch/white) | Trp361X (1173G→A) / Gly574Glu (1811G→A) | |
| 102c,d (U.S./white) | Trp361X (1173G→A) / … | |
| 84c (U.S./Amish-Mennonite) | Gly574Arg (1810G→A) / Gly574Arg (1810G→A) | |
| 108c (U.S./Amish-Mennonite) | Gly574Arg (1810G→A) / Gly574Arg (1810G→A) | |
| 135 (Columbian/white) | Trp536X (1698G→A) / Trp536X (1698G→A) | |
| 175 (French) | 1798_1800delTTC / Arg405His (1304G→A) | |
| 20 (Finnish) | Trp361X (1173G→A) / Trp361X (1173G→A) | |
| 154 (Finnish) | Trp361X (1173G→A) / … | |
| 116 (Norwegian) | Trp361X (1173G→A) / Trp361X (1173G→A) | |
| 163 (Swedish) | Trp361X (1173G→A) / Leu572Pro (1805T→C) | |
| 15 (U.S./white) | 1568_1572delTCTTT / IVS1 −2 A→G | |
| 143 (SA/Asian) | Arg164X (580C→T) / Arg121X (451C→T) | |
| 25 (SA/Asian) | Arg243X (876C→T) / Arg243X (876C→T) | |
| 40 (Japanese) | Arg419His (1396G→A) / Arg419His (1396G→A) | |
| 46 (Japanese) | Arg389His (1306G→A) / Arg389His (1306G→A) | |
| 63 (Japanese) | del exon 3 / del exon 3 | |
| 113 (Japanese) | Arg389His (1306G→A) / Arg389His (1306G→A) | |
| 132 (Japanese) | Arg419His (1396G→C) / Arg550Ser (1790A→C) | |
| 140 (Japanese) | Arg408X (1362C→T) / Arg408X (1362C→T) | |
| 146 (Japanese) | Arg389His (1306G→A) / Arg389His (1306G→A) | |
| 157 (U.S./white) | Arg419Pro (1396G→C) / Arg419Pro (1396G→C) | |
| 149 (African American) | Glu146Gln (576G→C) / … | |
| 1c,e (German/Swiss) | Trp361X / Trp361X | |
| 2c,e (U.S./Amish) | Gly574Arg / Gly574Arg | |
| 3c,e (U.S./white) | Trp361X / Tyr658X | |
| 5c,d (U.S./white) | Trp361X / Arg412X | |
| 6c,e (U.S./white) | Leu596Arg / … | |
| 7d (U.S./Hispanic) | Arg412X / del547C→191X | |
| 8c,e (New Zealand/white) | Trp361X / … | |
| 4e (Chinese) | Arg263Gln / Pro231Thr | |
| 9e (Chinese) | Arg408X / … | |
Table 3.
Frequency of Mutant Alleles in ABCG5 and ABCG8
| Gene and Mutation | No. of Alleles | Frequency | Restriction-Enzyme Recognition |
| ABCG5: | |||
| Glu146Gln | 1 | .05 | Gain of AlwNI |
| Arg243X | 2 | .10 | Gain of AlwNI |
| Arg389His | 6 | .30 | Loss of BstUI |
| Arg408X | 3 | .15 | Loss of AvaI |
| Arg419Pro | 2 | .10 | Loss of BstUI |
| Arg419His | 3 | .15 | Loss of BstUI |
| del exon 3 | 2 | .10 | … |
| Arg550Ser | 1 | .05 | … |
| Total | 20 | ||
| ABCG8: | |||
| Arg121X | 3 | .061 | Gain of DdeI |
| Arg164stop | 1 | .020 | … |
| Gln172X | 1 | .020 | Gain of BfaI |
| Arg184His | 1 | .020 | Gain of NalIII |
| Pro231Thr | 1 | .020 | Loss of NlaIV |
| Arg263Gln | 1 | .020 | Gain of AluI |
| Trp361X | 19 | .39 | … |
| Arg405His | 1 | .020 | … |
| Arg412X | 3 | .061 | Gain of DdeI |
| Leu501Pro | 1 | .020 | Loss of AluI |
| Trp536X | 2 | .041 | Gain of AhdI |
| Arg543Ser | 1 | .020 | … |
| Leu572Pro | 1 | .020 | Gain of FauI |
| Gly574Glu | 1 | .020 | Loss of MspI |
| Gly574Arg | 4 | .082 | Loss of MspI |
| Leu596Arg | 1 | .020 | Gain of MspI |
| Tyr658X | 2 | .041 | Gain of SfcI |
| IVS1 −2A→G | 3 | .061 | Gain of BtgI |
| 1798_1800delTTC | 1 | .020 | … |
| 1568_1572delTCTTT | 1 | .020 | … |
| Total | 49 |
A number of polymorphic changes, both silent and nonsilent, also were identified. These were found both in normal controls (table 4) and in some of the probands, parents, and unaffected siblings (data not shown).
