Summary
Antibacterial quinolones inhibit type II DNA topoisomerases by stabilizing covalent topoisomerase-DNA cleavage complexes, which are apparently transformed into double-stranded breaks by cellular processes such as replication. We used plasmid pBR322 and two-dimensional agarose gel electrophoresis to examine the collision of replication forks with quinolone-induced gyrase-DNA cleavage complexes in Escherichia coli. Restriction endonuclease-digested DNA exhibited a bubble arc with discrete spots, indicating that replication forks had been stalled. The most prominent spot depended upon the strong gyrase binding site of pBR322, providing direct evidence that quinolone-induced cleavage complexes block bacterial replication forks in vivo. We differentiated between stalled forks that do or do not contain bound cleavage complex by extracting DNA under different conditions. Resealing conditions allow gyrase to efficiently reseal the transient breaks within cleavage complexes, while cleavage conditions cause the latent breaks to be revealed. These experiments showed that some stalled forks did not contain a cleavage complex, implying that gyrase had dissociated in vivo and yet the fork had not restarted at the time of DNA isolation. Additionally, some branched plasmid DNA isolated under resealing conditions nonetheless contained broken DNA ends. We discuss a model for the creation of double-stranded breaks by an indirect mechanism after quinolone treatment.
Introduction
Topoisomerases are ubiquitous enzymes that regulate DNA topology. In a topoisomerase reaction cycle, an active site tyrosine residue attacks the phosphodiester backbone, creating a transient break in the DNA through formation of a covalent phosphotyrosine bond. For type II topoisomerases, the break consists of two staggered cuts covalently attached at their 5′ ends to the tyrosine residues of each subunit of the dimeric topoisomerase. This reaction intermediate is called the cleavage complex. Once the cleavage complex is formed, the enzyme passes another segment of double-stranded DNA through the transient break to change the DNA topology, and then reseals the DNA (reviewed in Wang, 2002).
In eukaryotes, the type II topoisomerase is called topoisomerase II (topo II; α and β in some eukaryotes), while prokaryotes contain DNA gyrase (gyrase) and topoisomerase IV (topo IV). These type II topoisomerases can be inhibited by important chemotherapeutic drugs that stabilize the cleavage complex. Bacterial gyrase and topo IV are inhibited by the class of synthetic antibacterial drugs called quinolones (including the fluoroquinolones such as norfloxacin) (for reviews see Drlica and Zhao, 1997; Levine et al., 1998). Although quinolones can inhibit both enzymes in Escherichia coli, gyrase is the major target with respect to cytotoxicity, because a single mutation in gyrA can confer drug resistance (Khodursky et al., 1995; Chen et al., 1996). The mammalian and bacteriophage T4 type II topoisomerases are inhibited by anti-tumour drugs such as aminoacridines, anthracyclines, ellipticines and epipodophyllotoxins (for reviews see Chen and Liu, 1994; Wang et al., 1997).
When drug-stabilized cleavage complexes are treated with SDS, the topoisomerase denatures, thereby revealing the latent DNA break. Subsequent digestion with proteinase K is generally used to remove the attached protein (see Cozzarelli, 1980; and Gellert, 1981). On the other hand, if the drug is diluted out or the cleavage complex is subjected to certain ‘resealing conditions’ before the addition of proteinase K, then the enzyme can reseal the staggered cuts in the cleavage complex. Resealing conditions generally consist of high temperatures (i.e. 65°C) and/or EDTA treatment (Sugino et al., 1977; Sander and Hsieh, 1983; Kreuzer and Alberts, 1984; Osheroff and Zechiedrich, 1987; Howard et al., 1994). Resealing of DNA strands after cleavage complex formation has also been seen on chromosomal DNA in mammalian cells in vivo (Hsiang and Liu, 1989).
A variety of results provide strong evidence that formation of the cleavage complex is central to the mechanism of cell killing by quinolones (reviewed in Drlica and Zhao, 1997). However, formation of the cleavage complex is not sufficient to cause cell death. Cytotoxicity likely results from the conversion of cleavage complexes into overt DNA breaks, but the mechanism of break formation is not known (Drlica and Zhao, 1997). Treatment of E. coli with quinolones induces the SOS response in a RecBC-dependent manner (McPartland et al., 1980). Because RecBC enzyme generally requires DNA ends as entry sites, this result argues that cleavage complexes can be converted into double-stranded breaks. Additionally, recombination-deficient mutants of E. coli exhibit quinolone hypersensitivity (McDaniel et al., 1978), indicating the importance of recombinational proteins in repairing quinolone-mediated damage. Furthermore, bactericidal concentrations of a quinolone cause free rotation of chromosomal DNA in the presence of ethidium bromide, which is characteristic of broken DNA (Chen et al., 1996). Important questions that have yet to be addressed are how these double-stranded breaks are formed and whether they are protein-free or protein-linked.
Although there is no direct evidence applying to type II topoisomerases, one model for the introduction of double-stranded breaks upon drug treatment is the replication run-off model (reviewed in Drlica, 1999). In this model, the replication fork disrupts the cleavage complex upon collision, detaching the DNA strand that is not covalently bound to the topoisomerase. The site of this break would therefore correspond precisely to the transient break created by the topoisomerase (Drlica, 1999 and references therein). Recently, this model was supported for eukaryotic type I topoisomerase inhibitors in vivo (Strumberg et al., 2000). With respect to type II topoisomerases, E. coli helicase II can disrupt a T4 topoisomerase cleavage complex in vitro, demonstrating that the model is reasonable (Howard et al., 1994). However, the E. coli replicative helicase DnaB does not cause DNA breaks when it encounters quinolone-induced topoisomerase cleavage complexes in vitro (Shea and Hiasa, 1999; also see Hiasa and Marians, 1996).
It is possible that the induced double-stranded breaks are not derived from the latent DNA breaks within the cleavage complex. Previous studies have shown that topoisomerase-DNA cleavage complexes can block replication in vitro without causing DNA breaks. For example, purified T7 DNA polymerase is stalled by a norfloxacin-induced gyrase-DNA cleavage complex without detectable DNA breakage (Wentzell and Maxwell, 2000). Likewise, both the replicative helicase DnaB and the complete E. coli replication complex are stalled by norfloxacin-induced topoisomerase-DNA cleavage complexes without generating a break (Hiasa et al., 1996; Shea and Hiasa, 1999). Furthermore, evidence suggests that double-stranded breaks are induced by an indirect pathway upon antitumour drug treatment of bacteriophage T4-infected E. coli (Hong and Kreuzer, 2003). In this system, the drug-stabilized cleavage complexes block replication forks in vivo; some of the cleavage complexes reverse in vivo and yet the forks remain stalled, indicating a need for replication restart (Hong and Kreuzer, 2000). These stalled forks can be cleaved in vitro by T4 endonuclease VII, a recombination endonuclease, and increased levels of stalled replication forks accumulate as a result of inactivating endonuclease VII in vivo (Hong and Kreuzer, 2003). These results suggest a ‘collateral damage’ model, in which the DNA breaks induced by topoisomerase inhibitors are created by recombination nucleases that act on stalled replication forks.
In this study, we examine replication fork dynamics of the plasmid pBR322 in norfloxacin-treated E. coli. A plasmid system was used because it allows us to visualize a relatively large amount of DNA, because the replicative intermediates of pBR322 have been well studied by two-dimensional agarose gel electrophoresis (Martin-Parras et al., 1991; 1998; Schvartzman et al., 1993; Lucas et al., 2001), and because the interaction of pBR322 DNA with gyrase and quinolones has been well studied (Fisher et al., 1981; 1986; Lockshon and Morris, 1985). We show that norfloxacin-induced gyrase cleavage complexes stall replication forks in vivo. Some of these stalled forks do not contain a cleavage complex at the time of isolation, indicating that the cleavage complex reversed in vivo and yet the fork did not immediately restart. Additionally, we found that norfloxacin treatment produces sigma-shaped DNA molecules that are present even after in vitro resealing of gyrase-induced cleavage complexes. These broken molecules may be generated by a collateral damage mechanism similar to the one described above.
Results
Norfloxacin treatment induces formation of cleavage complexes in vivo
We began by confirming the formation of norfloxacin-induced cleavage complexes on pBR322 DNA in vivo. Cells were lysed in the presence of SDS, proteinase K was added to degrade any covalently bound topoisomerase and reveal the latent double-stranded breaks within cleavage complexes, and the DNA was purified by phenol extraction and dialysis (see Experimental procedures). These DNA isolation conditions will be referred to as ‘cleavage conditions’. DNA from a drug-free control, cleaved with EcoRI, generated the expected linear band at 4.4 kb (Fig. 1A, lane 1). DNA from norfloxacin-treated cells included many sublinear fragments consistent with cleavage complex formation at expected gyrase binding sites (Fig. 1A, lane 2; see Lockshon and Morris, 1985). The major binding site for DNA gyrase on pBR322 is centred at 990 bp (Fisher et al., 1981), which corresponds to the prominent cleavage bands at 3.4 kb and 1.0 kb (upon digestion with EcoRI, arrows in Fig. 1B, lane 10). To confirm that these two cleavage bands corresponded to the gyrase binding site centred at 990 bp, we mutated this site to one that causes greatly reduced gyrase binding (pBR322MUT990 plasmid, see Experimental procedures; Fisher et al., 1986). As expected, the plasmid mutation resulted in loss of the bands at 3.4 kb and 1.0 kb, while the remaining pattern was unchanged (Fig. 1B, lane 11).
