Abstract
Pyrimethamine (Pyr) targets dihydrofolate reductase of Plasmodium vivax (PvDHFR) as well as other malarial parasites, but its use as antimalarial is hampered by the widespread high resistance. Comparison of the crystal structures of PvDHFR from wild-type and the Pyr-resistant (SP21, Ser-58 → Arg + Ser-117 → Asn) strain as complexes with NADPH and Pyr or its analog lacking p-Cl (Pyr20) clearly shows that the steric conflict arising from the side chain of Asn-117 in the mutant enzyme, accompanied by the loss of binding to Ser-120, is mainly responsible for the reduction in binding of Pyr. Pyr20 still effectively inhibits both the wild-type and SP21 proteins, and the x-ray structures of these complexes show how Pyr20 fits into both active sites without steric strain. These structural insights suggest a general approach for developing new generations of antimalarial DHFR inhibitors that, by only occupying substrate space of the active site, would retain binding affinity with the mutant enzymes.
Keywords: drug resistance, malaria, antifolates
A major share of the global malaria burden, with an estimated 80 million cases annually, is caused by Plasmodium vivax (Pv) (1, 2). The problem has recently been worsened by emergence of resistance of the parasite to chloroquine (3–6). Pyrimethamine (Pyr) and other antifolates are generally not used against vivax malaria, because of the resistance of the parasite, which has commonly been considered to be inherent (7). It recently has been shown, however, that wild-type (WT) P. vivax dihydrofolate reductase (PvDHFR), the target of antifolates, is susceptible to inhibition by Pyr and other antifolates (8–11), whereas for the mutant parasites, of which the common one is the double mutant (SP21, Ser-58 → Arg + Ser-117 → Asn), the affinities for binding with the antifolates are much reduced, rendering them ineffective. A similar, but less severe, situation was shown for Plasmodium falciparum (Pf), in which the homologous double mutant (K1, Cys-59 → Arg + Ser-108 → Asn) PfDHFR has reduced affinities for the antifolates (12–14). In the case of P. falciparum, there was only a moderate reduction in affinity (50- to 90-fold) relative to the WT enzyme, and a moderate level of resistance ensued, which was augmented by further mutations at other sites (14–17). The crystal structures of the bifunctional enzyme dihydrofolate reductase (DHFR)-thymidylate synthase (TS) of P. falciparum (PfDHFR-TS) revealed basic structural features of Plasmodial DHFR-TS, including the insert regions and the junction region (18). Comparison of the structures of the WT and mutant forms of PfDHFR-TS demonstrates that Pyr is involved in steric conflict with the side chain of Asn-108, resulting in antifolate resistance (13, 19, 20). However, the steric conflict did not appear to result in major displacement of Pyr in the active site of P. falciparum. In contrast, analogous mutations in P. vivax resulted in relatively larger reduction in binding affinity of Pyr. Because the crystal structure of PvDHFR, either of the WT or the SP21 double-mutant strain, had not been determined before, it was unclear how Pyr binds to the active site and how this binding is affected by the resistance mutations. Here, we report the structures of the 238-residue PvDHFR domain of the bifunctional PvDHFR-TS, of both the WT and double-mutant SP21, in complex with NADPH and inhibitor of either Pyr or Pyr20, an analog lacking the p-Cl atom. These structures allowed us to explore the relationship between the modes of drug binding and affinities to the active site, which are in turn linked with the levels of drug resistance. We show that, although the structures of the enzymes and the modes of binding are similar for both parasites, there are significant differences that explain the relatively higher effect of homologous mutations on PvDHFR on Pyr-binding affinity and generation of resistance. We show further that, unlike Pyr, Pyr20, which stays mainly in the confine of the space occupied by the substrate [dihydrofolate (DHF)], still largely retains its binding affinity to the mutant enzyme.
Materials and Methods
Protein Expression, Purification, and Crystallization. The protein expression and protein purification were carried out according to a protocol described in ref. 10. Briefly, Escherichia coli BL21(DE3)pLysS harboring pETPvDHFR was cultured in a LB broth with 100 μg·ml–1 ampicillin and 34 μg·ml–1 chloramphenicol at 18°C with isopropyl β-d-thiogalactoside induction. The WT PvDHFR and SP21 double-mutant PvDHFR were purified by a single-step affinity chromatograph on an methotrexate–Sepharose column and eluted by a solution of 4 mM DHF in 20 mM N-Tris(hydroxymethyl)methyl-2-aminoethane sulfonic acid (pH 8.0), 0.1 mM EDTA, and 10 mM DTT. The purified enzymes were desalted and concentrated to 10 mg·ml–1 by using a Centricon filter.