Table 4.
Polymorphisms Identified in ABCG5 and ABCG8
|
No. of Identified Polymorphisms |
||||||
| Geneand Region | Nucleotide (Amino Acid) Change | Restriction-Enzyme Change | +/+ | +/− | −/− | Hardy-Weinberg Equilibrium? |
| ABCG5: | ||||||
| 167C→T (Pro9Pro) | Gain of BstNI | … | … | … | … | |
| 1950C→G (Gln604Glu) | Loss of SmlI | 46 | 25 | 1 | Yes | |
| ABCG8: | ||||||
| Exon 1 | 141C→T (Pro17Pro) | … | 49 | 1 | 0 | Yes |
| Exon 1 | 145G→C (Asp19His) | Loss of BaeI | 74 | 3 | 1 | No |
| Exon 2 | 251G→A (Cys54Tyr) | Gain of SexAI | 21 | 23 | 23 | No |
| Exon 6 | 802G→A (Glu238Lys) | … | 10 | 1 | 0 | Yes |
| Exon 6 | 866C→T (Ala259Val) | Loss of HaeIII | 0 | 5 | 0 | … |
| Exon 8 | 1289C→A (Thr400Lys) | Gain of MseI | 116 | 53 | 4 | Yes |
| Exon 11 | 1785C→T (Ala565Ala) | Loss of MnlI | 20 | 7 | 1 | Yes |
| Exon 11 | 1813G→C (Gly575Arg) | Gain of HhaI | 188 | 6 | 4 | No |
| Exon 13 | 1984C→T (Ala632Val) | Loss of StyI | 107 | 50 | 13 | No |
| 5′ UTR | −15A→C | Gain of BstEII | 47 | 4 | 0 | Yes |
| 5′ UTR | −19T→G | Loss of Tsp451 | 38 | 42 | 29 | No |
| 5′ UTR | −41C→T | Loss of DdeI | 7 | 2 | 4 | No |
| Intron 1 | −7C→T | Loss of BsmAI | 21 | 23 | 4 | Yes |
| Intron 1 | −21C→A | Loss of MnlI | 20 | 23 | 4 | Yes |
| Intron 9 | −19C→T | … | 1 | 3 | 4 | … |
| Intron 10 | IVS10 +34delCC | … | 3 | 1 | 4 | … |
| Intron 10 | −50C→T | … | 4 | 4 | 2 | … |
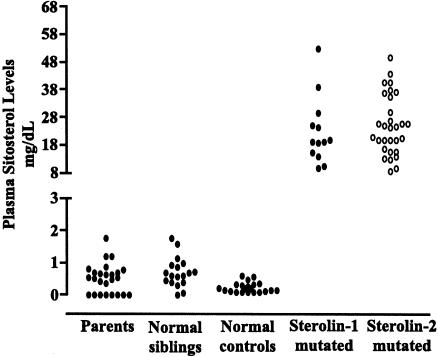
We examined whether being mutant sterolin-1 or mutant sterolin-2 resulted in a difference in plasma sitosterol levels (fig. 6). When the affected individuals were grouped on the basis of whether they have mutant sterolin-1 or mutant sterolin-2, no differences in plasma sitosterol levels were observed at diagnosis (fig. 6). Parents or siblings were not segregated on the basis of whether they are carriers for either of the mutant sterolins, since their plasma sitosterol levels are very close to the detection range.
Figure 6.
Effect of mutations, in either ABCG5 or ABCG8, on plasma sitosterol levels. Plasma sitosterol levels were grouped on the basis of whether affected individuals had mutant sterolin-1 (ABCG5) or mutant sterolin-2 (ABCG8) and were compared with those in their parents (obligate heterozygotes), in their unaffected siblings, and in controls. Plasma sitosterol levels in individuals with mutant sterolin-1 were not different from those in individuals with mutant sterol-2.