Fig. 1.

Norfloxacin treatment causes cleavage complex formation at gyrase binding sites. Cells containing pBR322 (lanes 1–10) or pBR322MUT990 (lane 11) were lysed under cleavage (C) or resealing (R) conditions; DNA was extracted and dialysed overnight into TE buffer and then digested with EcoRI. Samples were loaded onto a 1.2% agarose gel, which was analysed by Southern hybridization using a pBR322 probe.
A. JH39 (lanes 1–3), parCR (lanes 4–6), or gyrAR (lanes 7–9) cells were treated with norfloxacin (NOR) for 6 min (lanes 2–3, lanes 5–6, lanes 8–9) or were untreated (lanes 1, 4 and 7). The marker lane consisted of a mixture of restriction fragments derived from plasmids pBR322 and pSF5 (a pBR322 derivative with an approximately 13-kb fragment of phage lambda DNA; obtained from Dr M. Feiss, University of Iowa).
B. JH39 cells carrying pBR322 (lane 10) or pBR322MUT990 (lane 11) were treated with norfloxacin and DNA was extracted under cleavage lysis conditions. Arrows indicate cleavage complex bands resulting from cleavage at the major gyrase binding site centred at 990 bp. The marker lane consisted of a mixture of restriction fragments of pBR322.
As described in the Introduction, EDTA plus heat treatment (65°C) in the absence of a strong denaturant causes topoisomerases to religate and release their bound DNA. We confirmed that DNA purified from cells that were lysed under such ‘resealing conditions’ leads to the disappearance of nearly all the cleavage complex bands (Fig. 1A, lane 3). Using quantitative phosphorimager analysis, we estimate that 90% of cleavage complexes formed with norfloxacin treatment are resealed under these conditions. Isolating DNA under resealing conditions thereby allows us to differentiate between forms of DNA with and without a gyrase cleavage complex at the time of isolation.
To confirm that the effects of norfloxacin on cleavage complex formation are because of gyrase and not topo IV, we moved quinolone-resistant alleles of gyrA (S83L gyrase mutant) and parC (E83K topo IV mutant) (Khodursky et al., 1995) into the genetic background used in our experiments. We first examined cleavage complex formation after norfloxacin treatment and found that the parCR cells showed exactly the same pattern as wild-type cells (Fig. 1A, lanes 4–6). DNA from the gyrAR cells treated with norfloxacin showed only very small amounts of cleavage complexes when isolated under cleavage conditions, and these were eliminated under resealing conditions (Fig. 1A, lanes 7–9). Although the norfloxacin concentration used in this study, 1 μg ml−1, was 20-fold higher than the minimal inhibitory concentration for cells with wild-type gyrase (0.05 μg ml−1), it was similar to the minimal inhibitory concentration for cells with a mutated S83L gyrase (0.78 μg ml−1) (Yoshida et al., 1990). Therefore, it is not surprising that a small amount of cleavage complex formation was detected with the gyrAR cells.
Norfloxacin treatment causes stalled replication forks in vivo
We assayed for stalled replication forks by using neutral/neutral two-dimensional (2D) agarose gel electrophoresis, which separates replicating from non-replicating DNA. The first dimension was run under conditions that maximize size discrimination and minimize the contribution of shape. Each sample lane was then excised and cast across the top of a second-dimension gel that was run with conditions that maximize shape contributions (see Friedman and Brewer, 1995 for review). Figure 2 presents the expected 2D pattern after AlwNI or EcoRI digestion of pBR322 (also see Martin-Parras et al., 1991), which replicates unidirectionally. After restriction endonuclease digestion, the replicative intermediates form a characteristic 2D pattern of bubble (B) molecules until replication reaches the restriction endonuclease site, at which point the replicating molecules are shaped like double-Y (DY) molecules. This transition point is called the bubble-to-double-Y transition, and includes passage through a simple-Y (Y) intermediate. AlwNI digestion results in replicating molecules mostly in the form of bubbles, as AlwNI cleaves just behind the start site for unidirectional replication (Figs 2A and 3A).
Fig. 2.
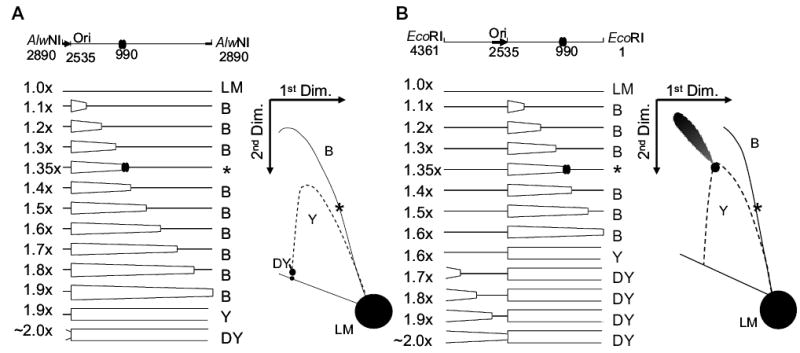
Diagrams of pBR322 DNA after cleavage with AlwNI (A) or EcoRI (B). Replication of pBR322 begins unidirectionally at position 2535 and is indicated by the thick black arrow. AlwNI cuts just behind the origin at position 2890 and EcoRI cuts midway around the plasmid at position 1, as indicated in the schematic on top. The major gyrase binding site is at position 990 and is indicated by two overlapping ovals. The map indicates the forms of DNA produced as replication proceeds from 1.0× to 2.0×. Non-replicating linear (LM) monomers are 4361 base pairs. As the replication fork crosses the location of the restriction endonuclease recognition site, the resulting molecules change from bubble-form (B) to double-Y form (DY), by passing through a simple-Y form [expected as a spot along the simple-Y arc (Y)]. The bubble-to-double-Y transition occurs at 1.9× for AlwNI and 1.6× for EcoRI. At 1.35×, the replication fork reaches the gyrase binding site centred at 990 bp, where stalling can occur if a gyrase cleavage complex is bound (molecules indicated by an asterisk). To the right of the maps are illustrations of the expected 2D gel electrophoresis patterns. The location of the entire simple-Y arc is shown as a dotted line; this arc is not expected for normal pBR322 replication (except for the discrete spot at the bubble-to-double-Y transition).
Fig. 3.
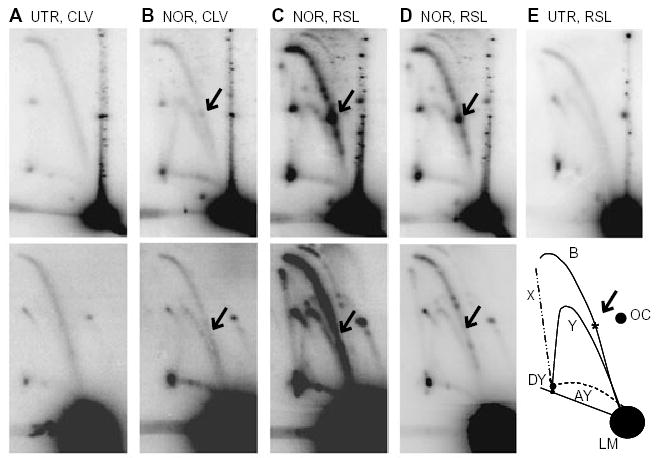
Norfloxacin treatment causes stalled replication forks at gyrase binding sites. DNA was isolated from JH39 cells carrying pBR322 (upper panels) or pBR322MUT990 (lower panels) and digested with AlwNI. The samples were as follows:
A. Cells were untreated (UTR) and DNA was isolated under cleavage conditions (CLV).
B. Cells were treated with norfloxacin (NOR) and DNA was isolated under cleavage conditions.
C. Cells were treated with norfloxacin and DNA was isolated under resealing conditions (RSL).
D. Shorter exposures of panel C.
E. Cells were untreated and DNA was isolated under resealing conditions (upper panel). The illustration (E, lower) indicates the various arcs seen after AlwNI digestion where B, bubble; LM, linear monomer; Y, simple-Y; AY, asymmetric-Y; DY, double-Y; X, X-shaped molecules; OC, open circular monomer. The arrows indicate the location that corresponds to the asterisk in the lower panel of (E).