Before crystallization, a 10 mg·ml–1 PvDHFR solution was mixed with 1 mM NADPH and 1 mM Pyr (Ki = 0.16 nM) and incubated overnight at 4°C. Suitable crystals of ternary PvDHFR–NADPH-inhibitor complexes were grown by the microbatch method (21) with a mixture of 1.5 μl of protein complex and the same volume of 30% polyethylene glycol 4000, 100 mM Tris·HCl buffer (pH 7.2), and 10% glycerol. PvDHFR crystals grown under these conditions have the typical dimensions of 0.10 × 0.15 × 0.20 mm and belong to the monoclinic space group C2. With one molecule of PvDHFR complex per asymmetric unit, the solvent content was 59%. Cocrystals of WT PvDHFR with Pyr20 (Ki = 1.55 nM) were prepared by ligand-exchange soaking of the WT PvDHFR–Pyr complex crystals in a stabilizing solution containing 3 mM Pyr20 for 2–3 days at 4°C. Complexes of SP21 PvDHFR with 1 mM Pyr or Pyr20 were cocrystallized in 100 mM Mes at pH 6.0. Before flash-cooling, the crystals were transferred into a reservoir solution supplemented with 20% glycerol and immediately frozen in the cryostream. All diffraction data sets were measured at 120 K on a generator (Rigaku, Tokyo) with an R-AXIS IV++ detector. All data were integrated and scaled by using d*trek (22). Data processing and refinement statistics are given in Table 1.
Table 1. Data collection and structure refinement statistics.
| Parameters | WT PvDHFR—Pyr | WT PvDHFR—Pyr20 | SP21—Pyr | SP21—Pyr20 |
|---|---|---|---|---|
| Space group | C2 | C2 | C2 | C2 |
| Unit cell a, b, and c, Å; and β, o | 134.28, 55.74, 45.72, 107.09 | 130.48, 55.57, 45.27, 106.76 | 135.27, 54.40, 46.09, 108.13 | 132.00, 56.31, 45.67, 107.50 |
| Resolution, Å | 20-1.90 | 20-3.00 | 20-2.50 | 20-2.25 |
| Completeness, % (final shell, %) | 99.1 (99.1) | 99.7 (99.9) | 98.9 (99.7) | 97.5 (97.5) |
| Unique reflections | 24,153 | 6,307 | 11,042 | 14,912 |
| Redundancy | 3.21 | 3.40 | 4.85 | 3.72 |
| Rsym, % (final shell)* | 4.4 (40.6) | 7.9 (31.5) | 9.2 (37.9) | 4.5 (30.4) |
| 〈I/σ〉 (final shell) | 14.8 (3.3) | 11.5 (3.8) | 10.1 (3.8) | 11.8 (2.0) |
| R factor/Rfree,† % | 20.79/25.82 | 19.08/29.99 | 19.39/28.18 | 21.33/27.60 |
| Average B factor, Å2 | 53.5 | 41.63 | 49.69 | 63.80 |
| No. of atoms | 2,094 | 1,907 | 2,005 | 2,031 |
| Nonhydrogen protein | 1763 | 1781 | 1769 | 1760 |
| Ligands | 65 | 77 | 76 | 76 |
| Solvents | 266 | 147 | 62 | 195 |
| rms deviation | ||||
| Bond lengths, Å | 0.0073 | 0.0096 | 0.0091 | 0.0079 |
| Bond angles, o | 1.187 | 1.204 | 1.190 | 1.160 |
Rsym = Σ|I — 〈I〉|/ΣI, where I is the observed intensity and 〈I〉 is the average intensity from multiple observations of symmetry-related reflections.
R factor = Σhkl|Fo(hkl) — Fc(hkl)|/Σhkl Fo(hkl), where Fo(hkl) and Fc(hkl) are the observed and calculated structure factor amplitudes for reflection hkl. Rfree is computed by using randomly selected 5% of the data that were excluded throughout refinement.
Structure Determination and Refinement. The structure of PvDHFR–NADPH–Pyr ternary complex was determined by using the molecular replacement method using the structure of the N-terminal DHFR domain of PfDHFR-TS [Protein Data Bank ID code 1J3J] as a search model with the program ccp4-amore (23). Electron density maps were inspected and models were manually built by using the graphics program xtalview (24). The refinement was carried out by using cns (25) and included individual B factor refinement, simulated annealing, and positional refinement using the parameters of Engh and Huber (26). The other three PvDHFR complex structures were solved with difference Fourier technique or with molecular replacement by using the structural model of the WT PvDHFR–NADPH–Pyr complex and refined with cns. Water molecules were inserted automatically and inspected manually. The model displays good stereochemistry with no disallowed ϕ/ψ angles for all of the final models (Table 1), as verified by procheck (27). Structural alignments were performed by ccp4-tops (28), and figures were prepared with pymol (www.pymol.org).
Kinetic Studies. Inhibition experiments were performed as described in ref. 10, and the inhibition constants (Ki) were calculated from a nonlinear least-square equation for competitive inhibitor by using kaleidagraph 3.5 (Synergy Software, Reading, PA).
