Abstract
Class switch recombination (CSR) is the process by which B cells alter the effector function properties of their Ig molecules. The decision to switch to a particular Ig isotype is determined primarily by the mode of B cell activation and cytokine exposure. More recent work indicates that the likelihood or probability of switching increases with successive cell divisions and is largely independent of time. We have analyzed different molecular features of CSR using cell division as a reference point in an attempt to gain insight into the mechanism of division-linked switching. Our results indicated that the accessibility of Ig heavy chain constant regions targeted for CSR was established after the cells had undergone a single cell division and did not vary significantly with subsequent cell divisions. In contrast, expression of activation-induced cytidine deaminase (AID) mRNA was found to increase with successive divisions, exhibiting a striking correlation with the frequency of CSR. Levels of AID in a given division remained constant at different time points, strongly suggesting that the regulation of AID expression was division-linked and independent of time. In addition, constitutive AID expression from a transgene accelerated division-linked CSR. Thus, we propose that the division-linked increase in AID expression provides an underlying molecular explanation for division-linked CSR.
Keywords: B lymphocyte, cell division
Class switch recombination (CSR) is a region-specific DNA recombination mechanism that occurs by exchanging the currently expressed Ig heavy chain constant region (CH) for another downstream CH region. CSR enables B cells to alter the effector properties of their Ig molecules without altering their antigen specificity. Conceptually, class switching can be divided into three stages of targeting, cleavage, and DNA repair/rejoining, with specific molecular events associated with each stage. For example, cytokine-induced CH region accessibility is marked by acetylation of histone proteins and germ-line transcription across targeted switch (S) regions (1, 2), highly repetitive sequences upstream of each CH gene except Cδ. S region cleavage involves the introduction of DNA lesions in S regions targeted for recombination (3–5), and finally, the rejoining of the cleaved ends requires a number of ubiquitously expressed DNA repair factors. In addition, CSR absolutely requires the activity of the activation-induced cytidine deaminase (AID) (6). Although originally speculated to be an RNA-editing enzyme, recent data strongly suggest that AID functions as a DNA deaminase (7, 8), acting in the context of transcription to deaminate cytosines on single-stranded DNA templates within S regions before the introduction of double-stranded lesions (9–11).
Class switching to particular Ig isotypes in vivo is determined by a combination of exposure to pathogenic stimuli, T cell help, and particular cytokines (reviewed in ref. 12). Induction of switching to all of the different Ig isotypes can be recapitulated ex vivo, which has enabled the characterization of the role of different variables in regulating CSR. Two of the most important variables are the method of B cell activation and the type of cytokine stimuli. Other work has identified cell division as another variable regulating CSR. By using CFSE to track cell proliferation, it was found that naïve B cells activated to switch to IgG1 and IgE started to express significant levels of each isotype after three to four and five to six divisions, respectively (13, 14). Importantly, the percentage of switched cells in a given division remained constant over three consecutive days of culture, suggesting that switching was cell division-linked and was largely independent of time. Further work indicated that a division-linked mechanism regulated CSR to all murine Ig isotypes (15, 16).
Although the cellular regulation of division-linked CSR is well understood, the molecular mechanism underlying the division-dependence is not. Using a well characterized ex vivo model, we examined several different key molecular events associated with CSR by using cell division as a reference point. Our findings indicated that expression of AID is regulated in a division-linked, time-independent manner, and is tightly correlated with the frequency of recombination at both the molecular and cellular level. Furthermore, overexpression of AID increased the frequency of division-linked switching. Thus, we propose that division-regulated expression of AID represents a primary mechanistic control for division-linked CSR.
Materials and Methods
Mice. Inbred strains of mice (C57BL/6 and BALB/c) were obtained from the National Cancer Institute (Bethesda), and AID transgenic mice were generously provided by T. Honjo (Kyoto University, Kyoto). Mice of between 6 and 10 weeks of age were used in accordance with Yale University animal experimentation guidelines.
B Cell Purification and ex Vivo Culture. For most experiments, naive splenic B cells were prepared by using a protocol described in detail elsewhere (17) with minor modifications. Briefly, homogenized, single-cell splenocytes were depleted of red blood cells (RBCs) and adherent cells by hypotonic lysis and incubation on plastic tissue culture dishes, respectively. T cells were subsequently depleted by complement lysis (Cedarlane Laboratories, Ontario, Canada) after incubation with purified CD4-, CD8-, and Thy1-specific mAbs (all from PharMingen). B cells were separated over a discontinuous Percoll gradient, and small, resting B cells were recovered from the 65/72% interface. For experiments comparing B cells from AID-transgenic (AID Tg) mice and non-Tg controls, naive B cells were obtained by negative selection using CD43 MACS Microbeads (Miltenyi Biotech, Auburn, CA) followed by FACS based on IgDhigh and B220+ surface expression.