When the cells were treated with norfloxacin, a discrete spot appeared on the bubble arc, corresponding to stalled replication forks (Fig. 3B, upper panel; multiple spots were observed on long exposures; also see Fig. S1; Fig. 3C and D). For DNA isolated under cleavage conditions (i.e. Fig. 3B), this spot represents unbroken stalled replication forks where in vivo reversal of the cleavage complexes must have occurred (see Discussion). The strong spot on the bubble arc in Fig. 3B (upper panel) migrated at approximately the position expected for replication forks blocked at the major gyrase binding site centred at 990 bp. We confirmed this interpretation by repeating this analysis with the pBR322MUT990 plasmid (Fig. 3B, lower panel; also see Fig. S1). The prominent spot disappeared, and we therefore conclude that gyrase cleavage complexes at the major gyrase cleavage site block replication forks upon norfloxacin treatment. Analysis of 2D gels with other single-cutting restriction enzymes verified this conclusion (see below for EcoRI digestion of pBR322, which shows the main spot at 1.35×, and data not shown for restriction enzyme digestion of pBR322MUT990, where the main spot at 1.35× is always absent). All of the DNA samples from norfloxacin treatments in this article were from cells exposed to norfloxacin for 6 min; very similar results were obtained with 3-, 10- and 20 min norfloxacin treatments.
We next isolated DNA under resealing conditions after treatment of the cells with norfloxacin, and found a very substantial increase in both the number and intensity of discrete spots (compare Fig. 3C with Fig. 3B). As both DNA samples came from the same norfloxacin-treated cells, we infer that the cells contained a substantial amount of replication forks blocked at ‘active’ cleavage complexes (i.e. ones that break upon SDS addition; Fig. 4A, arrows 1 and 2). Once again, the strongest spot on the bubble arc, in the position expected for the major gyrase binding site, disappeared with the pBR322MUT990 plasmid (Fig. 3C and D, lower panels).
Fig. 4.
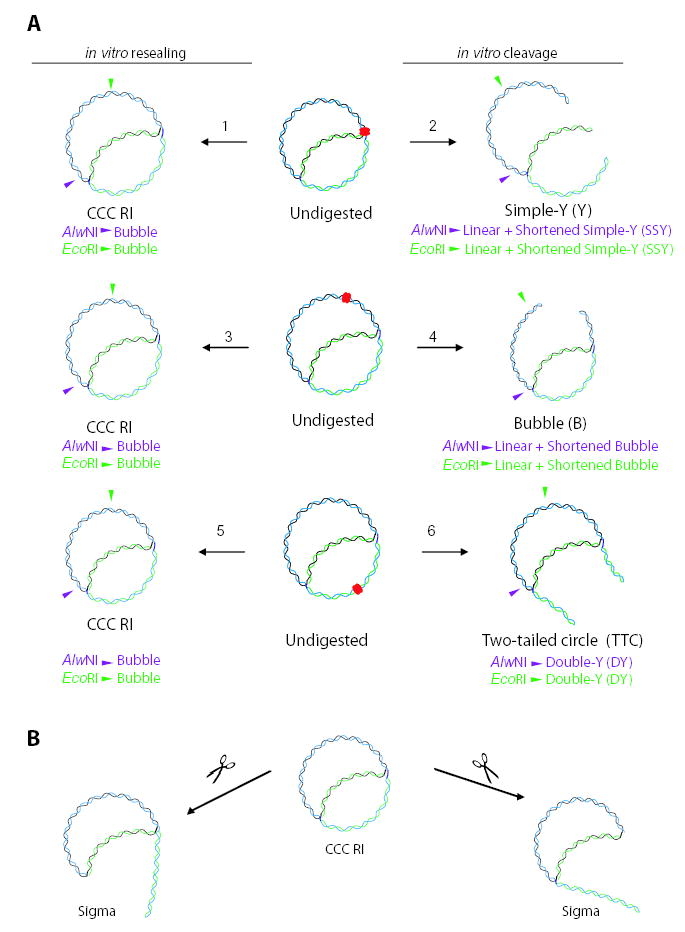
DNA molecules resulting from in vitro resealing or cleavage lysis conditions after norfloxacin treatment (A) and DNA molecules resulting from breakage of sigma molecules in vivo (B). The partially replicated plasmid considered in this figure is stalled at the major gyrase binding site (centred at 990 bp). The gyrase cleavage complex is indicated by red overlapping ovals. Newly synthesized DNA strands are shown in green. The purple and green arrowheads indicate the AlwNI and EcoRI cut sites respectively.
A. Reactions that occur during in vitro lysis are as follows. Under resealing lysis conditions, a gyrase cleavage complex at a blocked fork would reseal the latent DNA break, resulting in an intact covalently closed circular replication intermediate (CCC RI) (arrow 1); treatment of this molecule with either AlwNI or EcoRI yields a bubble. Under cleavage conditions, the latent DNA break within the gyrase cleavage complex at a stalled fork is revealed, resulting in a simple-Y (Y) molecule (arrow 2); treatment of this molecule with either AlwNI or EcoRI yields a linear fragment plus a shortened simple-Y (SSY) molecule. If a gyrase cleavage complex exists well ahead of the replication fork, isolation under resealing conditions results in an intact CCC RI (arrow 3); treatment of this molecule with either AlwNI or EcoRI yields a bubble. In this case, isolation under cleavage conditions results in a bubble (B) (arrow 4); treatment of this molecule with either AlwNI or EcoRI yields a linear fragment plus a shortened bubble. If a gyrase cleavage complex is located behind the replication fork, isolation under resealing conditions results in an intact CCC RI (arrow 5); treatment of this molecule with either AlwNI or EcoRI yields a bubble. In this case, isolation under cleavage conditions produces a two-tailed circle (TTC) (arrow 6); treatment of this molecule with either AlwNI or EcoRI yields a double-Y (DY) molecule.
B. A CCC RI without a gyrase cleavage complex can be broken in vivo at the fixed branch (left side) or the replicating fork (right side).
Several results argue that the discrete spots on the bubble arc are branched DNA molecules that accumulated from blockage of the replication fork by a gyrase cleavage complex. First and most importantly, DNA that appears along a bubble arc can only be produced by DNA replication. Second, the same samples that contained discrete spots on the bubble arc also contained norfloxacin-induced cleavage complexes (see Fig. 1A, lane 2). Third, the position of the most prominent spot on the bubble arc was consistent with DNA replication from the origin to the strongest gyrase binding site in pBR322 DNA, and mutation of the strong gyrase binding site eliminated both the intense cleavage complex bands (Fig. 1B, lane 11) and the discrete spot on the bubble arc (Fig. 3B–D, lower panels). We conclude that norfloxacin-induced gyrase cleavage complexes block replication forks in vivo.
As described above, resealing conditions substantially increased the amount of branched bubble forms of DNA, and we attribute this increase to replication forks blocked by ‘active’ cleavage complexes. If this interpretation is correct, we should be able to detect unique branched forms of DNA corresponding to these molecules under cleavage conditions, where the gyrase-mediated DNA breaks are revealed by SDS treatment during cell lysis (see Fig. 4A, top). The expected DNA molecules would be shortened simple-Y (SSY)-form DNA after AlwNI digestion, and such molecules would be difficult to detect because they are very similar to linear DNA (Fig. 4A, arrow 2). However, digestion with EcoRI readily revealed the expected Y-form DNA (see Fig. 2B for EcoRI 2D gel predictions, and Fig. 4A, arrow 2, for this specific DNA form). An additional arc always appeared when DNA from norfloxacin-treated cells was isolated under cleavage conditions but not resealing conditions (Fig. 5). This arc started at the bubble-to-double-Y transition and extended to the right with a downward slope towards (but above) the monomer spot (see Fig. 5B and diagram). This arc contained multiple discrete spots and was completely absent under resealing conditions (Fig. 5C). A similar arc was also seen with PstI, SalI and StyI restriction digests, at the expected locations for those enzymes (data not shown). We assign this arc as SSY molecules, in which replicating DNA is cleaved by both a gyrase cleavage complex (at the stalled fork) and the restriction enzyme (in the unreplicated region) (see Fig. 4A, arrow 2). To control for the possibility that resealing conditions affect replicating DNA structures in some way other than resealing gyrase cleavage complexes, we isolated DNA from untreated cells under resealing conditions. The pattern was identical to that with DNA isolated under cleavage conditions (Fig. 3E, upper panel for AlwNI, and data not shown for other digests).
Fig. 5.