Results and Discussion
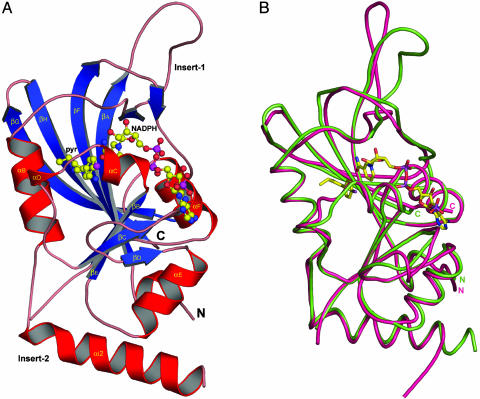
Overall Structure of P. vivax DHFR. The crystal structure of 238-residue N-terminal DHFR domain of the bifunctional PvDHFR-TS enzyme shows that PvDHFR is a monomer of ≈38 × 49 × 62 Å, whereas the full-length PfDHFR-TS exists as a homodimeric enzyme with extensive intersubunit contacts between the two TS domains. The PvDHFR domain, consisting of an eight-stranded mixed β-sheet (βA-H) flanked by four α-helices (αb,c,e,f) (Fig. 1A), shares a similar overall polypeptide fold to that characteristic of other DHFRs (29) with two additional short α-helices (αA and αD). Similarity can be noted in the overall folding with that previously modeled structure (30), although there are many significant differences, particularly in the insert regions. The DHFR domains of P. vivax and P. falciparum share 66% sequence identity, and their structures can be superimposed well with a rms deviation (rmsd) of 0.87 Å for 194 Cα atoms (Fig. 1B). Plasmodial DHFRs has two unique regions of insert sequences (in P. vivax; Insert-1, residues 19–35, and Insert-2, residues 63–107). In our PvDHFR structure, Insert-1 adopted a flexible loop connecting between βA and αB, with a different orientation from that of PfDHFR-TS structure by as much as 13–15 Å for residues 25–28 (Fig. 1B). Because the Insert-1 of PfDHFR has been shown to interact with the TS domain, the difference in the insert arrangement may be due to the lack of the intact TS domain in the PvDHFR structures presented or may indicate the presence of different interactions in the enzymes from the two species. Although crystals of PvDHFR-TS had been obtained, they gave diffraction quality with insufficient resolution to yield the structure of the bifunctional enzyme for direct comparison with PfDHFR-TS (P.K., unpublished results). Similar to that of PfDHFR, although distinct from the homology model, Insert-2 formed another α-helical stretch αi2 (residues 66–84). Conserved basic residues in the middle of αi2, pointing toward the interior of the protein, make extensive contacts with the globular structure and hence stabilize its orientation. Although Lys-75 forms polar contacts with the main-chain C O of Asp-6 and the carboxylate side chain of Asp-9, Arg-76 extensively interacts with the carboxylate group of Asp-9, main-chain carbonyl oxygen atoms of Val-7, Tyr-167, and Tyr-168. At the C terminus of αi2, the side chain of Glu-83 interacts with the amine group of the Lys-164. No electron density for residues 85–105 in the Insert-2, probably due to protein flexibility, was visible, and thus these residues were not included in the model. The flexible conformation was revealed by relatively higher B factor toward the visible region of Insert-2, of which conserved residues comprise several basic residues, suggesting a possible mRNA binding site and translational regulation of the protein (31).
O of Asp-6 and the carboxylate side chain of Asp-9, Arg-76 extensively interacts with the carboxylate group of Asp-9, main-chain carbonyl oxygen atoms of Val-7, Tyr-167, and Tyr-168. At the C terminus of αi2, the side chain of Glu-83 interacts with the amine group of the Lys-164. No electron density for residues 85–105 in the Insert-2, probably due to protein flexibility, was visible, and thus these residues were not included in the model. The flexible conformation was revealed by relatively higher B factor toward the visible region of Insert-2, of which conserved residues comprise several basic residues, suggesting a possible mRNA binding site and translational regulation of the protein (31).
Fig. 1.
Crystal structure of the DHFR domain of P. vivax. (A) Structure of P. vivax DHFR complexed with NADPH and Pyr; α-helices are in red, and β-strands are in blue, including the Insert-1 loop and the Insert-2 α-helix. The carbons, nitrogen, oxygen, and chlorine atoms of Pyr and NADPH are shown in yellow, blue, red, and magenta, respectively. (B) Comparison of the DHFR domains from P. vivax (green) and P. falciparum (magenta). The superimposed structures demonstrate overall structural similarity with major deviation in the insert regions.