Purified B cells were cultured in medium consisting of RPMI medium 1640 (Life Technologies, Grand Island, NY) supplemented with 2 mM l-glutamine, 0.1 mM nonessential amino acids, 10 mM Hepes (pH 7.4), 100 μg/ml streptomycin, 60 μg/ml penicillin, 5 × 10-5 M 2-mercaptoethanol (all from Sigma), 1 mM Na-pyruvate (Invitrogen), and 10% heat-inactivated FCS (Gemini Bioproducts, Woodland, CA). B cells were labeled with carboxy-fluorescein diacetate-succinimyl ester (CFSE) (Molecular Probes, Eugene, OR) before culture to monitor cell division. Cells were stimulated at a density of 5 × 105 cells per ml with anti-mouse CD40 mAb (HM40–3, 10 μg/ml, eBioscience, San Diego) or recombinant CD40L membranes (prepared from insect cells infected with CD40L-expressing baculovirus) in the presence or absence of mouse IL-4 (gift of C. A. Janeway, Yale University) at saturating concentrations (i.e., 400 units/ml) except where otherwise indicated.
Flow Cytometry. Stimulated B cells were harvested at various times and stained for surface expression of IgG1 in the presence of FcγRII blocking antibody (mAbs from PharMingen). For sorting experiments, cells were separated according to division number using a FACSVantage (BD Biosciences). Data were analyzed by using cellquest (BD Biosciences) and f lojo (TreeStar, Ashland, OR).
Digestion–Circularization (DC) PCR. Genomic DNA was isolated from sorted B cells and DC-PCR was performed as described (18). Briefly, DNA was digested with EcoRI (0.5–2 μg of DNA per 100 μl) before overnight ligation (180 ng DNA per 100 μl) with T4 ligase (400 units; New England Biolabs). Circularized DNA was subsequently dialyzed against deionized water before PCR. PCR oligos and conditions for acetylcholine receptor (AchR) and Sμ-Sγ1 were as described (19), except that Jumpstart TaqDNA polymerase (Sigma) was used. Plasmid standards P2A0 (AchR) and P4AP (Sμ-Sγ1) (18) were used as controls for PCR amplification and were used to determine the linear range of PCR amplification (data not shown). PCR products were detected by Southern blot using Sμ and AchR-specific probes (Table 1, which is published as supporting information on the PNAS web site).
Quantitative RT-PCR (qRT-PCR). RNA was extracted with RNAzol (Invitrogen) as per the manufacturer's instructions. RNA (0.5–1 μg) was reverse transcribed with random hexamers and SuperScript II reverse transcriptase (Invitrogen) at 50°C in a volume of 20 μl. Subsequent qRT-PCR was performed by using a MX3000P cycler (Stratagene) with Jumpstart Taq polymerase (Sigma) in a total volume of 50 μl for 40 cycles. Table 1 lists sequences of oligos (obtained from Invitrogen) and oligo probes (obtained from Biosearch Technologies, Novato, CA). Oligos for AID qRT-PCR have been described (20). Transcript levels were estimated by comparison to standard curves generated by PCR of control plasmids containing the relevant amplicon, and were normalized to levels of mb-1 mRNA.
Chromatin Immunoprecipitation (ChIP). ChIP for acetylated histone 3 (H3) (anti-acetyl H3 antibody, Upstate Biotechnology) was performed on 5 × 105 to 1 × 106 primary cells by using a commercially available kit (Upstate Biotechnology). qRT-PCR was performed by using Jumpstart Taq polymerase (Sigma) in a total volume of 50 μl for 40 cycles. Oligo and probe sequences can be found in Table 1. Results were analyzed as described for qRT-PCR.
Western Blot for AID Protein. AID protein expression by division-sorted BL6 and AID knock-out B cells activated with anti-CD40 or CD40L and IL-4 was performed on cell lysates of 0.5–1 × 106 cells, using an affinity-purified rabbit antibody raised against a peptide (CEDRKAEPEGLRRLHR) derived from human AID.
Results
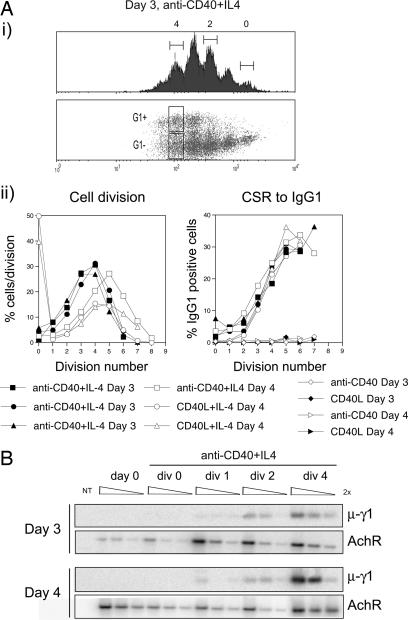
CSR Is Linked to Cell Division at the Molecular Level. For our analyses of the molecular basis of division-linked CSR, we used a well characterized ex vivo model of murine B cell stimulation. We activated naive, CFSE-labeled B cells with CD40L or anti-CD40 mAb in the presence of IL-4, which stimulated cell division and switching to IgG1. Fig. 1A illustrates division-linked switching by B cells, results of which are in accordance with published results (13, 14). Subsequently, we wanted to determine whether the division-linked surface expression of switched isotypes was mirrored by recombination at the DNA level. To do this, we examined CSR by DC-PCR on division-sorted cells from successive days. A similar experimental strategy was used for analyses of other molecular events associated with CSR.