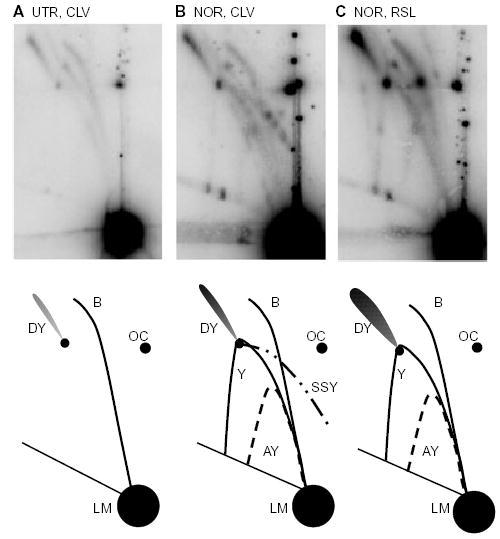
Norfloxacin treatment causes formation of shortened simple-Y molecules. DNA was isolated from JH39 cells carrying pBR322 and digested with EcoRI. The samples were as follows:
A. DNA was isolated under cleavage conditions (CLV) from untreated cells (UTR).
B. DNA was isolated under cleavage conditions from norfloxacin-treated cells (NOR).
C. DNA was isolated under resealing conditions (RSL) from norfloxacin-treated cells.
The illustrations below indicate the various arcs seen after EcoRI digestion, where B, bubble; LM, linear monomer; SSY, shortened simple-Y; Y, simple-Y; DY, double-Y; AY, asymmetric-Y; and OC, open circular monomer.
Examination of undigested DNA from norfloxacin-treated cells
To further analyse the DNA molecules produced after norfloxacin treatment, we examined undigested DNA on 2D agarose gels; the lower panels of Fig. 6 show the established migration patterns of various forms of undigested DNA (see Martin-Parras et al., 1998; and Lucas et al., 2001). As expected, we found that most plasmid DNA was in the form of non-replicating covalently closed circles (CCC and TopCCC), with significant amounts of linear monomers (LM), and open circles (OC). In addition, some replicating plasmid DNA was detected as CCC replication intermediates (Fig. 6A). Norfloxacin treatment led to dramatic changes in the 2D gel electrophoresis pattern (Fig. 6B). First, there was a large increase in linear DNA, consistent with in vitro SDS-induced cleavage of non-replicating CCC DNA bound by a single gyrase cleavage complex (also see Fig. 1). Second, a prominent simple-Y arc appeared, consistent with in vitro cleavage of replicating CCC DNA bound by a single gyrase cleavage complex at a blocked fork (note that the SDS-induced cleavage must have occurred very close to the fork position or the resulting molecules would not be Y-form; see below; also see Fig. 4A, arrow 2). Many discrete spots are located on the simple-Y arc, corresponding to replicating DNA blocked at different gyrase cleavage sites. As expected, the most prominent spot is at a location (roughly 1.35×) consistent with a replication block at the strongest gyrase binding site (centred at 990). Note that these simple-Y molecules are the same molecules that give rise to the SSY arc discussed above. If these simple-Y molecules are subsequently cut by a restriction endonuclease in their replicated portion, the resulting SSY molecules have the same size stem but shorter branches compared with the undigested simple-Y. If the restriction endonuclease cleaves in the unreplicated part of the simple-Y, the resulting SSY molecules will have a shorter stem but identical branches compared with the undigested simple-Y (see Fig. 4A, arrow 2, AlwNI and EcoRI digests).
Fig. 6.
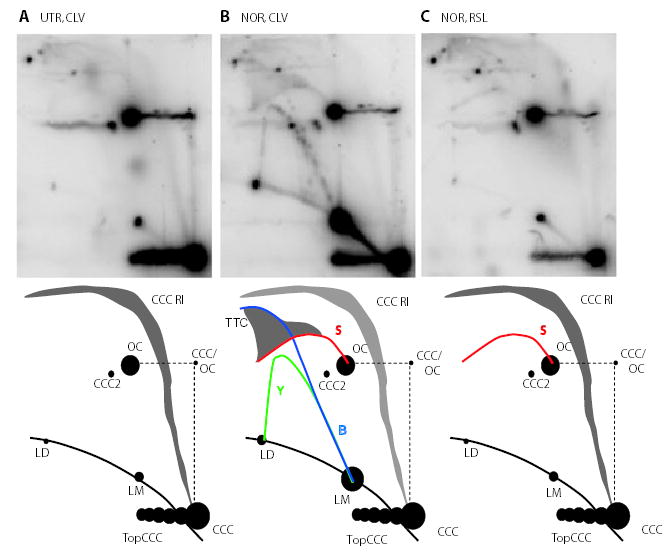
Norfloxacin treatment causes formation of a sigma arc in undigested DNA. pBR322 DNA was isolated from JH39 under the following conditions:
A. Cleavage conditions (CLV) from untreated cells (UTR).
B. Cleavage conditions from norfloxacin-treated cells (NOR).
C. Resealing conditions (RSL) from norfloxacin-treated cells.
None of the samples were treated with restriction enzymes.
The illustrations indicate the various arcs, where B, bubble (blue); LM, linear monomer; LD, linear dimer; Y, simple-Y (green); TTC, two-tailed circle; S, sigma (red); OC, open circular monomer; CCC, covalently closed circle; CCC RI, covalently closed circle replication intermediate; TopCCC, topoisomers of CCCs; CCC2, dimer of CCC; and CCC/OC, CCC in first dimension, OC in second dimension.
A third change induced by norfloxacin in the undigested DNA samples was the generation of sigma molecules (Fig. 6B; S arc). Sigma molecules form an eyebrow-shaped arc that begins at the OC on the 2D gel electrophoresis pattern (Belanger et al., 1996; Lucas et al., 2001). Sigma molecules can be created when replicating CCC DNA becomes nicked on one strand near the replication fork (Fig. 4B). We will argue below that these sigma molecules are created by fork breakage in vivo.
Two additional changes to the undigested 2D pattern upon norfloxacin treatment are the appearance of bubbles (B) and two-tailed circles (TTC). The TTC region occupies a funnel shape above the eyebrow–shaped S arc, while the bubble arc is in the same location as the bubble arc after restriction endonuclease treatment (Lucas et al., 2001; and see Fig. 6). Bubbles and TTC are presumably generated when CCC replicating DNA contains a gyrase cleavage complex some distance ahead or behind the replication fork respectively. During DNA isolation with cleavage lysis conditions, disruption of the gyrase cleavage complex away from the replication fork reveals the latent double-stranded break, thereby generating the B and TTC forms (Fig. 4A, arrows 4 and 6 respectively).
Many of the branched DNA forms that appeared in norfloxacin-treated samples after isolation under cleavage conditions did not appear when the DNA was isolated under resealing conditions (Fig. 6C). In fact, the 2D pattern of DNA isolated under resealing conditions after norfloxacin treatment was quite similar to that of DNA from drug-free samples isolated under cleavage conditions (compare Fig. 6A and C; note that DNA from drug-free samples isolated under resealing conditions generated the same pattern as that in Fig. 6A with cleavage conditions; data not shown). However, there was one important exception: DNA isolated under resealing conditions from norfloxacin-treated cells contained the sigma arc that was also present under cleavage conditions. Therefore, the breaks that cause sigma molecule formation are not latent DNA breaks of gyrase cleavage complexes revealed by SDS treatment. These results are therefore consistent with the possibility that the sigma molecules are created by norfloxacin-dependent fork breakage in vivo.
Eyebrow-shaped sigma arcs can be generated by rolling-circle replication (Belanger et al., 1996). To directly compare the sigma arc with bonafide rolling circle intermediates in our system, we examined the 2D gel pattern from recB− cells, which are known to replicate plasmid DNA by both theta and rolling-circle mechanisms (Cohen and Clark, 1986). In the 2D pattern of undigested DNA from recB− cells, the eyebrow-shaped arc was evident, but extended farther in an S-shaped curve (Fig. 7A). Thus, the norfloxacin-induced eyebrow arc in recB+ cells (Fig. 6B and C) has the same shape as the rolling circle arc in recB− cells, but does not extend beyond the dimer size. Upon examination of restriction endonuclease-digested DNA from recB− cells, we found a simple-Y arc with all restriction endonucleases tested (Fig. 7B and data not shown), as predicted for rolling-circle replication.
Fig. 7.
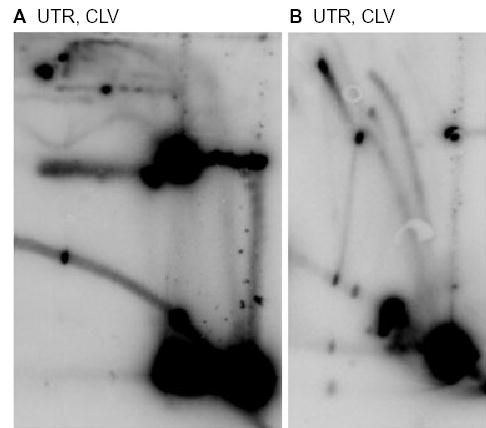
Rolling circle replication in recB cells in the absence of norfloxacin. pBR322 DNA was isolated under cleavage conditions (CLV) from recB− cells without drug treatment (UTR). DNA was undigested (A) or digested with EcoRI (B).