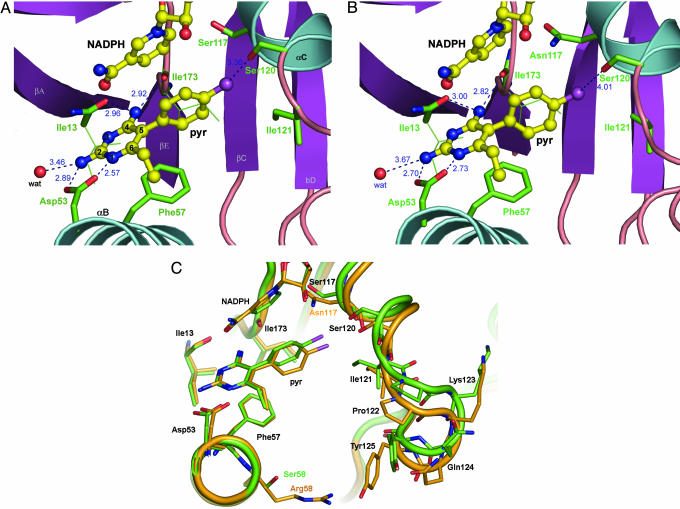
Pyr Binding with the Active Site. Pyr binds at the active site of PvDHFR with its pyrimidine ring buried in the interior of the deep cleft through hydrogen bonds (H-bonds) and van der Waals interactions (Fig. 2A). H-bonds play a significant role in the binding of the Pyr; N1 and 2-amino nitrogen atoms of Pyr are H-bonded to the carboxylate oxygen atoms of Asp-53 (distances of 2.57 and 2.89 Å, respectively), and the 4-amino nitrogen atom is H-bonded to the main-chain carbonyl oxygen atoms of Ile-13 and Ile-173 (distances of 2.96 and 2.92 Å, respectively). In addition, the N2 amino nitrogen atom interacts with a welldefined water molecule, which sits deep in the active site (Fig. 2 A). Although the pyrimidine ring is in van der Waals contact with Phe-57, Ile-13, and Leu-45, its p-chlorophenyl ring is in contact with the nicotinamide moiety of the NADPH cofactor. Interestingly, the p-Cl atom of Pyr lies ≈3.36 Å from the hydroxyl group of Ser-120 with an angle C-Cl···(H)O of 141.8° that can be classified as a H-bond between a weak acceptor and a strong donor, respectively (32, 33). Conformations of Pyr in its unbound and bound states are different such that upon binding to PvDHFR, the angle between the pyrimidine and the phenyl rings of Pyr changes from –116.7° in the free state to –66.3° in the bound form. Because the geometry of the pyrimidine moiety is locked in the binding pocket, the only possible free adjustment is by a simple rotation around a single bond connecting between the two aromatic rings to maximize van der Waals interactions of the p-chlorophenyl moiety to the nicotinamide group of the NADPH cofactor.
Fig. 2.
Pyr bound in the PvDHFR active site. The Pyr and NADPH cofactor are shown as balls and sticks with carbon, nitrogen, and chlorine colored yellow, blue, and magenta, respectively. (A) Pyr binding with the WT PvDHFR. Interactions between the enzyme and the pyrimidine ring of the inhibitor include electrostatic interactions and H-bonds indicated by dotted lines. Numbers next to the lines indicate distances in Å. (B) Pyr binding with the SP21 double-mutant PvDHFR. Interactions around the pyrimidine ring are similar to the WT enzyme. The mutation at codon 117 from Ser to Asn increases a steric factor in the active site, and, as a result, the positions of both NADPH and Pyr are perturbed from their optimum binding, reducing the efficiency of Pyr by as much as 300-fold. (C) Superposition of Pyr-binding sites in the WT PvDHFR (green) and the SP21 double-mutant enzyme (orange) with a rmsd of 0.53 Å. While the position of pyrimidine is held in the same place, the mutation at S117N causes the displacement of p-chlorophenyl moiety of Pyr from its optimal binding with the p-Cl atom shifted by 1.1 Å and the torsion plane between the two rings twisted by –32°. The mutation also caused a local main-chain movement of residues 118 –125 (0.60 –1.88 Å), with respect to the WT enzyme. The mutation at codon 58 from Ser to Arg is not in proximity with the Pyr-binding site.
Enzyme Changes Induced by Mutations. Fig. 2 A and B shows the structures of the active sites of the PvDHFR from the WT and SP21 mutant, complexed with Pyr. The amino acid residues equivalent to those implicated in substrate catalysis and inhibitor binding of PfDHFR (18, 34) also are found in the active sites of PvDHFR from both sources. The overall structures of WT and mutant enzymes are similar with rmsd fit for all atoms of 0.531 Å. The pyrimidine moiety of the inhibitors is held tightly in the same position in both PvDHFR complexes, whereas the position of the phenyl ring is perturbed by the mutation at codon 117. Nonetheless, distinguishable differences in the orientation of Pyr and NADPH in the active sites of the WT and mutant enzymes can be clearly observed in the binding pocket due to the mutation at Ser-117 → Asn. In the structure of SP21 mutant complexed with Pyr, the Ser-117 → Asn mutation caused steric conflict with the p-Cl atom, resulting in displacement of both the p-chlorophenyl ring of the inhibitor and a local conformational rearrangement of the peptide chain at residues 118–125, with consequent loss of favorable interactions between the enzyme and the inhibitor. In addition, there was a main-chain movement of αF (residues 174–178) by 0.3–0.9 Å in the SP21 structure in conjunction with displacement of NADPH. The nicotinamide moiety of the cofactor was both shifted away (0.52 Å) and slightly rotated (dihedral angle of –119.5° to –131.8°) from the optimum position found in the WT enzyme.