Fig. 1.
Molecular and cellular analysis of division-linked CSR. (A) CSFE-labeled B cells were activated under various conditions (see key) for 3 or 4 days, where-upon cells were harvested and their expression of IgG1 was analyzed. (i) FACS profiles of cell division (with divisions 0, 2, and 4 indicated), and acquisition of surface IgG1. (ii) (Left) The proportion of cells in each division. (Right) The acquisition of surface IgG1 expression. Results from three independent experiments are shown. (B) Semiquantitative DC-PCR for μ-γ1 and AchR was performed on genomic DNA purified from day 3 and 4 cultured cells that had been sorted according to division number. Data shown are representative of two experiments.
Fig. 1B shows the level of μ-γ1 recombination in division-sorted B cells. As a positive control for DC-PCR, we assayed for μ-γ1 recombination in division 4-sorted IgG1 positive cells (Fig. 5, which is published as supporting information on the PNAS web site). We also performed DC-PCR for the AchR as a control for DNA amount and ligation efficiency. Fig. 1B indicates that the frequency of switch events at the molecular level increased with successive cell divisions, but did not vary significantly with culture duration. Low levels of μ-γ1 recombination could be observed after a single cell division, suggesting that CSR at the molecular level can occur one to two divisions before significant surface expression of the switched isotype (Fig. 1A). In contrast, there was no evidence of recombination in undivided cells. Collectively, these results suggested that naïve B cells were required to complete at least one cell division to switch, and that the frequency of recombination at both the molecular and cellular level increased with successive cell divisions in a manner largely independent of time.
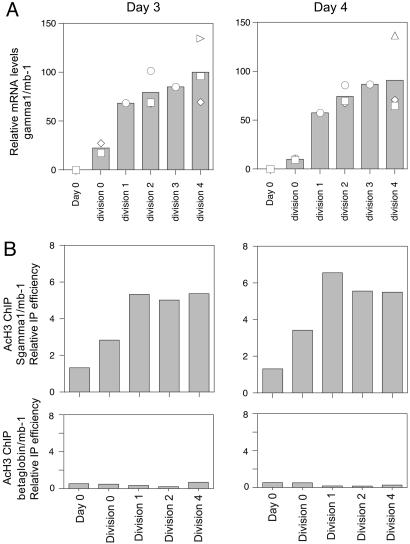
Accessibility of Sγ1 Is Induced After a Single Cell Division. Fig. 1 indicated that the division-requirement for CSR was manifest at both the cellular and molecular level. Consequently, we were interested in investigating whether various molecular events associated with CSR might also be regulated in a division-dependent manner, perhaps providing insight into the mechanistic controls of division-linked switching. Initially we sought to determine whether the accessibility of CH regions targeted for recombination was regulated by cell division. To do this, we examined the levels of H3 acetylation at Sγ1 by ChIP, as well as γ1 germ-line transcription in division-sorted cells harvested at different time points. Fig. 2A indicates that, upon activation, division 0 cells started to express γ1 transcripts. The level of these transcripts increased after cells had undergone one cell division, after which point transcript levels plateaued and remained similar in subsequent cell divisions. An identical pattern was observed for acetylation of histone 3 (H3) within Sγ1 (Fig. 2B). There was no change in the acetylation status of the β-globin gene upon activation or with division (Fig. 2B). Hence, by two different measures, Cγ1 accessibility was induced after cells had undergone one cell division, and maintained with subsequent cell divisions at a level that did not vary with the time of culture. Interestingly, the induction of S region accessibility did not correlate well with the division-dependent increase in the frequency of switched cells, suggesting that the molecular mechanism of CSR could involve molecular events associated with events downstream of S region targeting.
Fig. 2.
Maximal Sγ1 accessibility is established after a single cell division. (A) qRT-PCR for γ1 transcripts on RNA prepared from division-sorted B cells activated for 3 or 4 days with anti-CD40 and IL-4. γ1 mRNA levels from several independent experiments were normalized to the amount of mb-1 mRNA, and relative differences in γ1 transcript levels on both days are plotted relative to the average γ1 mRNA yield from day 3, division 4 sorted B cells, which was arbitrarily set to 100. Replicates for different samples: day 4 (n = 6), day 3 div 0 (n = 3), div 1 (n = 1), div 2(n = 3), div 3 (n = 1), div 4 (n = 3), day 4 div 0 (n = 3), div 1 (n = 1), div 2 (n = 3), div 3 (n = 1), div 4 (n = 3). Symbols indicate independent samples, with the mean of the samples represented by the column graph. (B) Levels of H3 acetylation at Sγ1. Division-sorted B cells activated for 3 or 4 days were subject to ChIP using an anti-acetylated H3 antibody or no antibody control. qPCR for Sγ1, β-globin (negative control) and mb-1 (positive control) were performed, and results are presented as: (Sγ1 or β-globin AcH3/Sγ1 or β-globin input)/(mb-1 AcH3/mb-1 input) versus division number. Data depict the average of two experiments.