Discussion
Selectivity for sterol absorption, which is a feature demonstrated in humans, rats, and dogs, may be more widely present in mammals. On the basis of the deficiencies in sitosterolemia, this pathway involves molecular mechanisms operating in both the liver and the intestine, controlling both sterol excretion into bile and selective sterol absorption from the diet.
After localizing the sitosterolemia locus to human chromosome 2p21, we identified two candidate genes, designated “ABCG5” and “ABCG8,” encoding two proteins—sterolin-1 and sterolin-2, respectively. These encode for two half-ABC transporters, since they each contain only 6 transmembrane domains, as opposed to the 12 transmembrane domains that characterize many of the ABC proteins (Klein et al. 1999). The term “transporter” is used loosely, since this function has not been formally assigned to these proteins.
Elsewhere (Lee et al. 2001b), we have reported that ABCG5 was mutated in nine probands, a finding also reported by (Berge et al. 2000), who, in addition, reported mutations in ABCG8 in a selection of probands with sitosterolemia, many of whom are also members of the pedigrees used in the present study. We report here a detailed characterization of a very large multiethnic cohort of patients with sitosterolemia. Our analyses indicate that all affected individuals, representing 37 families, carry a mutation in either ABCG5 or ABCG8 but not in both. Clinically, there are no readily apparent features that differentiate individuals mutated for sterolin-1 versus those mutated for sterolin-2. We interpret these data to suggest that these two proteins either act as functional heterodimers or are tightly coupled along a pathway that regulates dietary-sterol absorption. Thus, complete loss of any of the sterolins will lead to a functional deficiency. An individual who, hypothetically, carries a mutation in one copy of ABCG5 and in one copy of ABCG8 may not have sitosterolemia, since he or she would be predicted to have ⩾25% normally functioning sterolins. Consistent with this interpretation, the obligate heterozygous parents of our probands do not appear to manifest any clinical or biochemical features, although they are predicted to have 50% normally functioning sterolins. These predictions are amenable to both genetic and biochemical testing.
All of the Japanese probands appear to have mutations in ABCG5 only (Lee et al. 2001b). However, mutations in ABCG5 are not exclusively limited to the Japanese: one South African pedigree of Indian Asian origin and one South African pedigree of U.S. white origin were also found to be mutated in ABCG5. There is a very high degree of homozygosity for informative markers around the STSL locus in many of our probands, despite the presence of only one known consanguineous marriage (Lee et al. 2001a). Additionally, there is robust evidence for linkage disequilibrium (Lee et al. 2001a). Elsewhere (Lee et al. 2001a), we have hypothesized that the mutations giving rise to sitosterolemia are very old, perhaps >1,800 years. Two mutations—Arg389His in ABCG5 and Trp361X in ABCG8—are very common. Arg389His is found only in the Japanese population, although we did not detect this mutation a random sample of 82 Japanese subjects. In this context, it is noteworthy that more individuals had ABCG8 mutations than had ABCG5 mutations. An additional observation is that ABCG8 is more polymorphic than ABCG5, despite the relative proximity of these two genes and their apparent mutual requirement for function. Thus, if founder effects are responsible for some of the sitosterolemia mutations, the disparate nature of polymorphisms in ABCG8—but not in ABCG5—is difficult to explain at present.
The structure of ABCG5/ABCG8 is also worthy of comment. Although the exact start-transcription site for either gene has not been identified definitively, both on the basis of the presence of sequences in the cDNAs and by comparison to rodent cDNAs, the distance separating the two start-transcription sites is ⩽140 bp. These 140 bp do not contain a canonical TATA box, nor do they identify any motifs that suggest promoter sequences. The functioning of this promoter may be unique, especially if sterolins are functional heterodimers, and may need to be expressed coordinately. Berge et al. (2000) identified ABCG5 and ABCG8 as genes that showed alteration in RNA abundance after exposure to LXR agonists, and it is noteworthy that Repa et al. (2000) showed that manipulation of the LXR pathway modulates cholesterol absorption. Thus at least one transcriptional activator of these genes may be the LXR-RXR heterodimers. If confirmed, this may be a link to the coordinate regulation of both the bile acid–biosynthesis pathway and the sterol-absorption and sterol-excretion pathways.