Although the conjugated mutation at Ser-58 to Arg did not directly interact with the inhibitor, it might be involved in the substrate (DHF or dihydrofolylpolyglutamate) binding (18). This mutation, located on the αB helix, from Ser to Arg, created an additional positive charge near the entrance of the active-site cavity but did not perturb the orientation of the helix. In contrast to the equivalent mutation of C59R in PfDHFR-TS structure of which the mutated Arg pointed out into the solvent, the conformation of Arg-58 in the SP21 PvDHFR pointed toward the active-site cavity. Although the position of Arg-58 was too far to interact with the Pyr inhibitor, it in turn interacted with the bound 4-morpholinoethanesulfonate (Mes) molecule used as a crystallization buffer. The Mes molecule was situated in an equivalent position to the α-carboxylic group of the DHF substrate found in the E. coli DHFR complex (35), and its binding was mainly favored by salt-bridge interaction between the sulfonate group of Mes and the positive-charged side chain of Arg-131 and Arg-58.
Changes in Inhibitor Binding. Conformational changes of the loop 118–125 are necessary to accommodate Pyr binding in SP21. The dihedral angle of the two aromatic planes of Pyr in the two structures differed by 32° (–66.3° in WT and –98.2° in SP21). The p-chlorophenyl ring also was tilted from its original position with the distance difference at the chlorine atom of 1.14 Å. In contrast, a significant displacement is absent in Pyr binding with the equivalent double-mutant K1 PfDHFR (P.C., unpublished results). Moreover, upon accommodation of Pyr, a large conformational change was observed for the SP21–Pyr complex owing to a movement at a peptide region 118–125 by as much as 1.74–1.84 Å for the Cα atoms of Pro-122 to Gln-124, ascribed to repulsive interaction between Ile-121 side chain and the p-Cl group (Fig. 2C). Clearly, the change in the SP21–Pyr complex perilously affects the interaction between the strong H-bond donor hydroxyl side chain of Ser-120 and the weak H-bond acceptor p-Cl group of the Pyr with the distance, O(H)···Cl, of 3.36 Å in WT to 4.01 Å in SP21 (32, 33). In addition, the retention of the H-bond (≈3.30 Å) between the side chain of Ser-111 and the p-Cl group of Pyr in P. falciparum indicates that the mutation at Asn-108 in K1 perturbed the inhibitor less than that in P. vivax. This structural feature explains why the mutation at Ser-117 causes more reduction in enzyme-binding affinity and a higher level of resistance to Pyr in P. vivax than in P. falciparum.
The difference in energy caused by loss of the H-bond of Ser-120···p-Cl and the loss in van der Waals interaction, together with any entropic penalties paid by the conformational change of the peptide chain at 118–125, resulted in >300-fold loss of Pyr-binding affinity for the double-mutant SP21 PvDHFR, compared with 50- to 90-fold reduction in the K1 PfDHFR-TS (14, 17). The complex of Pyr with the mutant enzyme shows that both Pyr and NADPH are displaced with respect to the WT structure. This displacement of NADPH may contribute to the reduced Pyr-binding affinity, which corresponds to a difference of ≈3.4 kcal·mol–1 between mutant and WT enzymes (Table 2) (36, 37).
Table 2. Inhibition constant (Ki) of recombinant P. vivax DHFR with antifolate inhibitors.
|
Ki, nM
|
Δ(ΔG°)Pyr-Pyr20,† kcal·mol-1
|
||||
|---|---|---|---|---|---|
| Inhibitor | PvDHFR (WT) | PvDHFR SP21 (S58R+S117N) | Δ(ΔG°)wt-SP21,* kcal·mol-1 | WT | SP21 |
| Pyr | 0.16 ± 0.03 | 50 ± 4.0 | 3.40 | 1.34 | -1.14 |
| Pyr20 | 1.55 ± 0.12 | 7.33 ± 0.49 | 0.92 | ||
Δ(ΔG°)wt-SP21 is the relative binding affinity in comparison of WT—ligand complex with SP21—ligand complex, —RT In(Ki,wt/Ki,SP21).
Δ(ΔG°)Pyr-Pyr20 is the relative binding affinity of protein in complex with Pyr and Pyr20, —RT In(Ki,pyr/Ki,Pyr20).