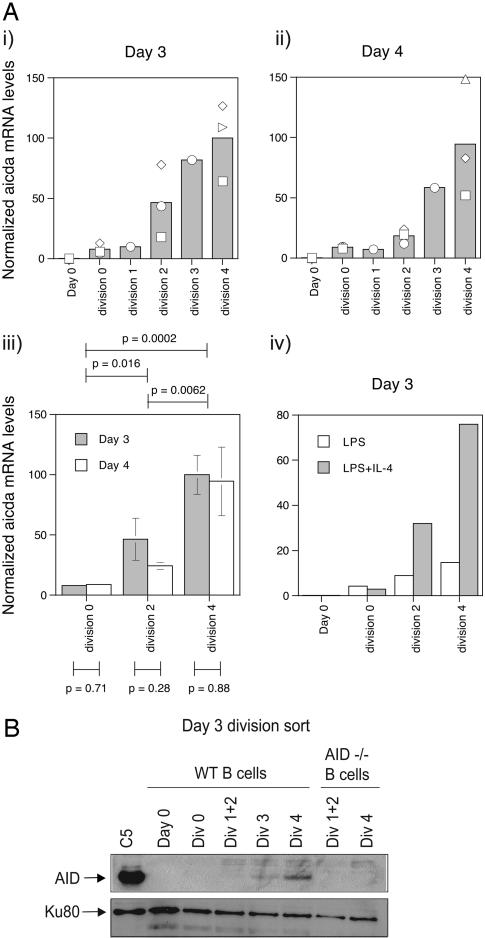
Division-Linked Expression of AID. The bulk of available evidence suggests that AID deaminates cytosine residues within S region substrates, the processing of which leads to the formation of S region associated DNA double strand breaks. Consequently, we were interested in examining the expression of AID in B cells activated to switch by using cell division as a reference point. We performed qRT-PCR for Aicda (the gene that encodes for AID) on RNA prepared from division-sorted B cells that had been stimulated to undergo CSR to IgG1. Fig. 3A i and ii indicates that there was a progressive increase in the level of AID mRNA with successive cell divisions on days 3 and 4. Importantly, AID transcript levels in B cells from a particular cell division on consecutive days were very similar (P > 0.05, Fig. 3Aiii), suggesting that AID expression was regulated in a division-linked, time-independent manner. Statistical analysis of AID levels between divisions indicated that there was a significant increase in transcript levels from divisions 0 to 2 to 4. Finally, a similar division-linked increase in AID expression was also observed for B cells activated to switch with LPS or LPS and IL-4 (Fig. 3Aiv), suggesting that division-linked regulation of AID levels may represent a common mechanism for controlling division-dependent CSR to different isotypes.
Fig. 3.
Division-linked expression of AID. (A) qRT-PCR was performed on RNA prepared from division sorted B cells activated with anti-CD40 and IL-4 for 3 (i) and 4(ii) days. RNA samples were the same as used for γ1 transcripts (see Fig. 2), and AID transcript levels were calculated as described for γ1 transcripts. Symbols indicate independent samples, with the mean of the samples represented by the column graph. (iii) Student's t test was used to evaluate the statistical significance of AID levels between divisions (i.e., by comparing AID levels in a given division to those in a different division, above graph; for this analysis, AID levels from a given division on days 3 and 4 were combined before statistical comparison with other divisions) or between different time points (i.e., by comparing AID levels in a given division on different days, below graph). Mean and standard error of data from Ai and ii are shown. (iv) Division-linked expression of AID of B cells activated for 3 days with LPS or LPS and IL-4. (B) An AID Western blot was performed on lysates prepared from WT and AID-deficient division-sorted B cells that had been cultured for 3 days with CD40L and IL-4. A Western blot for Ku80 was performed as a control for protein levels. C5 is a subclone of RAMOS selected for AID overexpression after infection with an AID-GFP expressing retrovirus.
We also investigated the expression of AID protein with cell division by performing Western blots on cell lysates prepared from division-sorted B cells after 3 days of culture. Fig. 3B indicates that, like AID mRNA, levels of AID protein increased with successive cell divisions (compare Fig. 3 Ai and B). A similar trend of a division-linked increase in AID protein levels was also seen in lysates prepared from day 4, division-sorted B cells (data not shown).