Several points about the eyebrow-shaped arcs are worth emphasizing. First, the beginning of the sigma arc in the DNA isolated from recB− cells comigrated with the eyebrow arc in the norfloxacin-treated samples, confirming the assignment of sigma molecules after norfloxacin treatment. Second, as expected for rolling circle replication, the sigma arc continued beyond the 2.0× length in the DNA isolated from recB− cells. This is precisely where the sigma arc ends in the norfloxacin-treated samples, consistent with broken forks rather than rolling circle replication. Third, norfloxacin-treated recB− cells yield essentially the same 2D gel pattern as norfloxacin-treated wild-type cells (data not shown). Finally, upon cleavage with EcoRI, a large majority of the eyebrow-shaped molecules from the recB− DNA were transformed into simple-Y molecules, with no evidence of the SSY molecules that were prominent after norfloxacin treatment of recB+ cells.
Novel DNA forms depend on DNA gyrase, not topo IV
The experiments presented above with the pBR322MUT990 plasmid strongly suggested that the norfloxacin effects we have uncovered are dependent on DNA gyrase. To further investigate the involvement of DNA gyrase versus topo IV we analysed DNA from cells carrying drug-resistance alleles of gyrA and parC. The 2D gel electrophoresis patterns of both undigested and EcoRI-digested DNA from the parCR cells with and without norfloxacin were essentially the same as those for wild-type cells (Fig. S2). Furthermore, the 2D patterns of DNA isolated from norfloxacin-treated gyrAR cells largely resembled the pattern seen from untreated cells, confirming the role of DNA gyrase in replication fork blockage (Fig. S3). The undigested DNA samples from norfloxacin-treated gyrAR cells showed a faint signal corresponding to simple-Y molecules in the cleavage conditions and sigma molecules in both cleavage and reversal conditions (Fig. S3, lower panels). In addition, restriction endonuclease digestion of these samples resulted in faint simple-Y arcs in cleavage and resealing conditions and faint spots on the bubble arc in resealing conditions (Fig. S3, upper panels). These very weak signals presumably reflect a low frequency of replication fork blockage from the reduced amount of cleavage complex formation in the gyrAR cells (see above; Fig. 1A, lane 8).
Discussion
Norfloxacin treatment causes stalled replication forks in vivo
In this study, we examined replication fork dynamics after norfloxacin treatment by analysing the accumulation of branched DNA using 2D gel electrophoresis. We detected a variety of discrete branched DNA forms along replicative arcs, including strong spots in Y- and bubble-arcs corresponding to the major gyrase binding site at position 990. The intensity of the bubble arc was consistently higher when the DNA was isolated under resealing conditions compared with cleavage conditions (Figs 3 and 5 and data not shown). This result implies that some of the branched molecules contained active gyrase cleavage complexes, such that the DNA was broken when cells were lysed under cleavage conditions (see Fig. 4A, arrow 2). The locations of these cleavage complexes were revealed when we analysed the DNA without restriction enzyme digestion (Fig. 6). In this case, norfloxacin clearly induced the production of TTC, bubbles and simple-Y molecules. As diagrammed in Fig. 4, TTC result when the cleavage complex is located behind the fork, bubbles when the cleavage complex is ahead of the fork, and simple-Y form DNA when the cleavage complex is very close to the branch point. Gyrase is thought to act ahead of the fork to help solve the topological problem during DNA replication (discussed in Postow et al., 1999; 2001; Zechiedrich et al., 2000), consistent with our finding of cleavage complexes ahead of the fork (bubbles and simple-Y molecules). We also found cleavage complexes behind the fork (TTC), and in some non-replicating molecules (linear spot). The generation of simple-Y form DNA is particularly interesting, because it implies that these molecules contained a cleavage complex extremely close to the site of the blocked replication fork. In these cases, disruption of the cleavage complex during lysis under cleavage conditions left a duplex DNA break that was so close to the branch point that it presumably allowed all three branches of the fork to dissociate from each other (Fig. 4, reaction 2). We infer that these branched molecules represent replication forks that have been blocked by physical encounter with the gyrase cleavage complex, and that the blocking cleavage complex was still present at the time of cell lysis.
Some norfloxacin-induced stalled replication forks reverse in vivo
The presence of discrete spots along the bubble arc when DNA was isolated under cleavage conditions (Figs 3B and 5B; Fig. S1) was informative. If the cleavage complex that blocked the fork was still present at the time of isolation, the latent double-stranded break would be revealed upon cell lysis under cleavage conditions. These DNA molecules would not migrate along the bubble arc. Therefore, the gyrase cleavage complex that stalled the replication fork apparently reversed in vivo before isolation (see Fig. 8). A similar conclusion was reached in an analysis of antitumour drug-stabilized topoisomerase cleavage complexes in the phage T4 system (Hong and Kreuzer, 2000).
Fig. 8.
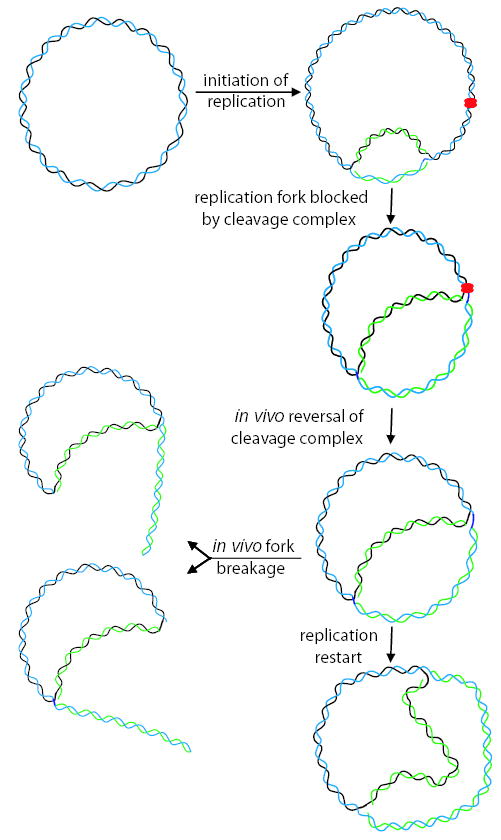
Model for fork blockage and breakage in vivo. Replication is initiated at the unidirectional origin and the fork encounters a norfloxacin-stabilized cleavage complex, resulting in fork blockage. Once the fork is blocked, the cleavage complex can reverse in vivo, presumably allowing for direct fork restart. Alternatively, the blocked replication fork can be processed by in vivo fork breakage (at either the fixed branch or the active fork).
Upon reversal of the cleavage complex in vivo, one might expect the replication fork to quickly resume (Fig. 8; replication restart). However, detection of spots on the bubble arc under cleavage conditions implies that restart is delayed. Replication restart could be delayed if stalling of the replication fork caused dissociation of an essential replication factor. The replicative helicase DnaB is a likely candidate, as loading of DnaB by PriA is required for multiple pathways of replication restart in E. coli (Jaktaji and Lloyd, 2003). The simplest model, direct fork restart, is based on elegant work with phage Mu, where the replicative transposition substrate closely resembles an arrested E. coli replication fork. In this system, DnaB is loaded directly on the substrate in a PriA-dependent reaction without further processing of the DNA (Jones and Nakai, 2000). Other models for fork restart involve regression of the stalled fork into a four-way junction, or formation of a d-loop from a broken intermediate (Marians, 2000; Michel, 2000; Gregg et al., 2002; Jaktaji and Lloyd, 2003).
Norfloxacin appears to trigger fork breakage in vivo
Examination of undigested DNA on 2D gels revealed sigma-shaped molecules after norfloxacin treatment (Fig. 6). These molecules can be created when one of the two parental strands is nicked at or very near a replication fork (see Fig. 4B). We detected sigma molecules when DNA was isolated under cleavage or resealing conditions, strongly suggesting that the breaks were generated in vivo. This is an important difference from the bubble (B) and two-tailed circle (TTC) arcs, which were seen only in DNA isolated under cleavage conditions.
The nature of the sigma molecules was clarified by further examination of restriction enzyme-digested DNA samples (AlwNI digestion in Fig. 3 and EcoRI digestion in Fig. 5). We detected two norfloxacin-dependent arcs when DNA was isolated under cleavage or resealing conditions, corresponding to Y-shaped DNA molecules. Some Y-shaped molecules migrated along an arc that was similar to the simple-Y arc, while a second arc contained asymmetric Y molecules that approached the ‘line of linears’ at approximately 1.9× monomer size for AlwNI-digested DNA and 1.6× monomer size for EcoRI-digested DNA (AY arc in Figs 3 and 5). Because the AY arcs return to the line of linears before reaching linear dimer size, they must be composed of Y molecules with arms of unequal (asymmetric) length. Considering the EcoRI digests, these molecules can be explained by in vivo breakage at the fixed branch near the replication origin and EcoRI digestion in vitro (i.e. EcoRI digestion of molecules in Fig. 4B, left side). In vivo breakage of the active fork that had been stalled by a cleavage complex would generate Y-shaped molecules that progress from monomer size up to the size of the bubble-to-double-Y transition (EcoRI digestion of molecules in Fig. 4B, right side). For AlwNI digests, breakage of the active fork yields the AY arc.