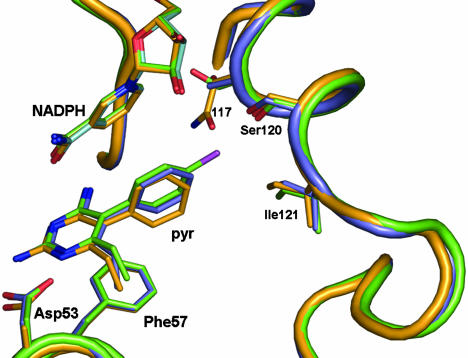
To clarify the effect of the p-Cl moiety of the Pyr, the structures of both WT and SP21 mutant enzymes in complex with Pyr20 were determined (Fig. 3). In WT PvDHFR complexed with Pyr20, the overall structure and the active site were very similar to those of the WT PvDHFR–Pyr complex (rmsd of all Cα atoms = 0.36 Å), and the two inhibitors were in almost identical positions with the dihedral angle between the two rings of –72.3°. However, the binding affinity was reduced by 1.34 kcal·mol–1, presumably equivalent to an internal H-bond in the former complex, apparently due to the lack of the p-Cl group on the phenyl substituent of Pyr20 (37).
Fig. 3.
Superposition of the Pyr binding-site of WT PvDHFR–Pyr20 (deschloropyrimethamine) complex (cyan) and SP21 PvDHFR–Pyr20 complex (orange) onto of the WT PvDHFR–Pyr complex (green). The steric hindrance of Asn-117 is lessened in the case of Pyr20-bound enzymes with neither movement of the nicotinamide moiety nor the local changes in residues 118–125 but only slight displacement of phenyl ring. Pyr20 binds with the WT enzyme similar to Pyr. Pyr, Pyr20, and NADPH are depicted as stick models.
In the ternary complex structure of SP21 with bound NADPH and Pyr20 (Fig. 3), the steric effect from the mutation of Asn-117 was greatly lessened because of the absence of the p-Cl atom in Pyr20. The side chain of Asn-117 readjusted itself to fill up the empty cavity resulting from the missing p-Cl atom, such that it interacted with the main-chain carbonyl groups of Ile-173 and Gly-174 as well as H-bonded to the hydroxyl group of Ser-120. Consequently, NADPH returned to its optimal position observed in the WT enzyme. Unlike that of the SP21–Pyr complex, there was neither conformational perturbation of the residues 118–125 nor the NADPH displacement observed in the SP21–Pyr20 complex, substantiating the conclusion that the p-Cl group was responsible for the conformational change upon the binding of inhibitor to release the unfavorable contact with the Ile-121 side chain. Nevertheless, the bound Pyr20 was rotated in a similar fashion to the bound Pyr seen in the SP21 mutant, with the dihedral angle of –84.1°, indicating that the larger side chain at codon 117 resulting from the mutation still affects the position of the phenyl ring even without the p-Cl atom, and hence the favorable van der Waals interaction has been only partly recovered.
In conclusion, different geometries between Pyr and Pyr20 in the WT and SP21 double mutant of PvDHFRs reflect the effect of the mutations in causing resistance to Pyr and the failure to cause resistance when the Cl atom is removed from the inhibitor. The steric constraint around the side chain of Asn-117 is mainly responsible for the loss of affinity of Pyr for PvDHFR, with consequent displacement of both the inhibitor and NADPH, and conformational changes to optimize binding. The detailed structural results we report for drug complexes of the WT and SP21-resistant strain of PvDHFR support a compelling explanation for the mechanism of molecular resistance.
Implications for the Molecular Mechanism of Resistance. Our observation leads to some important conclusion for the mechanism of resistance of P. vivax to antifolate drugs. First, because our results show that Pyr binds well with the WT PvDHFR, the WT parasite is likely sensitive to the drug and only becomes resistant on mutations that disturb drug binding, supporting earlier observations (10, 38, 39). Second, the poorer binding of Pyr to SP21 PvDHFR is caused by steric clash between Ser-117 → Asn and the p-Cl atom, whereas the Pyr20 (which fits approximately within the substrate envelope) binds relatively well to both WT (1.5 nM) and SP21 mutant enzyme (7 nM). Resistance therefore was generated through mutations that can occur to disrupt binding with the parts of the inhibitor that protrude beyond the envelope of the substrate. This general idea of developing resistance-avoiding inhibitors also is supported by our previous work on the Pf-DHFRTS–WR99210 complex (34), which showed that this flexible inhibitor binds well to both WT and Pyr-resistant strains by adopting a conformation that keeps it within the substrate envelope. A similar picture has been shown for resistance to HIV protease inhibitor drugs (40, 41). Structural studies in HIV protease show that the small flexible nature of the inhibitor such as tenofovir enables it to stay in the small substrate envelope and avoid the surveillance mechanism used by reverse-transcriptase inhibitor-resistance mutants, whereas those inhibitors protruding beyond the substrate envelope can develop mutations in the active site that confer resistance.