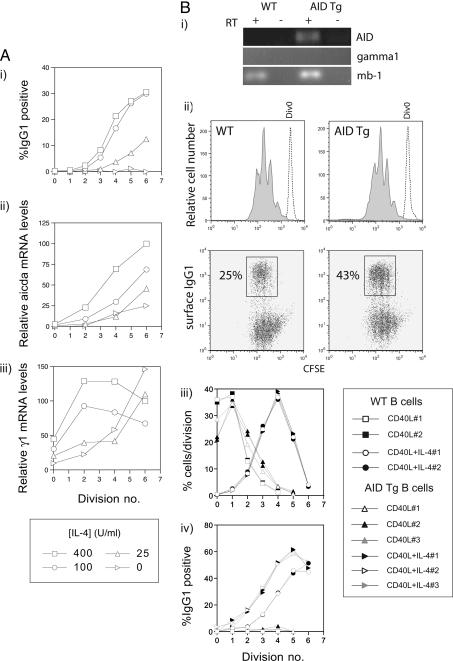
To further examine the correlation between division-linked AID levels and CSR, we measured AID expression in division-sorted B cells cultured with varying concentrations of IL-4, which alters the frequency of switching to IgG1 (13). Fig. 4A indicates that the progressive increase in the frequency of switched cells with successive divisions was mirrored by the expression of AID mRNA, confirming the strong correlation between division linked CSR and AID levels. Thus, sensitivity to IL-4 altered the probability that a B cell would switch to IgG1 primarily by regulating AID levels. At subsaturating IL-4 concentrations, expression of γ1 transcripts occurred during later divisions; however, it is unclear whether this would dramatically affect switching to IgG1, as previous work has indicated that inhibition of γ1 transcript levels by >80% had no affect on the frequency of μ-γ1 recombination (21).
Fig. 4.
AID expression correlates with CSR frequency. (A) B cells were cultured with a constant CD40L dose in the presence of varying concentrations of IL-4. Cells were sorted according to division number after day 3 and subject to qRT-PCR for AID γ1 transcripts, and mb-1. (i) Percentage of IgG1 positive cells with division number after three days of culture with CD40L and varying IL-4 dosage. (ii) Levels of AID transcripts with division were normalized to mb-1 mRNA levels and expressed relative to the amount of AID transcript in division 4 (in 400 units/ml IL-4 cultures). (iii) Levels of γ1 germ-line transcripts with division were normalized to mb-1 mRNA levels and expressed relative to the amount of γ1 transcript in division 4 (in 400 units/ml IL-4 cultures). (B) B cells from AID transgenic mice and wild-type mice were cultured with CD40L in the presence or absence of IL-4 for 3 days before FACS analysis. (i) RT-PCR of AID, γ1 germ-line transcripts and mb-1 from purified, naive AID Tg and control B cells. Amplifications (30 cycles) were performed by using oligos identical to those used for qRT-PCR. RT controls are indicated. (ii) Division profiles and division-linked expression of surface IgG1 of WT and AID AID Tg B cells. The proliferation profiles of undivided cells (hashed line) overlay the profiles of B cells stimulated with CD40L and IL-4 (gray fill). The expression of surface IgG1 with division is shown. (iii) The distribution of cells with division showing data from replicate cultures where B cells were activated with CD40L in the presence or absence of IL-4. Note that the figure legend applies to iii and iv.(iv) Acquisition of surface IgG1 expression with cell division by AID Tg and WT B cells from replicate cultures after harvest at day 3 after culture. Data are representative of two experiments.
We also examined the effect of overexpression of AID on division-linked switching, reasoning that enforced expression of AID before cell division should accelerate CSR. For these experiments, we made use of B cells purified from AID Tg mice in which AID expression is driven by the ubiquitous and constitutively active chicken β-actin promoter (22). Because these mice possess several lymphoid abnormalities, it is possible that B cells from these mice may have received additional stimuli in vivo before harvesting, which could potentially affect their ability to switch. To minimize this effect, we obtained spleens from young, healthy Tg and control mice for analysis. After purification of splenic B cells by negative selection, we sorted IgDhigh, B220positive cells for subsequent CFSE labeling and culture. RT-PCR analysis indicated that, as expected, unstimulated AID Tg B cells expressed AID, in contrast to control B cells (Fig. 4Bi). Neither wild-type or AID Tg naïve B cells expressed γ1 germ-line transcripts (Fig. 4Bi), further suggesting that B cells purified from both strains were naive.
Fig. 4Bii illustrates the proliferation profiles and acquisition of surface IgG1 of naive B cells from AID Tg and control mice stimulated for 3 days with CD40L and IL-4. Overexpression of AID had no effect on the progression of cells through cell division (Fig. 4B ii and iii); however, the frequency of switched B cells from AID transgenic mice was almost 2-fold higher compared with control cells. Plotting the frequency of IgG1 expressing cells with division indicated that, as predicted, a higher proportion of B cells from each division had switched (Fig. 4Biv). No switching was observed in Tg or control B cells stimulated only with CD40L. Thus, by modulating the expression of AID, it was possible to directly alter the division-linked frequency of CSR. This observation, together with the finding that AID levels are regulated in a division-linked manner, strongly indicates that division-linked regulation of AID expression provides a molecular basis for division-linked CSR.
Discussion
The relationship between cell division and CSR is well documented, with both human and murine B cells displaying evidence of a division-linked increase in the frequency of cell surface expression of switched Ig isotypes after stimulation (reviewed in ref. 23). However, despite detailed understanding of the cellular regulation of division-linked switching, there is little knowledge of the underlying molecular controls. In this study, we sought to gain insight into the molecular mechanism of division-linked switching by examining various molecular events associated with CSR by using cell division as a reference point.