We identified an alternatively spliced form of ABCG8, resulting from of a CAG repeat at the 3′ splice-acceptor site at the intron 7/exon 8 boundary. Although this type of splice site is very unusual, it is not without precedent. Three genes with similar behavior have been reported; CD3ζ, the gene for insulin-like growth factor–receptor 1 (IGFRI), and the Hoxd-11 gene have been shown to have, at the intron/exon boundaries, a CAGCAG repeat that results in both alternative splicing and the presence or absence of an extra amino acid (Yee et al. 1989; Moingeon et al. 1990; Rogina and Upholt 1995). To date, no functional consequences of such alterations have been reported. Smith et al. (1993) have suggested that the 3′ splice-site selection mechanism may involve scanning for the presence of AGs—and, thus, that the presence of two adjacent CAGs at the intron/exon boundary results in the inclusion/exclusion of a triplet codon. Mouse ABCG8 mRNA—and, perhaps, rat ABCG8 mRNA—also shows a similar alternative splicing pattern (K. Lu, M.-H. Lee, and S. B. Patel, unpublished data).
Recently, a number of other gene products have been implicated as playing a role in dietary-cholesterol transport. Repa et al. (2000) have suggested that ABC1 may play a crucial role, on the basis of (a) high levels of gene transcription in intestinal RNA of animals treated with LXR agonists and (b) correlative studies in which cholesterol absorption in LXR-deficient or agonist-treated animals was altered. Interestingly, Berge et al. (2000) had identified ABCG5 and ABCG8 as part of a screen for genes whose expression was altered by LXR-agonist treatments. We suggest that, in the experiments by Repa et al., expression of ABCG5/ABCG8 was altered and that the robust expression of ABC1 seen in their studies could be explained by expression in nonenterocytes, such as fibroblasts and lymphoid cells in the intestinal wall, but not the enterocytes. Lawn et al. (2001) have recently examined the expression of ABC1 by in situ expression in normal mouse tissues, and their data would support our interpretation. Whether enterocytes specifically express ABC1 after LXR activation has not been reported. Another protein implicated as playing a role is scavenger receptor 1 (SR-B1) (Hauser et al. 1998; Schulthess et al. 2000). There are both biochemical and localization data that support the role of SR-B1 in dietary-cholesterol absorption. It is unclear, however, whether the role of SR-B1 is limited to helping to off-load the sterol contents of the micelles that attach to the lumenal enterocytes or whether it also plays a role in regulating the types of sterols that are allowed to enter the enterocyte. That SR-B1 is not absolutely required for this process but may play a facilitatory role is supported by data from mice deficient in SR-B1 (Mardones et al. 2001). Such mice do not appear to manifest any deficiencies in dietary-cholesterol absorption.
Sterolins are likely involved both in the selective transport of dietary cholesterol in and out of enterocytes and in selective sterol excretion by the liver into bile, as evidenced by the consequences when it is deficient (Salen et al. 1992). The identification of these proteins now provides strong evidence for the hypothesis that dietary-cholesterol absorption is both selective and controlled by molecular mechanisms. Further study of the specific mechanisms by which these processes occur will be greatly facilitated by the identification of the genes found to be mutated in sitosterolemia.