These structural results point the way to the design of new generations of antimalarial antifolate drugs in which the basic structural rules for avoiding resistance are now postulated. One approach is to design inhibitors that stay approximately within the substrate space of the DHFR, such as Pyr20 (39, 42). However, because these inhibitors may still induce further resistance as shown in ref. 15, additional approaches, such as employing combination of inhibitors that require different sets of mutations to generate resistance as has been noted for Pyr and WR99210 (39, 42), are needed.
Acknowledgments
This work was supported by the Thailand Research Fund and the Thailand-Tropical Diseases Research Program (P. Kongsaeree), the Wellcome Trust (Y.Y. and M.D.W.), and the Medicines for Malaria Venture and European Union International Cooperation–Developing Countries (INCO-Dev) (Y.Y.). P. Khongsuk was supported by scholar-ships from the Postgraduate Education and Research Program in Chemistry and National Centre for Genetic Engineering and Biotechnology (Thailand).
Author contributions: P. Kongsaeree, U.L., P.C., M.D.W., and Y.Y. designed research; P. Kongsaeree, P. Khongsuk, U.L., and P.C. performed research; B.T. contributed new reagents/analytic tools; P. Kongsaeree, P. Khongsuk, U.L., P.C., and Y.Y. analyzed data; and P. Kongsaeree, U.L., P.C., M.D.W., and Y.Y. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DHF, dihydrofolate; DHFR, DHF reductase; Pf, Plasmodium falciparum; Pv, Plasmodium vivax; Pyr, pyrimethamine; rmsd, rms deviation; TS, thymidylate synthase.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2bl9, 2blb, 2bla, and 2blc).
References
- 1.Sina, B. (2002) Trends Parasitol. 18, 287–289. [DOI] [PubMed] [Google Scholar]
- 2.Mendis, K., Sina, B. J., Marchesini, P. & Carter, R. (2001) Am. J. Trop. Med. Hyg. 64, 97–106. [DOI] [PubMed] [Google Scholar]
- 3.Schwartz, I. K., Lackritz, E. M. & Patchen, L. C. (1991) New Engl. J. Med. 324, 927. [DOI] [PubMed] [Google Scholar]
- 4.Rieckmann, K. H., Davis, D. R. & Hutton, D. C. (1989) Lancet 2, 1183–1184. [DOI] [PubMed] [Google Scholar]
- 5.Kublin, J. G., Cortese, J. F., Njunju, E. M., Mukadam, R. A., Wirima, J. J., Kazembe, P. N., Djimde, A. A., Kouriba, B., Taylor, T. E. & Plowe, C. V. (2003) J. Infect. Dis. 187, 1870–1875. [DOI] [PubMed] [Google Scholar]
- 6.Hastings, I. M., Bray, P. G. & Ward, S. A. (2002) Science 298, 74–75. [DOI] [PubMed] [Google Scholar]
- 7.Peters, W. (1998) Adv. Parasitol. 41, 1–62. [DOI] [PubMed] [Google Scholar]
- 8.de Pecoulas, P. E., Tahar, R., Ouatas, T., Mazabraud, A. & Basco, L. K. (1998) Mol. Biochem. Parasitol. 92, 265–273. [DOI] [PubMed] [Google Scholar]
- 9.Imwong, M., Pukrittakayamee, S., Looareesuwan, S., Pasvol, G., Poirreiz, J., White, N. J. & Snounou, G. (2001) Antimicrob. Agents Chemother. 45, 3122–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leartsakulpanich, U., Imwong, M., Pukrittayakamee, S., White, N. J., Snounou, G., Sirawaraporn, W. & Yuthavong, Y. (2002) Mol. Biochem. Parasitol. 119, 63–73. [DOI] [PubMed] [Google Scholar]
- 11.Eldin de Pecoulas, P., Basco, L. K., Tahar, R., Ouatas, T. & Mazabraud, A. (1998) Gene 211, 177–185. [DOI] [PubMed] [Google Scholar]
- 12.Yuthavong, Y. (2002) Microbes Infect. 4, 175–182. [DOI] [PubMed] [Google Scholar]
- 13.Warhurst, D. C. (2002) Sci. Prog. 85, 89–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sirawaraporn, W., Sathitkul, T., Sirawaraporn, R., Yuthavong, Y. & Santi, D. V. (1997) Proc. Natl. Acad. Sci. USA 94, 1124–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chusacultanachai, S., Thiensathit, P., Tarnchompoo, B., Sirawaraporn, W. & Yuthavong, Y. (2002) Mol. Biochem. Parasitol. 120, 61–72. [DOI] [PubMed] [Google Scholar]