Initially, we examined CSR in division-sorted cells by using DC-PCR to determine how recombination at the molecular level varied with cell division. Our results indicated that division-linked CSR operated at both the molecular and cellular level, with the level of μ-γ1 recombination increasing with successive cell divisions in a time-independent manner (Fig. 1). We also failed to observe CSR in undivided cells even after 4 days of stimulation, despite the presence of low levels of AID mRNA. There are several explanations for the absence of recombination in division 0 cells. Levels of γ1 germ-line transcripts and H3 acetylation are lower in division 0 than in division 1 cells (Fig. 2A), indicating that full accessibility of the γ1 switch region has not been achieved in undivided cells. In addition, certain DNA repair pathways involved in CSR have been demonstrated to operate in particular stages of the cell cycle, which might prevent recombination from occurring until the cell enters and completes these stages. Our data are consistent with the notion that at least one round of DNA replication is required for CSR, which is in agreement with experiments using cell cycle inhibitors (ref. 24 and data not shown) and with the model that single strand lesions generated by the combined action of AID and the mismatch repair (MMR) and base-excision repair (BER) pathways are converted into double-strand breaks by replication of the IgH locus during S phase (25). However, it should be noted that DNA repair foci containing Nijmegen breakage syndrome protein (Nbs1) and phosphorylated histone H2AX (γ-H2AX) were found predominantly in switching B cells in the G1 phase of the cell cycle, arguing that double-strand break formation (and possibly repair) largely occurs during G1 (26). S region double-strand breaks may also be generated in a replication-independent manner if AID (and there-after the MMR/BER machineries) can act on both DNA strands. Improved understanding of these issues awaits a more lucid definition of the pathways involved in the generation and repair of S region double-strand breaks.
Although the above discussion provides one explanation for how CSR may be linked to the cell cycle, this framework cannot account for the observation that the levels of μ-γ1 recombination in cells from a given division are similar irrespective of whether these cells are harvested at day 3 or 4. Maintaining equivalent levels of recombination would suggest that the quantity and/or activity of factors that regulate the amount of recombination would remain relatively constant within a given division at different times. Consequently, we sought to examine whether various molecular events associated with CSR were regulated in a division-linked manner, and if so, whether they might represent the underlying molecular controls for division-linked switching.
First, we examined cytokine-induced germ-line transcripts and S-region-associated histone modifications as measures of targeting of specific S regions for recombination. We were particularly interested in investigating the division-linked regulation of S region targeting because previous results had indicated that expression of γ1 transcripts increased with successive cell divisions (27). This result suggested that the induction of S-region accessibility might regulate division-linked CSR. We were surprised to find that the induction of accessibility at Sγ1 did not correlate well with the division-linked increase in CSR to IgG1, as we did not observe a progressive increase in accessibility with consecutive cell divisions. The disparity between our results and those previously reported most likely reflects sensitivity differences intrinsic to each method, as we made use of qRT-PCR, whereas the previous study did not. We note that our conclusions regarding division-linked accessibility were supported by assessment of two distinct parameters. Thus, we would argue that, although required for CSR, the induction of accessibility at CH regions in IgH loci is not a primary mechanistic control of division-linked CSR.
Next we sought to investigate molecular events associated with DNA cleavage. Previous results have indicated that AID-dependent DNA lesions are associated with S regions of B cells activated to undergo CSR (3–5). One such study indicated that Sμ-associated DNA double-strand breaks could be detected in undivided cells, but that the frequency of these lesions increased dramatically after 4 days in culture, at which point a majority of the cells have divided an average of three to four times (4). Because the quantity of cells used for comparison of these different populations was equivalent, this result suggested that the level of Sμ double-strand breaks is greater in cells that have divided several times. Other work indicated that the frequency of Sμ mutation is higher in cells that had undergone more cell divisions (ref. 28 and data not shown). This result suggested that the expression and/or activity of factors involved in making or processing the S region-associated lesions may be regulated in a division-linked manner.
Consequently, we examined the regulation of AID expression with cell division and found that both mRNA and protein levels increased with successive divisions in B cells that had been activated to switch (Fig. 3). In addition, levels of AID correlated strongly with the level of recombination, and replicate experiments indicated that the amount of AID transcript in a given division did not vary at different time points. These results revealed that cell division represented a previously undescribed regulatory pathway for controlling AID expression.
Previous work investigating the regulation of AID expression focused primarily on how different B cell stimuli affect AID levels. For example, IL-4 and CD40 signals synergized to promote AID expression by B cells in a Stat-6- and NF-κB-dependent fashion, whereas either stimuli alone failed to induce AID protein expression (29). These findings prompted the suggestion that a threshold level of AID may be required for efficient CSR to occur. Our results suggest that this threshold level of AID is under division-linked control. Furthermore, IL-4 appears to regulate the level of AID mRNA in given division as suboptimal concentrations of IL-4 lead to a reduction in the frequency of division-linked switching to IgG1. In contrast, transgenic overexpression of AID resulted in an increased frequency of division-linked CSR in B cells stimulated to switch. Thus, modulating the expression of AID directly alters the frequency of division-linked switching, strongly suggesting that the regulation of AID expression with cell division is a primary molecular control of division-linked CSR.