Acknowledgments
We are grateful to Yuehua Zhou and Bernard Gerrard, for technical assistance; to the General Clinical Research Center, MUSC, for assistance with sequencing; and to the members of the Patel lab, for helpful discussions. This work was funded by American Heart Association Scientist Development Award 9730087N and National Institutes of Health grant HL60616 (both to S.B.P.), by MO1 RR01070-25 (MUSC GCRC), and by an intramural award from the University Research Committee, Medical University of South Carolina (to S.B.P.). The participation by the families with sitosterolemia is particularly acknowledged.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- GenBank Overview, http://www.ncbi.nlm.nih.gov/Genbank/GenbankOverview.html (for DNA and protein sequences of ABCG5/sterolin-1 [accession numbers AF312715, AF312713, and AF312714—for humans, mice, and rats, respectively], ABCG8/sterolin-2 [accession numbers AF324494, AF324495, and AF351785—for humans, mice, and rats, respectively), genomic-fragment sequences [accession numbers AF351812–AF351824]), and ESTs [accession numbers AA034046, AA700586, and T99836)
- Online Mendelian Inheritance of Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for sitosterolemia [MIM 210250], ABCG5 [MIM 605459], and ABCG8 [MIM 605460])
References
- Berge KE, Tian H, Graf GA, Yu L, Grishin NV, Schultz J, Kwiterovich P, Shan B, Barnes R, Hobbs HH (2000) Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science 290:1771–1775 [DOI] [PubMed] [Google Scholar]
- Best MM, Duncan CH, VanLoon EJ, Wathen JD (1955) The effects of sitosterol on lipids. Am J Med 19:61–70 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya AK, Connor WE (1974) Beta-sitosterolemia and xanthomatosis: a newly described lipid storage disease in two sisters. J Clin Invest 53:1033–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkhem I, Boberg KM (1995) Inborn errors in bile acid biosynthesis and storage of sterols other than cholesterol. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic basis of inherited disease. Vol 2. McGraw-Hill, New York, pp 2073–2102 [Google Scholar]
- Farquhar JW, Smith RE, Dempsey ME (1956) The effect of beta-sitosterol on serum lipids of young men with arteriosclerotic heart disease. Circulation 14:77–82 [DOI] [PubMed] [Google Scholar]
- Felsenstein J (1996) Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol 266:418–427 [DOI] [PubMed] [Google Scholar]
- Gould RG, Jones RJ, LeRoy GV, Wissler RW, Taylor CB (1969) Absorbability of beta-sitosterol in humans. Metabolism 18:652–662 [DOI] [PubMed] [Google Scholar]
- Gregg RE, Connor WE, Lin DS, Brewer H Jr (1986) Abnormal metabolism of shellfish sterols in a patient with sitosterolemia and xanthomatosis. J Clin Invest 77:1864–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser H, Dyer JH, Nandy A, Vega MA, Werder M, Bieliauskaite E, Weber FE, Compassi S, Gemperli A, Boffelli D, Wehrli E, Schulthess G, Phillips MC (1998) Identification of a receptor mediating absorption of dietary cholesterol in the intestine. Biochemistry 37:17843–17850 [DOI] [PubMed] [Google Scholar]
- Hidaka H, Nakamura T, Aoki T, Kojima H, Nakajima Y, Kosugi K, Hatanaka I, Harada M, Kobayashi M, Tamura A, Fujii T, Shigeta Y (1990) Increased plasma plant sterol levels in heterozygotes with sitosterolemia and xanthomatosis. J Lipid Res 31:881–888 [PubMed] [Google Scholar]
- Ikeda I, Tanaka K, Sugano M, Vahouny GV, Gallo LL (1988) Inhibition of cholesterol absorption in rats by plant sterols. J Lipid Res 29:1573–1582 [PubMed] [Google Scholar]
- Klein I, Sarkadi B, Varadi A (1999) An inventory of the human ABC proteins. Biochim Biophys Acta 1461:237–262 [DOI] [PubMed] [Google Scholar]
- Kuksis A, Huang TC (1962) Differential absorption of plant sterols in the dog. Can J Biochem Physiol 40:1493–1504 [Google Scholar]
- Lawn RM, Wade DP, Couse TL, Wilcox JN (2001) Localization of human ATP-binding cassette transporter 1 (ABC1) in normal and atherosclerotic tissues. Arterioscler Thromb Vasc Biol 21:378–385 [DOI] [PubMed] [Google Scholar]