- 16.Sirawaraporn, W., Yongkiettrakul, S., Sirawaraporn, R., Yuthavong, Y. & Santi, D. V. (1997) Exp. Parasitol. 87, 245–252. [DOI] [PubMed] [Google Scholar]
- 17.Tarnchompoo, B., Sirichaiwat, C., Phupong, W., Intaraudom, C., Sirawaraporn, W., Kamchonwongpaisan, S., Vanichtanankul, J., Thebtaranonth, Y. & Yuthavong, Y. (2002) J. Med. Chem. 45, 1244–1252. [DOI] [PubMed] [Google Scholar]
- 18.Yuvaniyama, J., Chitnumsub, P., Kamchonwongpaisan, S., Vanichtanankul, J., Sirawaraporn, W., Taylor, P., Walkinshaw, M. D. & Yuthavong, Y. (2003) Nat. Struct. Biol. 10, 357–365. [DOI] [PubMed] [Google Scholar]
- 19.Sardarian, A., Douglas, K. T., Read, M., Sims, P. F., Hyde, J. E., Chitnumsub, P., Sirawaraporn, R. & Sirawaraporn, W. (2003) Org. Biomol. Chem. 1, 960–964. [DOI] [PubMed] [Google Scholar]
- 20.Rastelli, G., Sirawaraporn, W., Sompornpisut, P., Vilaivan, T., Kamchonwongpaisan, S., Quarrell, R., Lowe, G., Thebtaranonth, Y. & Yuthavong, Y. (2000) Bioorg. Med. Chem. 8, 1117–1128. [DOI] [PubMed] [Google Scholar]
- 21.D'Arcy, A., Elmore, C., Stihle, M. & Johnston, J. E. (1996) J. Cryst. Growth 168, 175–180. [Google Scholar]
- 22.Pflugrath, J. W. (1999) Acta Crystallogr. D 55, 1718–1725. [DOI] [PubMed] [Google Scholar]
- 23.Navaza, J. (2001) Acta Crystallogr. D 57, 1367–1372. [DOI] [PubMed] [Google Scholar]
- 24.McRee, D. E. (1999) J. Struct. Biol. 125, 156–165. [DOI] [PubMed] [Google Scholar]
- 25.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 26.Engh, R. A. & Huber, R. (1991) Acta Crystallogr. A 47, 392–400. [Google Scholar]
- 27.Laskowski, R. A. (1993) J. Appl. Crystallogr. 26, 283. [Google Scholar]
- 28.Westhead, D. R., Slidel, T. W., Flores, T. P. & Thornton, J. M. (1999) Protein Sci. 8, 897–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matthews, D. A., Alden, R. A., Bolin, J. T., Freer, S. T., Hamlin, R., Xuong, N., Kraut, J., Poe, M., Williams, M. & Hoogsteen, K. (1977) Science 197, 452–455. [DOI] [PubMed] [Google Scholar]
- 30.Rastelli, G., Pacchioni, S. & Parenti, M. D. (2003) Bioorg. Med. Chem. Lett. 13, 3257–3260. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, K. & Rathod, P. K. (2002) Science 296, 545–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aullon, G., Bellamy, D., Orpen, G. A., Brammer, L. & Bruton, E. A. (1998) Chem. Commun., 653–654.
- 33.Brammer, L., Bruton, E. A. & Sherwood, P. (2001) Cryst. Growth Des. 1, 277–290. [Google Scholar]
- 34.Yuthavong, Y., Yuvaniyama, J., Chitnumsub, P., Vanichtanankul, J., Chusacultanachai, S., Tarnchompoo, B., Vilaivan, T. & Kamchonwongpaisan, S. (2005) Parasitology 130, 249–259. [DOI] [PubMed] [Google Scholar]
- 35.Sawaya, M. R. & Kraut, J. (1997) Biochemistry. 36, 586–603. [DOI] [PubMed] [Google Scholar]
- 36.Meyer, E. A., Castellano, R. K. & Diederich, F. (2003) Angew. Chem. Int. Ed. 42, 1210–1250. [DOI] [PubMed] [Google Scholar]
- 37.Myers, J. K. & Pace, C. N. (1996) Biophys. J. 71, 2033–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tahar, R., de Pecoulas, P. E., Basco, L. K., Chiadmi, M. & Mazabraud, A. (2001) Mol. Biochem. Parasitol. 113, 241–249. [DOI] [PubMed] [Google Scholar]
- 39.Hastings, M. D. & Sibley, C. H. (2002) Proc. Natl. Acad. Sci. USA 99, 13137–13141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larder, B. (2001) AIDS 15, S27–S34. [DOI] [PubMed] [Google Scholar]
- 41.King, N. M., Prabu-Jeyabalan, M., Nalivaika, E. A. & Schiffer, C. A. (2004) Chem. Biol. 11, 1333–1338. [DOI] [PubMed] [Google Scholar]
- 42.Ridley, R. G. (2002) Proc. Natl. Acad. Sci. USA 99, 13362–13364. [DOI] [PMC free article] [PubMed] [Google Scholar]