Both the asymmetric-Y (AY) arc and part of the simple-Y arc (for restriction endonuclease-digested DNA), as well as the sigma arc (for undigested DNA), have been described before as a result of breakage of the fixed branch or replicating fork of pBR322 (Martin-Parras et al., 1992; 1998). These authors found broken forks in plasmid DNA from untreated cells (i.e. DNA that was not expected to encounter frequent replication blocks) and concluded that breakage occurred during DNA isolation in vitro. However, we did not detect either the simple-Y or the asymmetric-Y arc (for restriction endonuclease-digested DNA) or the sigma arc (for undigested DNA) in untreated samples. Because the intensity of the bubble arc in our restriction endonuclease-digested samples is similar between untreated and norfloxacin-treated cells isolated under cleavage conditions (Fig. 3A and B), and yet the various arcs corresponding to broken forks appear only in norfloxacin-treated samples, we conclude that the simple-Y, asymmetric-Y and sigma arcs are not produced artifactually during isolation.
How are sigma molecules created?
Is the DNA break that creates sigma molecules from covalently closed circular replication intermediates created directly by DNA gyrase? One relatively trivial model along these lines is that gyrase sometimes leaves a nick at the fork upon resealing in vitro. That is, perhaps our in vitro resealing conditions allow incomplete resealing of gyrase-bound DNA breaks so that only one of the two gyrase monomers reseals its bound DNA break. Several results argue strongly against this model. First, sigma molecules are also produced under cleavage conditions, which would complicate the model and necessitate further assumptions. Second, we know of no convincing evidence that gyrase produces DNA nicks. Finally and perhaps most compelling, the detected breaks appear at both the stalled fork and the fixed branch back at the origin, where a gyrase cleavage complex is not expected.
Another possible model for creation of sigma molecules is replication fork run-off in vivo. In this model, a replication fork collides with a bound cleavage complex and detaches the parental DNA strand that is not covalently bound to the topoisomerase (the lagging strand template). In this model, the remaining gyrase cleavage complex would reseal the other parental template strand, either in vivo or upon lysis under resealing conditions. As with the nicking model described above, the strongest evidence against replication run-off is that fork breakage was detected at both the stalled fork and the fixed branch.
Our data strongly argue for a very different model of DNA breakage, namely that the break results from ‘collateral damage’ after the replication fork is stalled by the cleavage complex (Fig. 8; in vivo fork breakage). In this model, the collateral damage is likely a DNA break induced by a recombination nuclease after the cleavage complex has reversed but before replication has restarted. This model is similar to one proposed for antitumour drug action in the bacteriophage T4 model system, where blocked replication forks accumulated at topoisomerase cleavage complexes in vivo (Hong and Kreuzer, 2000). The amount of blocked forks increased markedly in endonuclease VII-deficient infections, and purified T4 endonuclease VII was shown to cleave purified blocked forks in vitro, arguing that this recombination nuclease is involved in generating the cytotoxic lesion in vivo (Hong and Kreuzer, 2003).
Which processing enzyme(s) might be involved in the creation of the norfloxacin-induced DNA breaks that generate sigma molecules? One candidate is the E. coli Ruv-ABC complex, which was shown to be required for breakage of E. coli replication forks debilitated by a mutation in DnaB or Rep helicase (Seigneur et al., 2000). Because RuvABC acts to resolve Holliday junctions, this chromosomal breakage was argued to occur after regression of stalled replication forks into the chicken foot structure (see Michel, 2000). Interestingly, RuvAB itself can disrupt a norfloxacin-induced topo IV-cleavage complex in vitro, causing reversal of the cleavage complex to yield intact DNA (Shea and Hiasa, 2003). This raises the possibility that RuvAB may play an additional role, dissociating the cleavage complex at the stalled replication fork before cleavage of the Holliday junction by RuvC. Another candidate for the processing endonuclease is SbcCD, which has been shown to selectively cleave branched molecules such as extruded base pairs (Connelly et al., 1998). In addition, an uncharacterized protein such as YqgF, which is related to RuvC by both sequence (Aravind et al., 2000) and structure (Liu et al., 2003) could be involved.
As discussed above, we detected molecules that were apparently broken at either the stalled fork or the fixed branch near the origin. The collateral damage model readily explains breakage of the stalled forks, but how does it explain breakage at the fixed branch near the origin? One possibility is that norfloxacin-induced blocking of plasmid replication forks greatly prolongs the lifetime of the branched intermediate at the origin, thereby exposing it to the same kind of collateral damage that can occur at a stalled fork. Note that chromosomal replication, being bidirectional, does not have the equivalent of such a fixed branch.
If sigma molecules are induced as proposed by the collateral damage model, they could represent the double-stranded breaks that cause cytotoxicity after quinolone treatment. As described in the Introduction, formation of a cleavage complex is necessary but not sufficient for cytotoxicity. The double-stranded end created by endonuclease cleavage could provide an entry site for RecBC, consistent with RecBC-dependent induction of the SOS response (McPartland et al., 1980). There is conflicting evidence as to whether DNA replication is required for induction of the SOS response after quinolone treatment (Gudas and Pardee, 1976; Sassanfar and Roberts, 1990), which would be predicted by the collateral damage model. A previous study inferred the existence of chromosomal DNA breaks in cells treated with bactericidal concentrations of quinolones (Chen et al., 1996), even when DNA was isolated without SDS (i.e. similar to our resealing conditions). These chromosomal breaks could be occurring by the same collateral damage mechanism that we are proposing.
Conclusion
In this study, we presented evidence that E. coli replication forks on plasmid pBR322 are stalled in vivo by norfloxacin-induced gyrase-DNA cleavage complexes. While some stalled forks contained bound gyrase cleavage complexes, others were not bound by gyrase cleavage complexes, indicating that the blocking cleavage complexes had reversed in vivo. We also found sigma molecules in undigested DNA after norfloxacin treatment, under conditions where any transient breaks from the quinolone-DNA-gyrase cleavage complex should have been resealed. Our data support a model in which the sigma molecules are created by ‘collateral damage’, namely endonuclease-mediated cleavage of forks that had been stalled by an encounter with gyrase cleavage complexes.
Experimental procedures
Materials
Stock solutions of norfloxacin (Sigma) at 0.25 mg ml−1 were made in ethanol and stored at −20°C until use. Oligonucleotides for site-directed mutagenesis were purchased from Qiagen Operon. Nylon-S Supercharge blotting membrane was purchased from Schleicher and Schuell. The random-primed labelling kit was obtained from Boehringer Mannheim. Restriction enzymes were purchased from New England Biolabs. Genomic DNA was isolated with Epicentre Technologies MasterPure Complete DNA and RNA Preparation Kit.
Bacterial strains
Strain JH39 [F− sfiA11, thr-1, leu-6, hisG4, argE3, ilv (ts), galK2, srl (?), rpsL31, lacΔU169, dinD1::MudI (Apr lac)] (O’Reilly and Kreuzer, 2004) was considered wild type for these studies. The JH39 recB strain was isolated in a screen for transposon insertion mutants deficient in SOS induction after nalidixic acid treatment (Newmark et al. submitted). The gyrAR (S83L) and parCR (E84K) derivatives of JH39 were created by P1 transduction from donor strain 1643 (Khodursky et al., 1995), with selection for nalidixic acid resistance and kanamycin resistance respectively. The desired mutations in gyrA and parC were confirmed by restriction digestion analysis of polymerase chain reaction (PCR) products from genomic DNA (the S83L mutation results in loss of a HinfI site in gyrA, while the E84K mutation results in addition of a PsiI site in parC). The desired plasmid (pBR322 or pBR322MUT990; see below) was introduced into each strain by transformation, selecting for resistance to tetracycline (10 μg ml−1). A colony that contained mostly monomeric (at least 80%) plasmid was chosen, grown overnight in Luria–Bertani, and aliquots of the overnight culture were frozen and stored at −80°C. These aliquots were used as innoculum for each experiment, so that all experiments involved cells with very similar amounts of monomeric plasmid DNA.
Plasmids
Monomeric plasmids (pBR322 and pBR322MUT990) were gel purified before transformation into desired strains. pBR322MUT990 DNA contains an altered gyrase binding site at the 990 location (ATGGCCTTCCC changed to ATG GCTTTTCC). This change, based on the work of Fisher et al. (1986), was created by site-directed mutagenesis using the Stratagene QuikChange mutagenesis kit and the following oligonucleotides: 5′-GAGGCTGGATGGCTTTTCCCATTATG ATTCTTCTCG-3′, 5′-CGAGAAGAATCATAATGGGAAAAGC CATCCAGCCTC-3′. The nucleotide changes were confirmed by sequencing.