- Lee M-H, Gordon D, Ott J, Lu K, Ose L, Miettinen T, Gylling H, Stalenhoef AF, Pandya A, Hidaka H, Brewer B Jr, Kojima H, Sakuma N, Pegoraro R, Salen G, Patel SB (2001a) Fine mapping of a gene responsible for regulating dietary cholesterol absorption: founder effects underlie cases of phytosterolaemia in multiple communities. Eur J Hum Genet 9:375–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M-H, Lu K, Hazard S, Yu H, Shulenin S, Hidaka H, Kojima H, Allikmets R, Sakuma N, Pegoraro R, Srivastava AK, Salen G, Dean M, Patel SB (2001b) Identification of a gene, ABCG5, important in the regulation of dietary cholesterol absorption. Nat Genet 27:79–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Lee M-H, Carpten JD, Sekhon M, Patel SB (2001) High-resolution physical and transcript map of human chromosome 2p21 containing the sitosterolemia locus. Eur J Hum Genet 9:364–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardones P, Quinones V, Amigo L, Moreno M, Miquel JF, Schwarz M, Miettinen HE, Trigatti B, Krieger M, VanPatten S, Cohen DE, Rigotti A (2001) Hepatic cholesterol and bile acid metabolism and intestinal cholesterol absorption in scavenger receptor class B type I-deficient mice. J Lipid Res 42:170–180 [PubMed] [Google Scholar]
- Moingeon P, Stebbins CC, D'Adamio L, Lucich J, Reinherz EL (1990) Human natural killer cells and mature T lymphocytes express identical CD3 zeta subunits as defined by cDNA cloning and sequence analysis. Eur J Immunol 20:1741–1745 [DOI] [PubMed] [Google Scholar]
- Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358 [DOI] [PubMed] [Google Scholar]
- Patel SB, Honda A, Salen G (1998a) Sitosterolemia: exclusion of genes involved in reduced cholesterol biosynthesis. J Lipid Res 39:1055–1061 [PubMed] [Google Scholar]
- Patel SB, Salen G, Hidaka H, Kwiterovich PO, Stalenhoef AF, Miettinen TA, Grundy SM, Lee MH, Rubenstein JS, Polymeropoulos MH, Brownstein MJ (1998b) Mapping a gene involved in regulating dietary cholesterol absorption: the sitosterolemia locus is found at chromosome 2p21. J Clin Invest 102:1041–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock OJ (1953) Reduction of blood cholesterol in man. Circulation 7:703–706 [DOI] [PubMed] [Google Scholar]
- Repa JJ, Turley SD, Lobaccaro JA, Medina J, Li L, Lustig K, Shan B, Heyman RA, Dietschy JM, Mangelsdorf DJ (2000) Regulation of absorption and ABC1-mediated efflux of cholesterol by RXR heterodimers. Science 289:1524–1529 [DOI] [PubMed] [Google Scholar]
- Rogina B, Upholt WB (1995) The chicken homeobox gene Hoxd-11 encodes two alternatively spliced RNA species. Biochem Mol Biol Int 35:825–831 [PubMed] [Google Scholar]
- Salen G, Shefer S, Nguyen L, Ness GC, Tint GS, Shore V (1992) Sitosterolemia. J Lipid Res 33:945–955 [PubMed] [Google Scholar]
- Schulthess G, Compassi S, Werder M, Han CH, Phillips MC, Hauser H (2000) Intestinal sterol absorption mediated by scavenger receptors is competitively inhibited by amphipathic peptides and proteins. Biochemistry 39:12623–12631 [DOI] [PubMed] [Google Scholar]
- Shulenin S, Schriml LM, Remaley A, Fojo S, Brewer B, Allikmets R, Dean M. A liver-specific ATP-binding cassette gene (ABCG5) from the ABCG (White) gene subfamily maps to human chromosome 2p21 in the region of the Sitosterolemia locus. Cytogenet Cell Genet (in press) [DOI] [PubMed] [Google Scholar]
- Smith CW, Chu TT, Nadal-Ginard B (1993) Scanning and competition between AGs are involved in 3′ splice site selection in mammalian introns. Mol Cell Biol 13:4939–4952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sossey-Alaoui K, Lyon JA, Jones L, Abidi FE, Hartung AJ, Hane B, Schwartz CE, Stevenson RE, Srivastava AK (1999) Molecular cloning and characterization of TRPC5 (HTRP5), the human homologue of a mouse brain receptor-activated capacitative Ca2+ entry channel. Genomics 60:330–340 [DOI] [PubMed] [Google Scholar]
- Swofford DL, Olsen GJ (1996) Phylogeny Reconstruction. In: Hillis DM, Moritz C (eds) Molecular systematics, 2d ed. Sinauer Associates, Sunderland, MA, 411–501 [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Zhu YH, Patel SB (1999) Cyclosporin-induced dyslipoproteinemia is associated with selective activation of SREBP-2. Am J Physiol 277:E1087–E1094 [DOI] [PubMed] [Google Scholar]
- Yee D, Lebovic GS, Marcus RR, Rosen N (1989) Identification of an alternate type I insulin-like growth factor receptor beta subunit mRNA transcript. J Biol Chem 264:21439–21441 [PubMed] [Google Scholar]
- Yu H, Tint GS, Salen G, Patel SB (2000) Detection of a common mutation in the RSH or Smith-Lemli-Opitz syndrome by a PCR-RFLP assay: IVS8 −1G→C is found in over sixty percent of US propositi. Am J Med Genet 90:347–350 [DOI] [PubMed] [Google Scholar]