Although cell division represents an underlying mechanistic regulator of AID expression, the molecular basis of division-linked AID expression remains unresolved. However, there are several possibilities. The expression and/or activity of transcription factors that directly regulate AID mRNA levels (e.g., Stat-6, NF-κB, or E-box proteins; refs. 20, 29, and 30) may be regulated in a division-linked manner, mirroring the expression of AID. It has also been demonstrated that phosphorylation of AID is required for its deaminase activity in vitro (31), and thus the regulation of this posttranslational modification may be controlled at the level of cell division, perhaps by regulating the activity of the relevant kinase(s). Recent results have suggested that somatic hypermutation and CSR may require distinct AID cofactors (32, 33), and thus it is possible that the expression/activity of these cofactors is division-linked or may affect the division-linked activity of AID. Another possibility is that proteins that negatively regulate AID levels might by progressively diluted with successive cell divisions, thereby enabling the division-linked increase in AID expression. Thus, there may be other factors involved in CSR in addition to AID whose expression or activity is regulated in a cell division linked manner. The existence of such factors is suggested by the fact that levels of CSR increase with successive cell divisions in AID transgenic B cells (Fig. 4Biv).
The AID transgenic mice used in this study were previously reported to exhibit a reduced life span, enlarged secondary lymphoid organs, and T cell lymphomas and other malignancies (22). No B cell defects were noted, and the effect of transgenic AID expression in B cells from these mice was not reported. Our data showing accelerated CSR in transgenic as compared to normal B cells indicates that the transgene encoded AID is active and can mediate CSR in stimulated B cells. It will now be of interest to determine whether transgenic AID can target the highly transcribed μ switch region in naive B cells, or whether, as discussed above, other factors/activities are induced upon B cell activation that are required for AID activity and/or targeting to Ig genes.
In conclusion, our findings implicate division-regulated AID expression as a major underlying molecular mechanism of division-linked CSR. Furthermore, our observation that levels of germ-line transcription and H3 acetylation at Sγ1 may also be regulated in a division-linked manner (albeit one that does not correlate well with CSR) suggest that it is possible that many of the molecular events associated with B cell differentiation may be linked to cell division. Consequently, we propose that cell division represents an additional level of mechanistic control for transcriptional regulation. Such a hypothesis is amenable to evaluation and would inform our thinking about how cell division and transcriptional regulation intersect to control cell fate decisions.
Supplementary Material
Acknowledgments
We thank Drs. T. Honjo and I. Okazaki (Kyoto University, Kyoto) for generously providing AID knockout and transgenic mice, Drs. M. Shlomchik and H. Wang (Yale University) for CD40L-expressing baculovirus and technical guidance, and Tom Taylor for cell sorting. All work described was supported by the Howard Hughes Medical Institute (HHMI). J.S.R. was an HHMI Postdoctoral Associate, and D.G.S. is an HHMI investigator.
Author contributions: J.S.R. and D.G.S. designed research; J.S.R., M.L., and V.H.O. performed research; V.H.O. and S.U. contributed new reagents/analytic tools; J.S.R. analyzed data; and J.S.R. and D.G.S. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CSR, class switch recombination; S region, switch region; CH, Ig heavy chain constant; DC, digestion–circularization; AchR, acetylcholine receptor; qRT-PCR, quantitative RT-PCR; Tg, transgenic; ChIP, chromatin immunoprecipitation.