DNA preparations
The method for DNA preparation was based on Hong and Kreuzer (2000) and Hong and Kreuzer (2003), with modifications. Frozen aliquots of the desired strain were thawed and added to fresh LB without antibiotic. Cells were grown with shaking at 37°C to log phase (0.5 < OD600 < 0.6). Norfloxacin (dissolved in ethanol) was added to a final concentration of 1 μg ml−1 (an equivalent volume of ethanol was added to the drug-free controls) and incubated at 37°C with shaking for 6 min.
For experiments involving only cleavage isolation conditions, cells from 1.5 ml of culture were collected by centrifugation and the pellet was frozen with dry ice and ethanol. The pellet was thawed with 300 μl of 1× cleavage lysis buffer [(50 mM Tris-HCl (pH 7.8), 10 mM EDTA, 100 mM NaCl, 0.2% SDS]. Proteinase K was added to 0.5 mg ml−1 and the solution was incubated at 65°C for 1 h. The total nucleic acids were then extracted sequentially with phenol, phenol/chloroform/isoamyl alcohol, and chloroform/isoamyl alcohol before dialysis overnight at 4°C into TE buffer [10 mM Tris-HCl (pH 7.8), 1 mM EDTA].
For all other experiments, cells from 1.5 ml of culture were collected in duplicate by centrifugation and the pellets were kept cold on ice. One set of samples was resuspended in 300 μl of 1× cleavage lysis buffer and incubated at 37°C for 20 min. Proteinase K was then added to 0.5 mg ml−1 and the samples were incubated at 65°C for 1 h. The other set of samples was resuspended in 300 μl of 1× resealing lysis buffer [50 mM Tris-HCl (pH 7.8), 10 mM EDTA, 1% Triton X-100, lysozyme at 1.8 mg ml−1] and incubated at 65°C for 20 min. Proteinase K and SDS were then added (to 0.5 mg ml−1 and 0.2%, respectively) and the samples were incubated at 65°C for 1 h. All samples were then extracted and dialysed as above.
Agarose gel electrophoresis
Nucleic acid samples were treated with the indicated restriction enzymes overnight or undigested samples were loaded directly onto agarose gels. Cleavage complex gels (Fig. 1) contained 1.2% agarose and were run in 0.5× TBE (45 mM Tris-HCl, 45 mM borate, 1 mM EDTA) at 2.8 V cm−1 for 17 h at room temperature. Two-dimensional gel electrophoresis was performed according to the method of Friedman and Brewer (1995) with modifications. Briefly, the first dimension used a 0.4% agarose gel run in 0.5× TBE buffer for 29 h at 1.2 V cm−1 at room temperature. This gel was stained in 0.5× TBE with ethidium bromide (final concentration 0.3 μg ml−1). The marker lanes were cut out and photographed under ultraviolet (UV) light and the sample lanes were cut out based on the migration of the bands in the marker lanes. In this way, sample lanes were not exposed to UV light between dimensions to minimize possible DNA breakage. The sliced sample lanes were cast across the top of a second dimension 1% agarose gel in 0.5× TBE containing 0.3 μg ml−1 ethidium bromide, which was run at 6 V cm−1 for 16 h at 4°C.
Southern hybridization
The gels were treated with 0.25 N HCl for 15–20 min, then denatured in a solution of 3 M NaCl and 0.4 M NaOH for at least 30 min. The gels were transferred to Nylon-S Supercharge membrane as adapted from the downward transfer methods of Koetsier et al. (1993) and Zhou et al. (1994). Briefly, the following items were layered on a tray from bottom to top: 2–3 inches of dry paper towels, 4 sheets of dry thick electrophoresis blotting paper, 4 sheets of dry 3M Whatman paper, 1 sheet of wet 3M Whatman paper, wet nylon membrane, agarose gel, 3 sheets of wet 3M Whatman paper, and a sponge saturated with transfer buffer (1.5 M NaCl, 0.4 M NaOH). Wet items were soaked in transfer buffer briefly before assembly. Downward transfer was allowed to proceed at room temperature for at least 4 h. The nylon membrane was then washed with 4× SSC and crosslinked with a 120 mJ/cm2 UV exposure. The membrane was then blocked with prehybridization buffer (6× SSC, 5× Denhardt’s Solution, 1% SDS, 50% formamide, sonicated salmon sperm DNA at 0.1 mg ml−1) before the probe was added and allowed to hybridize at 42°C for 12–20 h. The probe was a 2.5 kb linear fragment of pBR322 (AlwNI/BamHI fragment) labelled with [α-32P]-dATP by a random-primed DNA labelling kit. This probe avoids hybridization to a chromosomal bla gene present in the JH39 strains. All blots were visualized by both autoradiography and phosphorimager; phosphorimager results are presented in all figures.
Acknowledgments
This work was supported by NIH Grant R01 G072089.
Footnotes
Supplementary material
The following material is available from http://www.blackwellpublishing.com/products/journals/suppmat/mmi/mmi4638/mmi4638sm.htm
Fig. S1. Lysis under in vitro cleavage conditions after norfloxacin treatment causes accumulation of discrete spots on the bubble arc.
Fig. S2. 2D gel electrophoresis pattern of DNA from parCR cells.
Fig. S3. 2D gel electrophoresis pattern of DNA from gyrAR cells.
References
- Aravind L, Makarova KS, Koonin EV. Survey and summary: Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000;28:3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger K, Mirzayan C, Kreuzer H, Alberts B, Kreuzer K. Two-dimensional gel analysis of rolling circle replication in the presence and absence of bacteriophage T4 primase. Nucleic Acids Res. 1996;24:2166–2175. doi: 10.1093/nar/24.11.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AY, Liu LF. DNA topoisomerases: essential enzymes and lethal targets. Annu Rev Pharmacol Toxicol. 1994;34:191–218. doi: 10.1146/annurev.pa.34.040194.001203. [DOI] [PubMed] [Google Scholar]
- Chen CR, Malik M, Snyder M, Drlica K. DNA gyrase and topoisomerase IV on the bacterial chromosome: quinolone-induced DNA cleavage. J Mol Biol. 1996;258:627–637. doi: 10.1006/jmbi.1996.0274. [DOI] [PubMed] [Google Scholar]
- Cohen A, Clark AJ. Synthesis of linear plasmid multimers in Escherichia coli K-12. J Bacteriol. 1986;167:327–335. doi: 10.1128/jb.167.1.327-335.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JC, Kirkham LA, Leach DRF. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc Natl Acad Sci USA. 1998;95:7969–7974. doi: 10.1073/pnas.95.14.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzarelli NR. DNA gyrase and the supercoiling of DNA. Science. 1980;207:953–960. doi: 10.1126/science.6243420. [DOI] [PubMed] [Google Scholar]
- Drlica K. Mechanism of fluoroquinolone action. Curr Opin Microbiol. 1999;2:504–508. doi: 10.1016/s1369-5274(99)00008-9. [DOI] [PubMed] [Google Scholar]
- Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher LM, Mizuuchi K, O’Dea MH, Ohmori H, Gellert M. Site-specific interaction of DNA gyrase with DNA. Proc Natl Acad Sci USA. 1981;78:4165–4169. doi: 10.1073/pnas.78.7.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher LM, Barot HA, Cullen ME. DNA gyrase complex with DNA: determinants for site-specific DNA breakage. EMBO J. 1986;5:1411–1418. doi: 10.1002/j.1460-2075.1986.tb04375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman KL, Brewer BJ. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 1995;262:613–627. doi: 10.1016/0076-6879(95)62048-6. [DOI] [PubMed] [Google Scholar]
- Gellert M. DNA topoisomerases. Annu Rev Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- Gregg AV, McGlynn P, Jaktaji RP, Lloyd RG. Direct rescue of stalled DNA replication forks via the combined action of PriA and RecG helicase activities. Mol Cell. 2002;9:241–251. doi: 10.1016/s1097-2765(02)00455-0. [DOI] [PubMed] [Google Scholar]
- Gudas LJ, Pardee AB. DNA synthesis inhibition and the induction of protein X in Escherichia coli. J Mol Biol. 1976;101:459–477. doi: 10.1016/0022-2836(76)90240-0. [DOI] [PubMed] [Google Scholar]
- Hiasa H, Marians KJ. Two distinct modes of strand unlinking during θ-type DNA replication. J Biol Chem. 1996;271:21529–21535. doi: 10.1074/jbc.271.35.21529. [DOI] [PubMed] [Google Scholar]
- Hiasa H, Yousef DO, Marians KJ. DNA strand cleavage is required for replication fork arrest by a frozen topoisomerase-quinolone-DNA ternary complex. J Biol Chem. 1996;271:26424–26429. doi: 10.1074/jbc.271.42.26424. [DOI] [PubMed] [Google Scholar]