References
- 1.Nambu, Y., Sugai, M., Gonda, H., Lee, C. G., Katakai, T., Agata, Y., Yokota, Y. & Shimizu, A. (2003) Science 302, 2137-2140. [DOI] [PubMed] [Google Scholar]
- 2.Stavnezer, J., Radcliffe, G., Lin, Y. C., Nietupski, J., Berggren, L., Sitia, R. & Severinson, E. (1988) Proc. Natl. Acad. Sci. USA 85, 7704-7708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arudchandran, A., Bernstein, R. M. & Max, E. E. (2004) J. Immunol. 173, 3223-3229. [DOI] [PubMed] [Google Scholar]
- 4.Rush, J. S., Fugmann, S. D. & Schatz, D. G. (2004) Int. Immunol. 16, 549-557. [DOI] [PubMed] [Google Scholar]
- 5.Wuerffel, R. A., Du, J., Thompson, R. J. & Kenter, A. L. (1997) J. Immunol. 159, 4139-4144. [PubMed] [Google Scholar]
- 6.Muramatsu, M., Kinoshita, K., Fagarasan, S., Yamada, S., Shinkai, Y. & Honjo, T. (2000) Cell 102, 553-563. [DOI] [PubMed] [Google Scholar]
- 7.Di Noia, J. & Neuberger, M. S. (2002) Nature 419, 43-48. [DOI] [PubMed] [Google Scholar]
- 8.Petersen-Mahrt, S. K., Harris, R. S. & Neuberger, M. S. (2002) Nature 418, 99-103. [DOI] [PubMed] [Google Scholar]
- 9.Chaudhuri, J., Tian, M., Khuong, C., Chua, K., Pinaud, E. & Alt, F. W. (2003) Nature 422, 726-730. [DOI] [PubMed] [Google Scholar]
- 10.Dickerson, S. K., Market, E., Besmer, E. & Papavasiliou, F. N. (2003) J. Exp. Med. 197, 1291-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramiro, A. R., Stavropoulos, P., Jankovic, M. & Nussenzweig, M. C. (2003) Nat. Immunol. 4, 452-456. [DOI] [PubMed] [Google Scholar]
- 12.Stavnezer, J. (1996) Adv. Immunol. 61, 79-146. [DOI] [PubMed] [Google Scholar]
- 13.Hasbold, J., Lyons, A. B., Kehry, M. R. & Hodgkin, P. D. (1998) Eur. J. Immunol. 28, 1040-1051. [DOI] [PubMed] [Google Scholar]
- 14.Hodgkin, P. D., Lee, J. H. & Lyons, A. B. (1996) J. Exp. Med. 184, 277-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deenick, E. K., Hasbold, J. & Hodgkin, P. D. (1999) J. Immunol. 163, 4707-4714. [PubMed] [Google Scholar]
- 16.Hasbold, J., Hong, J. S., Kehry, M. R. & Hodgkin, P. D. (1999) J. Immunol. 163, 4175-4181. [PubMed] [Google Scholar]
- 17.Hasbold, J., Gett, A. V., Rush, J. S., Deenick, E., Avery, D., Jun, J. & Hodgkin, P. D. (1999) Immunol. Cell Biol. 77, 516-522. [DOI] [PubMed] [Google Scholar]
- 18.Chu, C. C., Paul, W. E. & Max, E. E. (1992) Proc. Natl. Acad. Sci. USA 89, 6978-6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Casellas, R., Nussenzweig, A., Wuerffel, R., Pelanda, R., Reichlin, A., Suh, H., Qin, X. F., Besmer, E., Kenter, A., Rajewsky, K. & Nussenzweig, M. C. (1998) EMBO J. 17, 2404-2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sayegh, C. E., Quong, M. W., Agata, Y. & Murre, C. (2003) Nat. Immunol. 4, 586-593. [DOI] [PubMed] [Google Scholar]
- 21.Dunnick, W. A., Shi, J., Graves, K. A. & Collins, J. T. (2004) J. Immunol. 173, 5531-5539. [DOI] [PubMed] [Google Scholar]
- 22.Okazaki, I. M., Hiai, H., Kakazu, N., Yamada, S., Muramatsu, M., Kinoshita, K. & Honjo, T. (2003) J. Exp. Med. 197, 1173-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tangye, S. G. & Hodgkin, P. D. (2004) Immunology 112, 509-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lundgren, M., Strom, L., Bergquist, L. O., Skog, S., Heiden, T., Stavnezer, J. & Severinson, E. (1995) Eur. J. Immunol. 25, 2042-2051. [DOI] [PubMed] [Google Scholar]
- 25.Rooney, S., Chaudhuri, J. & Alt, F. W. (2004) Immunol. Rev. 200, 115-131. [DOI] [PubMed] [Google Scholar]
- 26.Petersen, S., Casellas, R., Reina-San-Martin, B., Chen, H. T., Difilippantonio, M. J., Wilson, P. C., Hanitsch, L., Celeste, A., Muramatsu, M., Pilch, D., et al. (2001) Nature 414, 660-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCall, M. N. & Hodgkin, P. D. (1999) Biochim. Biophys. Acta 1447, 43-50. [DOI] [PubMed] [Google Scholar]
- 28.Reina-San-Martin, B., Difilippantonio, S., Hanitsch, L., Masilamani, R. F., Nussenzweig, A. & Nussenzweig, M. C. (2003) J. Exp. Med. 197, 1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dedeoglu, F., Horwitz, B., Chaudhuri, J., Alt, F. W. & Geha, R. S. (2004) Int. Immunol. 16, 395-404. [DOI] [PubMed] [Google Scholar]
- 30.Gonda, H., Sugai, M., Nambu, Y., Katakai, T., Agata, Y., Mori, K. J., Yokota, Y. & Shimizu, A. (2003) J. Exp. Med. 198, 1427-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaudhuri, J., Khuong, C. & Alt, F. W. (2004) Nature 430, 992-998. [DOI] [PubMed] [Google Scholar]
- 32.Barreto, V., Reina-San-Martin, B., Ramiro, A. R., McBride, K. M. & Nussenzweig, M. C. (2003) Mol. Cell 12, 501-508. [DOI] [PubMed] [Google Scholar]
- 33.Ta, V. T., Nagaoka, H., Catalan, N., Durandy, A., Fischer, A., Imai, K., Nonoyama, S., Tashiro, J., Ikegawa, M., Ito, S., et al. (2003) Nat. Immunol. 4, 843-848. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.