- Hong G, Kreuzer KN. An antitumor drug-induced topoisomerase cleavage complex blocks a bacteriophage T4 replication fork in vivo. Mol Cell Biol. 2000;20:594–603. doi: 10.1128/mcb.20.2.594-603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong G, Kreuzer KN. Endonuclease cleavage of blocked replication forks: an indirect pathway of DNA damage from antitumor drug-topoisomerase complexes. Proc Natl Acad Sci USA. 2003;100:5046–5051. doi: 10.1073/pnas.0835166100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard M, Neece S, Matson S, Kreuzer K. Disruption of a topoisomerase-DNA cleavage complex by a DNA helicase. Proc Natl Acad Sci USA. 1994;91:12031–12035. doi: 10.1073/pnas.91.25.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiang Y, Liu L. Evidence for the reversibility of cellular DNA lesion induced by mammalian topoisomerase II poisons. J Biol Chem. 1989;264:9713–9715. [PubMed] [Google Scholar]
- Jaktaji RP, Lloyd RG. PriA supports two distinct pathways for replication restart in UV-irradiated Escherichia coli cells. Mol Microbiol. 2003;47:1091–1100. doi: 10.1046/j.1365-2958.2003.03357.x. [DOI] [PubMed] [Google Scholar]
- Jones JM, Nakai H. PriA and phage T4 gp59: factors that promote DNA replication on forked DNA substrates. MicroReview. Mol Microbiol. 2000;36:519–527. doi: 10.1046/j.1365-2958.2000.01888.x. [DOI] [PubMed] [Google Scholar]
- Khodursky A, Zechiedrich E, Cozzarelli N. Topoisomerase IV is a target of quinolones in Escherichia coli. Proc Natl Acad Sci USA. 1995;92:11801–11805. doi: 10.1073/pnas.92.25.11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koetsier PA, Schorr J, Doerfler W. A rapid optimized protocol for downward alkaline Southern blotting of DNA. Biotechniques. 1993;15:260–262. [PubMed] [Google Scholar]
- Kreuzer KN, Alberts BM. Site-specific recognition of bacteriophage T4 DNA by T4 type II DNA topoisomerase and Escherichia coli DNA gyrase. J Biol Chem. 1984;259:5339–5346. [PubMed] [Google Scholar]
- Levine C, Hiasa H, Marians KJ. DNA gyrase and topoisomerase IV: biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochim Biophys Acta. 1998;1400:29–43. doi: 10.1016/s0167-4781(98)00126-2. [DOI] [PubMed] [Google Scholar]
- Liu D, Wang YS, Wyss DF. Solution structure of the hypothetical protein YqgF from Escherichia coli reveals an RNAse H fold. J Biomol NMR. 2003;27:389–392. doi: 10.1023/a:1025840121177. [DOI] [PubMed] [Google Scholar]
- Lockshon D, Morris DR. Sites of reaction of Escherichia coli DNA gyrase on pBR322 in vivo as revealed by oxolinic acid-induced plasmid linearization. J Mol Biol. 1985;181:63–74. doi: 10.1016/0022-2836(85)90324-9. [DOI] [PubMed] [Google Scholar]
- Lucas I, Germe T, Chevrier-Miller M, Hyrien O. Topoisomerase II can unlink replicating DNA by precatenane removal. EMBO J. 2001;20:6509–6519. doi: 10.1093/emboj/20.22.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel LS, Rogers LH, Hill WE. Survival of recombination-deficient mutants of Escherichia coli during incubation with nalidixic acid. J Bacteriol. 1978;134:1195–1198. doi: 10.1128/jb.134.3.1195-1198.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPartland A, Green L, Echols H. Control of recA gene RNA in E. coli: regulatory and signal genes. Cell. 1980;20:731–737. doi: 10.1016/0092-8674(80)90319-0. [DOI] [PubMed] [Google Scholar]
- Marians KJ. PriA-directed replication fork restart in Escherichia coli. Trends Biochem Sci. 2000;25:185–189. doi: 10.1016/s0968-0004(00)01565-6. [DOI] [PubMed] [Google Scholar]
- Martin-Parras L, Hernandez P, Martinez-Robles ML, Schvartzman JB. Unidirectional replication as visualized by two-dimensional agarose gel electrophoresis. J Mol Biol. 1991;220:843–853. doi: 10.1016/0022-2836(91)90357-c. [DOI] [PubMed] [Google Scholar]
- Martin-Parras L, Hernandez P, Martinez-Robles M, Schvartzman J. Initiation of DNA replication in ColE1 plasmids containing multiple potential origins of replication. J Biol Chem. 1992;267:22496–22505. [PubMed] [Google Scholar]
- Martin-Parras L, Lucas I, Martinez-Robles M, Hernandez P, Krimer D, Hyrien O, Schvartzman J. Topological complexity of different populations of pBR322 as visualized by two-dimensional agarose gel electrophoresis. Nucleic Acids Res. 1998;26:3424–3432. doi: 10.1093/nar/26.14.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B. Replication fork arrest and DNA recombination. Trends Biochem Sci. 2000;25:173–178. doi: 10.1016/s0968-0004(00)01560-7. [DOI] [PubMed] [Google Scholar]
- O’Reilly EK, Kreuzer KN. Isolation of SOS constitutive mutants of Escherichia coli. J Bacteriol. 2004;186:7149–7160. doi: 10.1128/JB.186.21.7149-7160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheroff N, Zechiedrich EL. Calcium-promoted DNA cleavage by eukaryotic topoisomerase II: trapping the covalent enzyme-DNA complex in an active form. Biochemistry. 1987;26:4303–4309. doi: 10.1021/bi00388a018. [DOI] [PubMed] [Google Scholar]
- Postow L, Peter BJ, Cozzarelli NR. Knot what we thought before: the twisted story of replication. Bioessays. 1999;21:805–808. doi: 10.1002/(SICI)1521-1878(199910)21:10<805::AID-BIES1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Postow L, Crisona NJ, Peter BJ, Hardy CD, Cozzarelli NR. Topological challenges to DNA replication: conformations at the fork. Proc Natl Acad Sci USA. 2001;98:8219–8226. doi: 10.1073/pnas.111006998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M, Hsieh T. Double strand DNA cleavage by type II DNA topoisomerase from Drosophila melanogaster. J Biol Chem. 1983;258:8421–8428. [PubMed] [Google Scholar]
- Sassanfar M, Roberts JW. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J Mol Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- Schvartzman J, Martinez-Robles M, Hernandez P. The migration behavior of DNA replicative intermediates containing an internal bubble analyzed by two-dimensional agarose gel electrophoresis. Nucleic Acids Res. 1993;21:5474–5479. doi: 10.1093/nar/21.23.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigneur M, Ehrlich SD, Michel B. RuvABC-dependent double-strand breaks in dnaBts mutants require RecA. Mol Microbiol. 2000;38:565–574. doi: 10.1046/j.1365-2958.2000.02152.x. [DOI] [PubMed] [Google Scholar]
- Shea ME, Hiasa H. Interactions between DNA helicases and frozen topoisomerase IV-quinolone-DNA ternary complexes. J Biol Chem. 1999;274:22747–22754. doi: 10.1074/jbc.274.32.22747. [DOI] [PubMed] [Google Scholar]
- Shea ME, Hiasa H. The RuvAB branch migration complex can displace topoisomerase IV-quinolone-DNA ternary complexes. J Biol Chem. 2003;278:48485–48490. doi: 10.1074/jbc.M304217200. [DOI] [PubMed] [Google Scholar]
- Strumberg D, Pilon AA, Smith M, Hickey R, Malkas L, Pommier Y. Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5′-phosphorylated DNA double-strand breaks by replication runoff. Mol Cell Biol. 2000;20:3977–3987. doi: 10.1128/mcb.20.11.3977-3987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino A, Peebles CL, Kreuzer KN, Cozzarelli NR. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc Natl Acad Sci USA. 1977;74:4767–4771. doi: 10.1073/pnas.74.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- Wang HK, Morris-Natschke SL, Lee KH. Recent advances in the discovery and development of topoisomerase inhibitors as antitumor agents. Med Res Rev. 1997;17:367–425. doi: 10.1002/(sici)1098-1128(199707)17:4<367::aid-med3>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Wentzell LM, Maxwell A. The complex of DNA gyrase and quinolone drugs on DNA forms a barrier to the T7 DNA polymerase replication complex. J Mol Biol. 2000;304:779–791. doi: 10.1006/jmbi.2000.4266. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Bogaki M, Nakamura M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother. 1990;34:1271–1272. doi: 10.1128/aac.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechiedrich EL, Khodursky AB, Bachellier S, Schneider R, Chen D, Lilley DMJ, Cozzarelli NR. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J Biol Chem. 2000;275:8103–8113. doi: 10.1074/jbc.275.11.8103. [DOI] [PubMed] [Google Scholar]
- Zhou MY, Xue D, Gomez-Sanchez EP, Gomez-Sanchez CE. Improved downward capillary transfer for blotting of DNA and RNA. Biotechniques. 1994;16:58–59. [PubMed] [Google Scholar]


